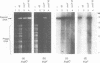Abstract
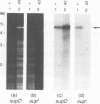
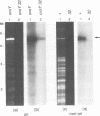
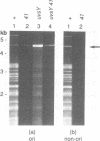
Bacteriophage T4 DNA replication initiates from origins at early times of infection and from recombinational intermediates as the infection progresses. Plasmids containing cloned T4 origins replicate during T4 infection, providing a model system for studying origin-dependent replication. In addition, recombination-dependent replication can be analyzed by using cloned nonorigin fragments of T4 DNA, which direct plasmid replication that requires phage-encoded recombination proteins. We have tested in vivo requirements for both plasmid replication model systems by infecting plasmid-containing cells with mutant phage. Replication of origin and nonorigin plasmids strictly required components of the T4 DNA polymerase holoenzyme complex. Recombination-dependent plasmid replication also strictly required the T4 single-stranded DNA-binding protein (gene product 32 [gp32]), and replication of origin-containing plasmids was greatly reduced by 32 amber mutations. gp32 is therefore important in both modes of replication. An amber mutation in gene 41, which encodes the replicative helicase of T4, reduced but did not eliminate both recombination- and origin-dependent plasmid replication. Therefore, gp41 may normally be utilized for replication of both plasmids but is apparently not required for either. An amber mutation in gene 61, which encodes the T4 RNA primase, did not eliminate either recombination- or origin-dependent plasmid replication. However, plasmid replication was severely delayed by the 61 amber mutation, suggesting that the protein may normally play an important, though nonessential, role in replication. We deleted gene 61 from the T4 genome to test whether the observed replication was due to residual gp61 in the amber mutant infection. The replication phenotype of the deletion mutant was identical to that of the amber mutant. Therefore, gp61 is not required for in vivo T4 replication. Furthermore, the deletion mutant is viable, demonstrating that the gp61 primase is not an essential T4 protein.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alfano C., McMacken R. Ordered assembly of nucleoprotein structures at the bacteriophage lambda replication origin during the initiation of DNA replication. J Biol Chem. 1989 Jun 25;264(18):10699–10708. [PubMed] [Google Scholar]
- Arluke A. The ethical thinking of animal researchers: problems and prospects. New Biol. 1991 Jan;3(1):1–2. [PubMed] [Google Scholar]
- Baker T. A., Sekimizu K., Funnell B. E., Kornberg A. Extensive unwinding of the plasmid template during staged enzymatic initiation of DNA replication from the origin of the Escherichia coli chromosome. Cell. 1986 Apr 11;45(1):53–64. doi: 10.1016/0092-8674(86)90537-4. [DOI] [PubMed] [Google Scholar]
- Barth K. A., Powell D., Trupin M., Mosig G. Regulation of two nested proteins from gene 49 (recombination endonuclease VII) and of a lambda RexA-like protein of bacteriophage T4. Genetics. 1988 Oct;120(2):329–343. doi: 10.1093/genetics/120.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breschkin A. M., Mosig G. Multiple interactions of a DNA-binding protein in vivo. I. Gene 32 mutations of phage T4 inactivate different steps in DNA replication and recombination. J Mol Biol. 1977 May 15;112(2):279–294. doi: 10.1016/s0022-2836(77)80144-7. [DOI] [PubMed] [Google Scholar]
- Cha T. A., Alberts B. M. Effects of the bacteriophage T4 gene 41 and gene 32 proteins on RNA primer synthesis: coupling of leading- and lagging-strand DNA synthesis at a replication fork. Biochemistry. 1990 Feb 20;29(7):1791–1798. doi: 10.1021/bi00459a018. [DOI] [PubMed] [Google Scholar]
- Cha T. A., Alberts B. M. Studies of the DNA helicase-RNA primase unit from bacteriophage T4. A trinucleotide sequence on the DNA template starts RNA primer synthesis. J Biol Chem. 1986 May 25;261(15):7001–7010. [PubMed] [Google Scholar]
- Cha T. A., Alberts B. M. The bacteriophage T4 DNA replication fork. Only DNA helicase is required for leading strand DNA synthesis by the DNA polymerase holoenzyme. J Biol Chem. 1989 Jul 25;264(21):12220–12225. [PubMed] [Google Scholar]
- Das S. K., Fujimura R. K. Processiveness of DNA polymerases. A comparative study using a simple procedure. J Biol Chem. 1979 Feb 25;254(4):1227–1232. [PubMed] [Google Scholar]
- De Waard A., Paul A. V., Lehman I. R. The structural gene for deoxyribonucleic acid polymerase in bacteriophages T4 and T5. Proc Natl Acad Sci U S A. 1965 Oct;54(4):1241–1248. doi: 10.1073/pnas.54.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean F. B., Bullock P., Murakami Y., Wobbe C. R., Weissbach L., Hurwitz J. Simian virus 40 (SV40) DNA replication: SV40 large T antigen unwinds DNA containing the SV40 origin of replication. Proc Natl Acad Sci U S A. 1987 Jan;84(1):16–20. doi: 10.1073/pnas.84.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr L. K., Kreuzer K. N. Expression and function of the uvsW gene of bacteriophage T4. J Mol Biol. 1990 Aug 5;214(3):643–656. doi: 10.1016/0022-2836(90)90283-R. [DOI] [PubMed] [Google Scholar]
- Dewey M. J., Frankel F. R. Two suppressor loci for gene 49 mutations of bacteriophage T4. I. Genetic properties and DNA synthesis. Virology. 1975 Dec;68(2):387–401. doi: 10.1016/0042-6822(75)90281-0. [DOI] [PubMed] [Google Scholar]
- Dodson M., McMacken R., Echols H. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda. Protein association and disassociation reactions responsible for localized initiation of replication. J Biol Chem. 1989 Jun 25;264(18):10719–10725. [PubMed] [Google Scholar]
- Dodson M., Roberts J., McMacken R., Echols H. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: complexes with lambda O protein and with lambda O, lambda P, and Escherichia coli DnaB proteins. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4678–4682. doi: 10.1073/pnas.82.14.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EDGAR R. S., DENHARDT G. H., EPSTEIN R. H. A COMPARATIVE GENETIC STUDY OF CONDITIONAL LETHAL MUTATIONS OF BACTERIOPHAGE T4D. Genetics. 1964 Apr;49:635–648. doi: 10.1093/genetics/49.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. S., Wood W. B. Morphogenesis of bacteriophage T4 in extracts of mutant-infected cells. Proc Natl Acad Sci U S A. 1966 Mar;55(3):498–505. doi: 10.1073/pnas.55.3.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund P. T. The initial step of in vitro synthesis of deoxyribonucleic acid by the T4 deoxyribonucleic acid polymerase. J Biol Chem. 1971 Sep 25;246(18):5684–5687. [PubMed] [Google Scholar]
- Formosa T., Alberts B. M. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell. 1986 Dec 5;47(5):793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- Gauss P., Krassa K. B., McPheeters D. S., Nelson M. A., Gold L. Zinc (II) and the single-stranded DNA binding protein of bacteriophage T4. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8515–8519. doi: 10.1073/pnas.84.23.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern M. E., Mattson T., Kozinski A. W. Origins of phage T4 DNA replication as revealed by hybridization to cloned genes. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6137–6141. doi: 10.1073/pnas.76.12.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern M., Mattson T., Kozinski A. W. Late events in T4 bacteriophage DNA replication. III. Specificity of DNA reinitiation as revealed by hybridization to cloned genetic fragments. J Virol. 1982 May;42(2):422–431. doi: 10.1128/jvi.42.2.422-431.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlett N. V., Berger H. Mutations altering genetic recombination and repair of DNA in bacteriophage T4. Virology. 1975 Feb;63(2):539–567. doi: 10.1016/0042-6822(75)90326-8. [DOI] [PubMed] [Google Scholar]
- Hinton D. M., Nossal N. G. Bacteriophage T4 DNA primase-helicase. Characterization of oligomer synthesis by T4 61 protein alone and in conjunction with T4 41 protein. J Biol Chem. 1987 Aug 5;262(22):10873–10878. [PubMed] [Google Scholar]
- Hinton D. M., Silver L. L., Nossal N. G. Bacteriophage T4 DNA replication protein 41. Cloning of the gene and purification of the expressed protein. J Biol Chem. 1985 Oct 15;260(23):12851–12857. [PubMed] [Google Scholar]
- King G. J., Huang W. M. Identification of the origins of T4 DNA replication. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7248–7252. doi: 10.1073/pnas.79.23.7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krell H., Dürwald H., Hoffmann-Berling H. A DNA-unwinding enzyme induced in bacteriophage-T4-infected Escherichia coli cells. Eur J Biochem. 1979 Jan 15;93(2):387–395. doi: 10.1111/j.1432-1033.1979.tb12835.x. [DOI] [PubMed] [Google Scholar]
- Kreuzer K. N., Alberts B. M. A defective phage system reveals bacteriophage T4 replication origins that coincide with recombination hot spots. Proc Natl Acad Sci U S A. 1985 May;82(10):3345–3349. doi: 10.1073/pnas.82.10.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer K. N., Alberts B. M. Characterization of a defective phage system for the analysis of bacteriophage T4 DNA replication origins. J Mol Biol. 1986 Mar 20;188(2):185–198. doi: 10.1016/0022-2836(86)90303-7. [DOI] [PubMed] [Google Scholar]
- Kreuzer K. N., Engman H. W., Yap W. Y. Tertiary initiation of replication in bacteriophage T4. Deletion of the overlapping uvsY promoter/replication origin from the phage genome. J Biol Chem. 1988 Aug 15;263(23):11348–11357. [PubMed] [Google Scholar]
- Kreuzer K. N., Yap W. Y., Menkens A. E., Engman H. W. Recombination-dependent replication of plasmids during bacteriophage T4 infection. J Biol Chem. 1988 Aug 15;263(23):11366–11373. [PubMed] [Google Scholar]
- Liu C. C., Burke R. L., Hibner U., Barry J., Alberts B. Probing DNA replication mechanisms with the T4 bacteriophage in vitro system. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):469–487. doi: 10.1101/sqb.1979.043.01.053. [DOI] [PubMed] [Google Scholar]
- Luder A., Mosig G. Two alternative mechanisms for initiation of DNA replication forks in bacteriophage T4: priming by RNA polymerase and by recombination. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1101–1105. doi: 10.1073/pnas.79.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald P. M., Mosig G. Regulation of a new bacteriophage T4 gene, 69, that spans an origin of DNA replication. EMBO J. 1984 Dec 1;3(12):2863–2871. doi: 10.1002/j.1460-2075.1984.tb02221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkens A. E., Kreuzer K. N. Deletion analysis of bacteriophage T4 tertiary origins. A promoter sequence is required for a rifampicin-resistant replication origin. J Biol Chem. 1988 Aug 15;263(23):11358–11365. [PubMed] [Google Scholar]
- Morrical S. W., Alberts B. M. The UvsY protein of bacteriophage T4 modulates recombination-dependent DNA synthesis in vitro. J Biol Chem. 1990 Sep 5;265(25):15096–15103. [PubMed] [Google Scholar]
- Mosig G., Luder A., Garcia G., Dannenberg R., Bock S. In vivo interactions of genes and proteins in DNA replication and recombination of phage T4. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):501–515. doi: 10.1101/sqb.1979.043.01.056. [DOI] [PubMed] [Google Scholar]
- Mosig G. The essential role of recombination in phage T4 growth. Annu Rev Genet. 1987;21:347–371. doi: 10.1146/annurev.ge.21.120187.002023. [DOI] [PubMed] [Google Scholar]
- Nossal N. G., Peterlin B. M. DNA replication by bacteriophage T4 proteins. The T4 43, 32, 44--62, And 45 proteins are required for strand displacement synthesis at nicks in duplex DNA. J Biol Chem. 1979 Jul 10;254(13):6032–6037. [PubMed] [Google Scholar]
- Oishi M. Studies of DNA replication in vivo. 3. Accumulation of a single-stranded isolation product of DNA replication by conditional mutant strains of T4. Proc Natl Acad Sci U S A. 1968 Jul;60(3):1000–1006. doi: 10.1073/pnas.60.3.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno J. R., Alberts B. M. An ATP stimulation of T4 DNA polymerase mediated via T4 gene 44/62 and 45 proteins. The requirement for ATP hydrolysis. J Biol Chem. 1978 Jul 25;253(14):5174–5179. [PubMed] [Google Scholar]
- Richardson R. W., Nossal N. G. Characterization of the bacteriophage T4 gene 41 DNA helicase. J Biol Chem. 1989 Mar 15;264(8):4725–4731. [PubMed] [Google Scholar]
- Selick H. E., Kreuzer K. N., Alberts B. M. The bacteriophage T4 insertion/substitution vector system. A method for introducing site-specific mutations into the virus chromosome. J Biol Chem. 1988 Aug 15;263(23):11336–11347. [PubMed] [Google Scholar]
- Silver L. L., Nossal N. G. DNA replication by bacteriophage T4 proteins: role of the DNA-delay gene 61 in the chain-initiation reaction. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):489–494. doi: 10.1101/sqb.1979.043.01.054. [DOI] [PubMed] [Google Scholar]
- Sinha N. K., Snustad D. P. DNA synthesis in bacteriophage T4-infected Escherichia coli: evidence supporting a stoichiometric role for gene 32-product. J Mol Biol. 1971 Nov 28;62(1):267–271. doi: 10.1016/0022-2836(71)90145-8. [DOI] [PubMed] [Google Scholar]
- Tessman E. S., Peterson P. K. Suppression of the ssb-1 and ssb-113 mutations of Escherichia coli by a wild-type rep gene, NaCl, and glucose. J Bacteriol. 1982 Nov;152(2):572–583. doi: 10.1128/jb.152.2.572-583.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J. I., Anraku N., Iwama Y. Molecular mechanisms of genetic recombination in bacteriophage. VI. A mutant defective in the joining of DNA molecules. J Mol Biol. 1966 Nov 14;21(2):247–253. doi: 10.1016/0022-2836(66)90095-7. [DOI] [PubMed] [Google Scholar]
- Venkatesan M., Silver L. L., Nossal N. G. Bacteriophage T4 gene 41 protein, required for the synthesis of RNA primers, is also a DNA helicase. J Biol Chem. 1982 Oct 25;257(20):12426–12434. [PubMed] [Google Scholar]
- Warner H. R., Barnes J. E. Deoxyribonucleic acid synthesis in Escherichia coli infected with some deoxyribonucleic acid polymerase-less mutants of bacteriophage T4. Virology. 1966 Jan;28(1):100–107. doi: 10.1016/0042-6822(66)90310-2. [DOI] [PubMed] [Google Scholar]
- Warner H. R., Hobbs M. D. Incorporation of uracil-14C into nucleic acids in Escherichia coli infected with bacteriophage T4 and T4 amber mutants. Virology. 1967 Nov;33(3):376–384. doi: 10.1016/0042-6822(67)90113-4. [DOI] [PubMed] [Google Scholar]
- Wiekowski M., Dröge P., Stahl H. Monoclonal antibodies as probes for a function of large T antigen during the elongation process of simian virus 40 DNA replication. J Virol. 1987 Feb;61(2):411–418. doi: 10.1128/jvi.61.2.411-418.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. R., Yeh Y. C. Requirement of a functional gene 32 product of bacteriophage T4 in UV, repair. J Virol. 1973 Oct;12(4):758–765. doi: 10.1128/jvi.12.4.758-765.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R., Geiduschek E. P. The role of replication proteins in the regulation of bacteriophage T4 transcription. I. Gene 45 and hydroxymethyl-C-containing DNA. J Mol Biol. 1975 Aug 25;96(4):513–538. doi: 10.1016/0022-2836(75)90137-0. [DOI] [PubMed] [Google Scholar]
- Yee J. K., Marsh R. C. Locations of bacteriophage T4 origins of replication. J Virol. 1985 May;54(2):271–277. doi: 10.1128/jvi.54.2.271-277.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegian C. D., Mueller M., Selzer G., Russo V., Stahl F. W. Properties of the DNA-delay mutants of bacteriophage T4. Virology. 1971 Dec;46(3):900–919. doi: 10.1016/0042-6822(71)90090-0. [DOI] [PubMed] [Google Scholar]