Abstract
The alcohol dehydrogenase (Adh) gene family is much more complex in Pinus banksiana than in angiosperms, with at least seven expressed genes organized as two tightly linked clusters. Intron number and position are highly conserved between P. banksiana and angiosperms. Unlike angiosperm Adh genes, numerous duplications, as large as 217 bp, were observed within the noncoding regions of P. banksiana Adh genes and may be a common feature of conifer genes. A high frequency of duplication over a wide range of scales may contribute to the large genome size of conifers.
It is well-established that nuclear genomes are much larger in conifers than in angiosperms. In conifers, 2C values (where C is the total amount of DNA in a haploid nucleus) typically range from 20 to 50 pg (1), whereas the corresponding values in angiosperms are typically less than 10 pg and often less than 2 pg (2). However, relatively little is known about the organization of these large conifer genomes. While the proportion of repeated DNA is higher than in angiosperms, there is little relationship between total genomic and repetitive DNA (3). Nuclear rDNA repeat units are much longer in conifers (4–6) and Southern blot analyses of pine DNA with cDNA probes often reveal complex band patterns, suggesting larger gene families and/or larger genes than in angiosperms (7–10).
One of the best characterized plant gene families codes for alcohol dehydrogenases (ADH; EC 1.1.1.1), enzymes that play a central role in anaerobic metabolism (11, 12). Most angiosperms express two or three ADH isozymes (13) and Southern blots generally display a corresponding level of complexity (14, 15). Sequence data are available for Adh genes of a number of angiosperm species, but relatively little is known about this gene family in conifers, with the only published sequences being three partial Pinus radiata Adh cDNAs, likely representing two loci (7). Typically, two to four ADH isozymes are observed in pines (16), but P. radiata Adh cDNA clones hybridize to many fragments of P. radiata and Pinus taeda DNA (7) and many diverse genomic Adh clones were recovered from these species (17). These observations suggest that the Adh gene family is larger in pines than in angiosperms and/or that pine Adh genes are larger. We characterized Adh genes in Pinus banksiana Lamb. (jack pine) to examine these hypotheses. Also, due to the successful development of segregating PCR-based gene-specific markers, we were able to examine linkage relationships among Adh loci.
METHODS
Isozyme Analysis.
P. banksiana seeds were germinated on distilled (d) H2O-moistened filter paper in Petri dishes at room temperature for 8 days, yielding seedling root lengths of 1–2 cm. The germinating seeds were then given either anaerobic treatment (immersed for 20 hr in dH2O under filter paper) or aerobic treatment (seeds remained on moist filter paper). Starch gel electrophoresis (18) was used to examine ADH enzyme activity in the haploid megagametophytes and diploid roots of these seedlings.
cDNA Cloning and Characterization.
Total RNA was extracted (19) from pooled aerobically treated megagametophytes of a single tree and from pooled anaerobically treated seedling roots. Adh cDNAs were cloned by a strategy similar to a previously described 3′-RACE scheme (20). First-strand cDNA synthesis was initiated from a NotI primer-adaptor and amplification by the polymerase chain reaction (PCR) was performed using this primer and F1, a sense primer corresponding to the first 20 bp of coding sequence of P. radiata Adh cDNA RCS1025 (7). Amplification products were cloned into pGEM-T (Promega). A small number of the clones were partially sequenced and antisense primers were designed within the 3′ untranslated region (UTR) of each cDNA class identified. These class-specific primers, paired with F1, were then used to screen additional transformants for inserts unlike those previously encountered. Novel inserts were partially sequenced and one cDNA from each of the classes identified was sequenced entirely (both strands).
Linkage Analysis.
We identified 10 trees that were heterozygous at the Adh2 isozyme locus and extracted DNA of individual megagametophytes using proteinase K and CTAB (21). We then used the class-specific primers to screen these megagametophyte DNAs for segregating PCR-based markers of Adh loci (for detailed information about the primer pairs used, see GenBank accession nos. U48366–U48372U48366U48367U48368U48369U48370U48371U48372). Seven trees that were heterozygous for at least three PCR-based markers each were included in an analysis of linkage. Jointly segregating trees were available for all 28 two-locus combinations, except AdhC2/AdhC4. For each tree, 29–33 haploid megagametophytes were divided in two, with one half of the tissue used for Adh2 isozyme genotyping and the other half for DNA extraction. Recombination was analyzed in megagametophyte arrays of single trees using a procedure equivalent to the double backcross method (22) and heterogeneity of recombination frequencies among trees was examined using a heterogeneity χ2 test (23). In the absence of heterogeneity, pooled estimates were used.
Characterization of Genomic Sequences.
We attempted to amplify larger genomic products corresponding to each of the cDNA classes by using touchdown PCR (24) with the class-specific primers and primer F1. These products would include the complete coding sequence except for the estimated first 20 bp. We directly sequenced these genomic products using independently amplified templates for each strand and a primer walking sequencing strategy. All sequencing templates were amplified from one tree, from single megagametophytes that expressed the common ADH2 allozyme and were nonrecombinant with respect to segregating Adh PCR-based markers.
Phylogenetic Analysis.
The region shared between the completely sequenced P. banksiana Adh cDNAs, 19 angiosperm Adh genes, and one human Adh (used as an outgroup) was aligned using clustalv (25), with minor adjustments performed manually. A neighbor-joining tree (26) based on Jukes–Cantor distances (27) was constructed using mega version 1.02 (28). The relative support for groups was determined based upon 1000 bootstrap trees.
RESULTS AND DISCUSSION
Isozyme analysis of megagametophyte tissue revealed a single intensely stained protein band (ADH2), encoded by a single locus (Adh2), as has been reported for several other pines (29–32). No ADH activity was observed in extracts of aerobically treated roots, but anaerobic treatment resulted in the induction of ADH2 and a more anodal zone of activity (ADH1), for which the genetic control is unknown. Similar tissue-specific anaerobically inducible expression of ADH has been found in other plants, including P. radiata (33).
All 112 megagametophyte cDNA clones evaluated were very similar and were assigned to one class (AdhC1). Of 152 anaerobic root-derived Adh cDNAs, one was classified as AdhC1 and the rest were assigned to six additional classes (AdhC2, 1 clone; AdhC3, 48 clones; AdhC4, 91 clones; AdhC5, 7 clones; AdhC6, 3 clones; AdhC7, 1 clone). Since the efficiency by which different cDNA species were amplified likely varied, the relative numbers of clones in each of these cDNA classes cannot be expected to reflect relative mRNA abundances. The 5′ end of the cDNA sequences obtained corresponds to position 21 of the P. radiata Adh cDNA RCS1025 (7). If the length of the additional undetermined coding sequence of each is also 20 bp, each would encode a protein of 382 amino acids, except for the cDNA representing AdhC7, which would encode only 381 amino acids due to an insertion of a single thymidine that results in a TGA codon immediately preceding the original termination codon. These lengths are similar to those of angiosperm ADHs. Nucleotide (and amino acid) identities among the coding regions of these P. banksiana genes are high, ranging from 81.2% (75.4%) between AdhC1 and AdhC6 to 98.7% (98.4%) between AdhC3 and AdhC5. Divergence among the 3′ UTRs is generally too great to allow meaningful alignment.
We successfully amplified large genomic products representing AdhC2, AdhC3, AdhC4, and AdhC5. Efforts to directly sequence the AdhC4 product yielded ambiguous results within several introns, suggesting that this product, although appearing as a single 2.6-kb fragment, actually contained two or more AdhC4-like sequences. The genomic sequence of AdhC6 was obtained as a composite of three overlapping PCR products (for details, see comments in GenBank accession no. U48376U48376). Attempts to obtain AdhC1 and AdhC7 genomic amplification products that extended 5′ of exon 8 were not successful.
The genomic sequences of AdhC2, AdhC3, AdhC5, and AdhC6 are each interrupted by nine (A+T)-rich (68.2%) introns, positioned exactly as the nine introns in other characterized plant Adh genes and ranging in size from 86 to 1926 bp (mean = 263 bp, SEM = 363 bp) (Fig. 1). If two uncharacteristically large AdhC6 introns (intron 1, 1926 bp; intron 4, 1418 bp) are excluded, the average intron size falls to 180 bp (SEM = 92 bp), similar to that observed in angiosperms. These data do not support the hypothesis that pine Adh genes have more or larger introns (7, 17). However, uncharacterized family members may differ in intron structure. Our inability to amplify AdhC1 and AdhC7 genomic products that extended 5′ of exon 8 is noteworthy because one possibility is that extremely long introns were encountered.
Figure 1.
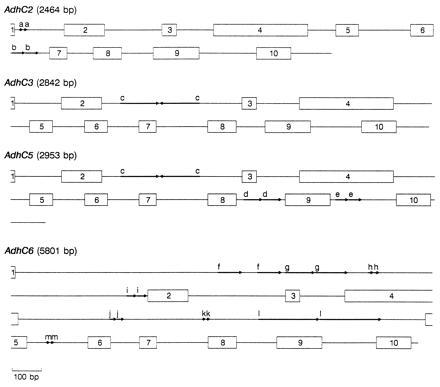
Schematic representation of four P. banksiana Adh genes. Numbered boxes represent exons and lines represent introns and 3′ UTRs. Arrows delineate 10- (a) and 43-bp (b) direct repeats in AdhC2, 141-bp (c) inverted repeat in AdhC3 and AdhC5, 65- (d) and 49-bp (e) direct repeats in AdhC5, and 86- (f), 106- (g), 17- (h), 40- (i), 21- (j), 12- (k), 217- (l), and 11- (m) bp direct repeats in AdhC6. Gene lengths are estimated from the presumed beginning of the initiation codon through the termination codon.
Duplications, the largest of which is 217 bp, are a common feature in many of the P. banksiana Adh introns (Fig. 1). Such repeats appear to be rare in angiosperm Adh genes. Except for a 104-bp element that is tandemly repeated three times in Zma1 (ref. 34, see Fig. 3 for abbreviations of gene names), we found no other large duplications in genomic angiosperm Adh sequences (Ath, Fan, Hvu2, Hvu3, Phy1, Psa1, and Zma1). To determine whether large repeats may be common in other conifer genes, we examined seven conifer nuclear gene sequences found in GenBank. Large direct repeats (>38 bp) are present in Larix laricina rbcS (GenBank accession no. X16039X16039), Pinus sylvestris BBS (X60753X60753), a P. taeda protochlorophyllide reductase gene (X66727X66727), a P. taeda 4-coumarate:CoA ligase gene (U39405U39405), and Pinus thunbergii cab (X61915X61915), but not in Pinus contorta cab (X67714X67714) nor P. sylvestris CHS (X60754X60754). The observation of large repeats in five of these seven sequences suggests that their occurrence may be widespread in conifer genes, at least within the Pinaceae.
Figure 3.
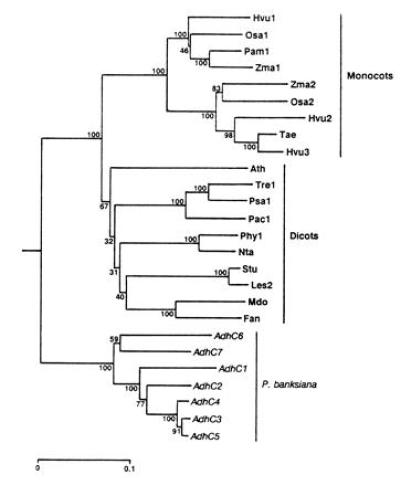
Phylogeny of Adh cDNA sequences inferred by the neighbor-joining method (26) from Jukes–Cantor distances (27) and rooted using human Adh χ (GenBank accession no. M30471M30471). Numbers represent the relative support for groupings based upon 1000 bootstrap trees and the scale bar represents 0.1 substitutions per site. The angiosperm genes included are Arabidopsis thaliana Adh (Ath; GenBank accession no. M12196M12196); Fragaria ananassa Adh (Fan; X15588X15588); Hordeum vulgare Adh1 (Hvu1; X07774X07774), Adh2 (Hvu2; X12733X12733), and Adh3 (Hvu3; X12734X12734); Lycopersicon esculentum Adh2 (Les2; M86724M86724); Malus domestica Adh (Mdo; Z48234Z48234); Nicotiana tabacum Adh (Nta; X81853X81853); Oryza sativa Adh1 (Osa1; X16296X16296) and Adh2 (Osa2; X16297X16297); Phaseolus acutifolius Adh1-F (Pac1; Z23170Z23170); Pennisetum americanum Adh1-S (Pam1; X16547X16547); Petunia hybrida Adh1 (Phy1; X54106X54106); Pisum sativum Adh1 (Psa1; X06281X06281); Solanum tuberosum Adh (Stu; X53242X53242); Trifolium repens Adh1 (Tre1; X14826X14826); Triticum aestivum Adh (Tae; ref. 37); and Zea mays Adh1-S (Zma1; X04049X04049) and Adh2 (Zma2; X01965X01965).
The high frequency of duplications within these pine Adh genes proved advantageous in the development of segregating PCR-based markers. Codominant markers were found for three loci (AdhC1, AdhC2, and AdhC7) by simply amplifying a portion of their 3′ ends from the DNA of a small panel of individuals. Sequence analysis revealed that all of these codominant polymorphisms involved large repeats within noncoding regions. Of three AdhC1 alleles, one (AdhC1-2) differed primarily by a tandem direct repeat of 42 bp in intron 9. In this same region, AdhC1-3 had an insertion of about 168 bp, containing a large degenerate inverted repeat, accompanied by the loss of about 49 bp of flanking sequence. The difference between the two AdhC2 alleles involved a large duplication (>80 bp) within the 3′ UTR that included the gene-specific PCR primer site. Although in AdhC2-2 the sequence of the internal primer site was exactly complementary to the AdhC2-specific primer, priming at the distal site was favored in PCR amplification. In AdhC2-1, either this duplication did not exist or the internal primer site was favored. The two AdhC7 alleles differed by a single direct repeat of 116 bp found in intron 9 of AdhC7-2. Dominant markers were found for AdhC3, AdhC4, and AdhC5, while AdhC6 had both dominant and codominant allele pairs. Amplifications using AdhC4- or AdhC6-specific primers produced complex band patterns, suggesting that each of these classes actually represents more than one genomic sequence, at least one of which is expressed. Due to their presence/absence or complex nature, dominant polymorphisms were not characterized.
The availability of these segregating PCR-based markers permitted an analysis of linkage, allowing us to further examine the organization of this gene family. Segregation of the individual PCR-based markers and ADH2 allozymes (Fig. 2) did not deviate significantly from 1:1 expectations and χ2 tests revealed no heterogeneity of recombination frequencies among trees. The loci comprised two linkage groups (AdhC1/AdhC2/AdhC3/AdhC4/AdhC5 and Adh2/AdhC6/AdhC7), with no recombination observed within groups and a recombination frequency of 0.0376 (SEM = 0.0130; 8 recombinants out of 213 megagametophytes examined) between groups. Pairs of ADH isozyme loci have been found to be tightly linked in other pines (16, 35, 36). It is not known which, if any, of these cDNAs encodes the ADH2 isozyme, but of the loci we identified, only AdhC6 and AdhC7 are candidates.
Figure 2.
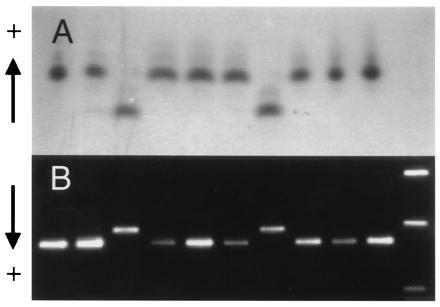
Joint segregation of Adh2 allozymes and a PCR-based marker of AdhC7. (A) Adh2 allozymes in 10 haploid megagametophytes of a single tree as detected using starch gel electrophoresis and enzyme staining. (B) PCR amplification products of AdhC7 from the same set of 10 megagametophytes revealed by agarose gel electrophoresis and ethidium bromide staining. Size markers shown in the right hand lane are the 1636-, 1018-, and 506-bp fragments of a 1-kb DNA ladder (GIBCO/BRL).
To examine the evolutionary relationships among members of this gene family, we constructed a phylogenetic tree (Fig. 3) that clearly shows three primary lineages corresponding to monocot, dicot, and pine genes. Gene duplications have taken place independently in each of these lineages. This general topology is in agreement with other published phylogenetic trees of Adh genes (7, 38–40). However, one tree (41) that was constructed using a progressive alignment of amino acid sequences did not show a clear separation of monocot and dicot lineages. In the present tree, P. banksiana AdhC1, AdhC2, AdhC3, AdhC4, and AdhC5, all members of the same linkage group, form a strongly supported lineage. AdhC6 and AdhC7, members of the other linkage group, fall outside of this lineage. This topology is corroborated by genomic sequences. Intron sequences of AdhC3 and AdhC5 were readily aligned, only some intron sequences of these genes could be aligned with AdhC2, and all of the AdhC6 introns were too divergent to be included in this alignment. The P. radiata cDNAs (7) were not included in the phylogenetic analysis because they lacked much of the region considered, but RCS1025 is most like AdhC3 (98.4% identity) and RCS1029 most closely resembles AdhC7 (99.2% identity). These data suggest that an initial duplication, predating the P. radiata–P. banksiana separation, formed the progenitors of the two linkage groups and was followed by duplication within the groups.
The fact that we could amplify markers of all seven cDNA classes from the DNA of a single haploid megagametophyte directly supports the existence of at least seven expressed Adh loci in P. banksiana, providing conclusive evidence of a much larger gene family size in pines. Clearly, the complex Adh Southern blots of P. radiata and P. taeda (7) and diversity of Adh genomic clones from these species (17) are due to the large size of this gene family. Since only two tissues were considered in the present study and the amplification primer was nondegenerate, other expressed loci likely remain undetected. Furthermore, genomic amplifications demonstrated that some cDNA classes may represent more than one locus. Whether all members of these compound classes are functional is unknown, but exons appear conserved relative to introns in AdhC4-like sequences, suggesting selective constraint. Possible explanations for the difference between the observed numbers of isozymes and expressed genes include isozymes with coincident electrophoretic migration, low concentrations, and/or low catalytic efficiencies under assay conditions. Why such a large diverse group of Adh genes is maintained in pines while as few as one is sufficient in angiosperms is unknown.
Duplication is a recurrent theme within the P. banksiana Adh gene family. Not only are there more Adh genes than have been found in angiosperms, but repeated sequences within the genes are much more common. If this pattern is typical of other conifer genes and gene families, it would represent one component contributing to the unusually large genome sizes of conifers.
Acknowledgments
David Harry shared his advice and unpublished P. radiata and P. taeda Adh sequence data. Glenn Howe provided helpful advice and Pauline Perry assisted in the laboratory. This research was supported by the College of Natural Resources, Agricultural Experiment Station, and Graduate School, University of Minnesota and is published as Minnesota Agricultural Experiment Station paper 22,276.
Footnotes
References
- 1.Ohri D, Khoshoo T N. Plant Syst Evol. 1986;153:119–132. [Google Scholar]
- 2.Arumuganathan K, Earle E D. Plant Mol Biol Rep. 1991;9:208–218. [Google Scholar]
- 3.Miksche J P. For Chron. 1985;61:449–453. [Google Scholar]
- 4.Bobola M S, Smith D E, Klein A S. Mol Biol Evol. 1992;9:125–137. doi: 10.1093/oxfordjournals.molbev.a040702. [DOI] [PubMed] [Google Scholar]
- 5.Beech R N, Strobeck C. Plant Mol Biol. 1993;22:887–892. doi: 10.1007/BF00027373. [DOI] [PubMed] [Google Scholar]
- 6.Karvonen P, Karjalainen M, Savolainen O. Genetica. 1993;88:59–68. [Google Scholar]
- 7.Kinlaw C S, Harry D E, Sederoff R R. Can J For Res. 1990;20:1343–1350. [Google Scholar]
- 8.Devey M E, Jermstad K D, Tauer C G, Neale D B. Theor Appl Genet. 1991;83:238–242. doi: 10.1007/BF00226257. [DOI] [PubMed] [Google Scholar]
- 9.Ahuja M R, Devey M E, Grover A T, Jermstad K D, Neale D B. Theor Appl Genet. 1994;88:279–282. doi: 10.1007/BF00223632. [DOI] [PubMed] [Google Scholar]
- 10.Kinlaw C S, Gerttula S M, Carter M C. Plant Mol Biol. 1994;26:1213–1216. doi: 10.1007/BF00040702. [DOI] [PubMed] [Google Scholar]
- 11.Harry D E, Kimmerer T W. For Ecol Manage. 1991;43:251–272. [Google Scholar]
- 12.Dolferus R, DeBruxelles G, Dennis E S, Peacock W J. Ann Bot (London) 1994;74:301–308. [Google Scholar]
- 13.Gottlieb L D. Science. 1982;216:373–380. doi: 10.1126/science.216.4544.373. [DOI] [PubMed] [Google Scholar]
- 14.Trick M, Dennis E S, Edwards K J R, Peacock W J. Plant Mol Biol. 1988;11:147–160. doi: 10.1007/BF00015667. [DOI] [PubMed] [Google Scholar]
- 15.Gregerson R, McLean M, Beld M, Gerats A G M, Strommer J. Plant Mol Biol. 1991;17:37–48. doi: 10.1007/BF00036804. [DOI] [PubMed] [Google Scholar]
- 16.Conkle M T. In: Isozymes of North American Forest Trees and Forest Insects. Conkle M T, editor. U.S. Dept. Agric.; 1981. For. Serv. Gen. Tech. RePSW481117. [Google Scholar]
- 17.Harry, D. E., Mordecai, K. S., Kinlaw, C. S., Loopstra, C. A. & Sederoff, R. R. (1989) in Proceedings of the 20th Southern Forest Tree Improvement Conference, June 26–30, 1989, Charleston, SC, Sponsored Publ. No. 42 of the Southern Forest Tree Improvement Committee, pp. 373–380.
- 18.Conkle M T, Hodgskiss P D, Nunnally L B, Hunter S C. Starch Gel Electrophoresis of Conifer Seeds: A Laboratory Manual. U.S. Dept. Agric.; 1982. For. Serv. Gen. Tech. RePSW64. [Google Scholar]
- 19.Coleman G D, Chen T H H, Ernst S G, Fuchigami L. Plant Physiol. 1991;96:686–692. doi: 10.1104/pp.96.3.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Froshman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perry, D. J. (1996) Ph.D. dissertation (Univ. of Minnesota, St. Paul).
- 22.Bailey N T J. Introduction to the Mathematical Theory of Genetic Linkage. London: Oxford Univ. Press; 1961. [Google Scholar]
- 23.Eckert R T, Joly R J, Neale D B. Can J For Res. 1981;11:573–579. [Google Scholar]
- 24.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higgins D G, Bleasby A J, Fuchs R. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 26.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 27.Jukes T H, Cantor C R. In: Mammalian Protein Metabolism. Munro H N, editor. New York: Academic; 1969. pp. 21–132. [Google Scholar]
- 28.Kumar S, Tamura K, Nei M. mega, Molecular Evolutionary Genetics Analysis. University Park: The Pennsylvania State Univ.; 1993. Version 1.01. [Google Scholar]
- 29.O’Malley D M, Allendorf F W, Blake G M. Biochem Genet. 1979;17:233–250. doi: 10.1007/BF00498965. [DOI] [PubMed] [Google Scholar]
- 30.Millar C I. Biochem Genet. 1985;23:933–946. doi: 10.1007/BF00499938. [DOI] [PubMed] [Google Scholar]
- 31.Furnier G R, Knowles P, Aleksiuk M A, Dancik B P. Can J Genet Cytol. 1986;28:601–604. [Google Scholar]
- 32.Strauss S H, Conkle M T. Theor Appl Genet. 1986;72:483–493. doi: 10.1007/BF00289530. [DOI] [PubMed] [Google Scholar]
- 33.Harry D E, Kinlaw C S, Sederoff R R. In: Genetic Manipulation of Woody Plants. Hanover J W, Keathley D E, editors. New York: Plenum; 1988. pp. 275–290. [Google Scholar]
- 34.Dennis E S, Gerlach W L, Pryor A J, Bennetzen J L, Inglis A, Llewellyn D, Sachs M M, Ferl R J, Peacock W J. Nucleic Acids Res. 1984;12:3983–4000. doi: 10.1093/nar/12.9.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudin D, Ekberg I. Silvae Genet. 1978;27:1–12. [Google Scholar]
- 36.Szmidt A E, Muona O. Hereditas. 1989;111:91–97. [Google Scholar]
- 37.Mitchell L E, Dennis E S, Peacock W J. Genome. 1989;32:349–358. doi: 10.1139/g89-454. [DOI] [PubMed] [Google Scholar]
- 38.Yokoyama S, Yokoyama R, Kinlaw C S, Harry D E. Mol Biol Evol. 1990;7:143–154. doi: 10.1093/oxfordjournals.molbev.a040593. [DOI] [PubMed] [Google Scholar]
- 39.Gregerson R G, Cameron L, McLean M, Dennis P, Strommer J. Genetics. 1993;133:999–1007. doi: 10.1093/genetics/133.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yokoyama S, Harry D E. Mol Biol Evol. 1993;10:1215–1226. doi: 10.1093/oxfordjournals.molbev.a040073. [DOI] [PubMed] [Google Scholar]
- 41.Sun H-W, Plapp B V. J Mol Evol. 1992;34:522–535. doi: 10.1007/BF00160465. [DOI] [PubMed] [Google Scholar]


