Summary
After export from the nucleus it turns out that all mRNAs are not treated equally. Not only is mRNA subject to translation, but through RNA binding proteins and other trans-acting factors, eukaryotic cells interpret codes for spatial sorting within the mRNA sequence. These codes instruct the cytoskeleton and translation apparatus to make decisions about where to transport and when to translate the intended protein product. Signaling pathways decode extra-cellular cues and can modify transport and translation factors in the appropriate cytoplasmic space to achieve translation locally. Identifying regulatory sites on transport factors as well as novel physiological functions for well known translation factors have provided significant advances in how spatially controlled translation impacts cell function.
Introduction
Spatial regulation of translation within the cytoplasm results in the accumulation of newly synthesized proteins in discrete locations of the cell. The most widely studied mechanism to spatially restrict protein synthesis is through active sorting of the template for translation within the cytoplasm, often termed mRNA localization [1]. mRNA localization allows cells the flexibility to determine the exact place and time of protein synthesis in the absence of de novo transcription providing a mechanism to quickly respond to changes in their extra-cellular environment. The information required for localization is contained within the mRNA sequence. There are many potential mechanisms to explain how a nucleic acid sequence distributes an mRNA within the cytoplasm [1]. It is possible that sequences within the mRNA interact directly with cytoskeletal elements [2], although most current models for localization suggest that mRNA sequences interact with subsets of RNA binding proteins, forming a Localizing Ribonucleo-Protein (L-RNP) complex. The L-RNP localizes through interactions with cytoskeletal elements either directly or indirectly. Therefore, in addition to carrying the information required for protein synthesis, an mRNA contains sequences whose purpose is to select the appropriate complement of trans-acting factors to achieve proper spatio-temporal regulation of translation.
Many localized mRNAs are translationally repressed, and it has been hypothesized that this is to prevent ectopic synthesis during transport [1]. Localized mRNAs need to associate with localizing factors as well as reversible translational repressors that are responsive to spatial cues in the cytoplasm. To ensure repression in the cytoplasm, translational regulatory factors join the mRNA as soon as it is synthesized in the nucleus [3]. The activities of these localization and translational repression factors need to be coordinated to achieve the proper timing of events and can be contained within a single factor or provided by interacting factors. As a consequence, a great deal of study has been devoted to the formation and function of the components of RNP complexes. Global analysis of mRNA associated with RNP components has observed that many of them associate with multiple functionally related mRNAs [4]. Thus the cell's ability to respond to extra-cellular signals may be coordinately regulated through RNPs by initiating the translation of many members of a multi-protein complex at the same time and place [4]. This central role of RNP complexes in spatial control of translation will be illustrated through one well-known mammalian mRNA localization factor, ZBP1. Other systems, such as yeast and Drosophila operate through parallel mechanisms and will not be detailed here due to space restrictions [5,6].
ZBP1 is a Src dependent translational repressor
ZBP1 is an RNA binding protein isolated from chick embryo fibroblasts based on its affinity for a cis-acting 54 nucleotide cytoplasmic localization element within the 3′UTR of β-actin mRNA known as the zipcode [7]. The zipcode sequence was necessary and sufficient for peripheral targeting of RNA [8]. ZBP1 functions as a translational inhibitor by preventing 80S ribosomal complex formation [3]. Importantly, Src phosphorylation of ZBP1 at tyrosine 396 resulted in translational derepression [3]. A non-phosphorylatable ZBP1 mutant prevented translational derepression resulting in decreased peripheral actin and aberrant neurite outgrowth [3]. Interestingly, β-actin translation sites were redistributed to the perinuclear cytoplasm in myoblast cells containing a transfected β-actin mRNA lacking the zipcode, supporting the hypothesis that interaction between ZBP1 and the zipcode prevents precocious translation [9]. IMP1 (the human ortholog to ZBP1) RNP complexes, biochemically isolated from HEK293 cells contain exon junction complex components and lack eIF4E, eIF4G, and 60S ribosomal subunits suggesting that IMP1 associated mRNAs have not undergone translation [10]. In addition, a mouse ortholog of ZBP1 represses the translation of insulin-like growth factor II mRNA in a developmentally regulated manner [11]. Altogether these data demonstrate that an interaction between ZBP1 and the zipcode is required to regulate β-actin mRNP complexes at the level of localization and translation. In this case, the localization and translational repression activities for β-actin mRNP complexes are present within a single trans-acting factor, and phosphorylation of this factor coordinates these activities.
β-actin mRNA is targeted in a Rho-dependent manner
In chicken embryo fibroblasts, β-actin mRNP complex targeting to the cell periphery was induced with serum or PDGF implicating signal transduction pathways in this process [12]. Inhibiting tyrosine kinase activity prevented PDGF induced β-actin RNP complex targeting [12]. Rho GTPases were similarly involved in localization as Rho inhibitors and a dominant negative RhoA reduced serum induced peripheral targeting of β-actin mRNP complexes while peripheral targeting increased in the presence of constitutively active RhoA [13]. In addition, ROCK inhibition reduced β-actin mRNP complex targeting while overexpression of p160ROCK increased targeting [13]. These data indicate RhoA and its downstream effector ROCK are required for β-actin mRNP complex targeting to discrete cytoplasmic sites. Consistent with the hypothesis that functionally related mRNAs may be coordinately regulated, all seven mRNAs of the Arp 2/3 complex are targeted to cellular protrusions in what is thought to be a Rho GTPase dependent manner [14]. Thus RhoA and ROCK signaling is required for peripheral targeting of RNP complexes.
ZBP1, adhesion and metastasis
ZBP1 levels in motile tumor cells collected in an in vivo collection assay were reduced 10 fold compared to the levels in cells remaining in the tumor, inversely correlating ZBP1 levels with metastatic potential [15,16]. Contrasted with this, high levels of IMP1 correlated with poor prognosis in ovarian carcinomas and with metastasis in colon cancer [17,18]. Given ZBP1's role as a translational regulator and localization factor, it is not surprising that ZBP1 expression could result in disparate effects since different substrate mRNAs can be found within ZBP1 containing RNP complexes in different cell backgrounds. Thus ZBP1 may act as an RNA regulon serving to integrate signals required for mRNA targeting and local translation of RNP complexes containing functionally related transcripts [4,19].
Areas with high RhoA activity and high Src activity are likely sites of ZBP1 RNP complex translational derepression establishing a local translation signature for ZBP1 containing RNP complexes [3,13] This local translation signature is found at cell-cell and cell-substrate adhesion complexes suggesting that ZBP1 mediated local β-actin translation may occur at these sites. In fact, full-length β-actin mRNA is locally translated and accumulated at cell-cell contacts in myoblast cells. In contrast, β-actin mRNA lacking the zipcode caused mislocalization of β-actin translation sites resulting in a significant reduction in the amount of N-cadherin targeted to adherens junctions [9]. Several studies support a role for ZBP1 mediated local translation in regulating cellular adhesions[9,20-23]. Depletion of IMP1 from HeLa adenocarcinoma cells resulted in a decrease in cell-cell contacts, reduced invadopod formation and delayed cell spreading [20], and a ZBP1 paralog was found at spreading initiation centers following replating in culture [21]. Moreover, β-actin, N-cadherin, β-catenin and other members of adherens junction complexes contain putative zipcode sequences suggesting that all of these mRNAs may be coordinately regulated. These data provide a physiological context for localized translation and may explain how ZBP1 functions as a metastasis suppressor in certain cell types [16,22]. Loss of ZBP1 expression in these cells may weaken cell-cell contacts at the level of adherens and tight junctions resulting in cells that no longer have an intrinsic polarity and are not attached as strongly to their neighbors making it easier for these cells to orient and move toward chemo-attractant gradients that entice cells to move out of the tumor.
Localized translation in Neurons
The mammalian nervous system has emerged as a particularly influential system for studies of localized translation and significant progress in our understanding the impact of spatially regulating translation has come from studies in neurons. At least two events in differentiated neurons have been proposed to involve localized translation within distinct domains of the cytoplasm. The first role for localized mRNA translation in developing neurons is within the growth cones of axons and is involved in axon guidance in response to guidance cues as well as during axon regeneration after injury [24-26]. Interestingly, the ZBP1 dependent localization system may play a role in this process similar to the one it plays in cell motility since β-actin mRNA and the Xenopus ZBP1 homolog, Vg1RBP/Vera, have both been recently demonstrated to localize to growth cones of Xenopus retinal axons [24,25]. Previously it was also demonstrated that ZBP1 and localization of β-actin mRNA played a role in dendritic spine formation of rodent hippocampal neurons [27,28].
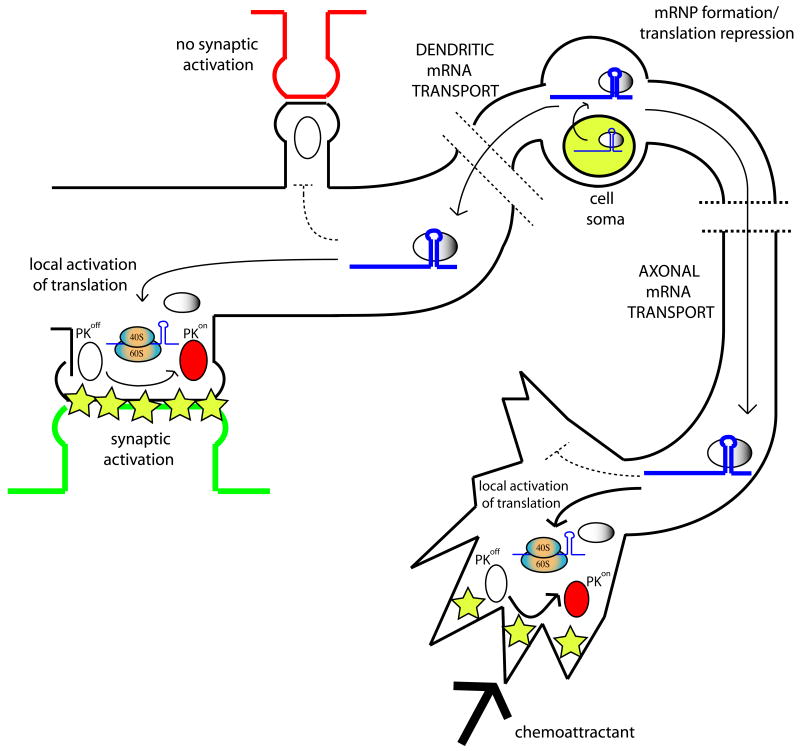
A second role for localized translation in neuron cytoplasm is in synaptic plasticity (Fig. 1). Early studies examining the long-term changes that occur at synapses following stimulation recognized that new protein synthesis was required for later phases of these changes to occur [29,30]. Modification of specific translation factors in response to synaptic activity have been defined recently and these studies have begun to reveal the molecular mechanisms by which activity influences translation in neurons. There have been two major avenues of investigation into the influence of synaptic activity on the translation machinery, one focused on the targeting of mRNAs specifically to the axons or dendrites of neurons and the other focused on the regulation of the general translation machinery due to synaptic activity. These two pathways likely operate together to achieve local protein synthesis within the processes of neurons.
Figure 1. Spatial control of mRNA translation in neurons.
Localizing mRNA to be transported to the neuronal processes begins RNP (L-RNP) formation within the nucleus, helping to ensure stringent translational repression. In the cell soma after export the mRNA (blue line) and associated RNA binding proteins (gray circle) form the L-RNP. In one pathway, an L-RNP is actively transported along the axon (axonal mRNA transport) to the growth cone where guidance cues activate local signal transduction pathways (yellow stars). This activates kinases (open circle; inactive kinase, red circle; activated kinase) that modify components of the transport and/or translation machinery, resulting in local translation (represented by 40S and 60S ribosomal subunits) of the mRNA toward the direction of the guidance cues to aid in navigation of the growth cone. In another pathway an L-RNP is actively transported into the dendrites (dendritic mRNA transport). At the post-synaptic region of activated synapses (green pre-synaptic terminal) synaptic activity activates signal transduction pathways, activating kinases that modify components of the transport and/or translation machinery, resulting in local translation of the mRNA in the vicinity of the guidance cues. The lack of activated kinases at non-stimulated synapses (red pre-synaptic terminal) does not cause translational de-repression at these sites.
Targeting mRNA to active synapses
In mature neurons, specific targeting of mRNAs to distinct locations within the cytoplasm provides the cell with a very powerful way to rapidly affect the concentration of particular proteins at regions of the neuron quite distal to the nucleus, a particularly important function when the lengths of some neuronal processes can reach several orders of magnitude over the length of the cell soma [31,32]. Based on global analyses of mRNA content within the processes of neurons it is clear that not all cellular mRNAs are present at these distal sites [33-35]. Because the mRNA content within neurites is not equivalent to the population of mRNA present within the whole cell, it is reasonable to hypothesize that mRNAs are under active sorting mechanisms in the neuronal cytoplasm.
Recruitment of individual mRNAs directly to active synapses has not been demonstrated directly, but many results indicate that synaptic activity influences the distribution of mRNA as well as mRNA binding factors within the processes of neurons [27,36-39]. Several RNA binding proteins demonstrate enrichment within microscopically observable punctate structures, both in fixed cells as well as in living cells using fluorescent protein chimeras [40,41]. Fluorescent mRNAs capable of transport into neuronal processes can be found in similar formations after microinjection, and general RNA staining dyes also show punctate staining [42,43]. Based on all of these observations, RNPs have been proposed to transport within entities that have been called RNA granules [40]. The heterogeneous nature of these RNA containing entities within the cytoplasm has made it challenging to gather information on specific mRNA transport pathways by studying them. Despite this, it is abundantly clear that synaptic activity influences the distribution and motility of these entities, and based on this it has been proposed that mRNA localizes to active synapses through the RNA binding factors and activities associated with RNA granules.
Micro RNAs (miRNAs) are a very recent addition to the repertoire of trans-acting factors that are involved in recognizing mRNA sequence. miRNAs are endogenous small RNAs (21 nt) that have complementarity to sites within subsets of mRNAs and as a result provide sequence specific binding to those mRNAs [44]. miRNA targeted mRNAs are post-transcriptionally silenced, through translational repression and perhaps enhanced mRNA turnover [45]. The miRNAs are part of a much larger multi-protein complex, and the mRNA-miRNA interaction functions to target this complex to an mRNA [45]. Several RNA binding proteins implicated in the transport of mRNA as well as translational control and stability of mRNA have been found among the components of these miRNA-associated complexes [46]. Moreover, components of miRNA RNP complexes are found in processes and at synapses, and one particular interaction (miR134-Limk1 mRNA) is important for controlling the size of dendritic spines in a synaptic activity sensitive mechanism [47,48]. Although it has not been shown that mRNA-miRNA interaction functions to localize mRNAs to the processes of neurons, this is evidence that mRNA-miRNA interactions might repress translation of mRNAs that do get localized.
Global activity, local effect
Recent work involving the translation machinery in neurons has also provided novel insights into how regulation of the general machinery may participate in spatial control of translation within the cytoplasm. The example most relevant to neuron function in learning and memory comes from studies of GCN2, an eIF2α kinase. eIF2α phosphorylation status appears to play a central role in controlling expression of the CREB-antagonizing ATF4 transcription factor [49,50]. ATF4 acts to repress memory formation that is stimulated by CREB mediated transcription, and GCN2-/- mice make less ATF4, therefore memory formation is enhanced [49]. The effect of GCN2 ablation in this process being due to eIF2α is supported by mice harboring a mutated eIF2α allele, that prevents the inhibitory GCN2 phosphorylation, also showing enhanced memory formation [50]. This suggests an attractive model for spatial control of translation where at the activated synapses in the neuronal memory circuit, local changes in eIF2α phosphorylation lead to the effects on ATF4 protein production. It has not been demonstrated directly that eIF2α is only modified locally or that ATF4 is translated at active synapses. However, given the strong spatial partitioning of synapses within the cytoplasmic volume of a neuron it is feasible that controlling a general factor by local activation from individual synapses can provide spatial control of translation of specific mRNAs. Recent publications have also explored responsiveness of the general translation machinery to synaptic activity suggesting that multiple mechanisms of translational control may be impacted by synaptic activity [51-54].
Concluding Remark
In this review we examined data on the physiological consequences of mRNA localization and local translation in both somatic and neuronal cells. Emerging evidence indicates that RNP complexes containing translational silencing factors are key mediators of a cell's initial response to extracellular environmental changes. In certain carcinoma cells, ZBP1 mediated regulation of β-actin translation sites may be required to prevent progression to metastasis. In neurons, regulated local translation occurs within growth cones to impact guidance as well as at synapses to effect plasticity in learning and memory. These examples underscore the importance of mRNA targeting and local translation on the physiology of multiple cell types.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Czaplinski K, Singer RH. Pathways for mRNA localization in the cytoplasm. Trends Biochem Sci. 2006;31:687–693. doi: 10.1016/j.tibs.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Rom I, Faicevici A, Almog O, Neuman-Silberberg FS. Drosophila Dynein light chain (DDLC1) binds to gurken mRNA and is required for its localization. Biochim Biophys Acta. 2007;1773:1526–1533. doi: 10.1016/j.bbamcr.2007.05.005. [DOI] [PubMed] [Google Scholar]
- ••3.Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]; This manuscript demonstrated that ZBP1 acts as a translational repressor by preventing 80S ribosome formation. Importantly, ZBP1 mediated translational repression begins in the nucleus at transcription sites. In addition, translational repression is relieved by Src phosphorylation of a specific tyrosine residue on ZBP1.
- •4.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]; This review discusses the concept of RNA binding proteins functioning as regulators of multiple mRNAs encoding functionally related proteins.
- 5.Paquin N, Menade M, Poirier G, Donato D, Drouet E, Chartrand P. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol Cell. 2007;26:795–809. doi: 10.1016/j.molcel.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 6.Geng C, Macdonald PM. Imp associates with squid and Hrp48 and contributes to localized expression of gurken in the oocyte. Mol Cell Biol. 2006;26:9508–9516. doi: 10.1128/MCB.01136-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••9.Rodriguez AJ, Shenoy SM, Singer RH, Condeelis J. Visualization of mRNA translation in living cells. J Cell Biol. 2006;175:67–76. doi: 10.1083/jcb.200512137. [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript showed that local β-actin translation required the zipcode sequence. In addition, it was demonstrated that mis-targeting these translation sites resulted in reduced accumulation of cadherin at cell-cell contacts.
- 10.Jonson L, Vikesaa J, Krogh A, Nielsen LK, Hansen T, Borup R, Johnsen AH, Christiansen J, Nielsen FC. Molecular composition of IMP1 ribonucleoprotein granules. Mol Cell Proteomics. 2007;6:798–811. doi: 10.1074/mcp.M600346-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19:1262–1270. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latham VM, Jr, Kislauskis EH, Singer RH, Ross AF. Beta-actin mRNA localization is regulated by signal transduction mechanisms. J Cell Biol. 1994;126:1211–1219. doi: 10.1083/jcb.126.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Latham VM, Yu EH, Tullio AN, Adelstein RS, Singer RH. A Rho-dependent signaling pathway operating through myosin localizes beta-actin mRNA in fibroblasts. Curr Biol. 2001;11:1010–1016. doi: 10.1016/s0960-9822(01)00291-3. [DOI] [PubMed] [Google Scholar]
- •14.Mingle LA, Okuhama NN, Shi J, Singer RH, Condeelis J, Liu G. Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J Cell Sci. 2005;118:2425–2433. doi: 10.1242/jcs.02371. [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript demonstrates that the mRNA encoding all seven members of the Arp2/3 complex target to the cell periphery. This is the first demonstration that the localization of all the mRNAs encoding components of a multi-protein complex can be coordinately localize.
- 15.Wang W, Wyckoff JB, Frohlich VC, Oleynikov Y, Huttelmaier S, Zavadil J, Cermak L, Bottinger EP, Singer RH, White JG, et al. Single cell behavior in metastatic primary mammary tumors correlated with gene expression patterns revealed by molecular profiling. Cancer Res. 2002;62:6278–6288. [PubMed] [Google Scholar]
- 16.Wang W, Goswami S, Lapidus K, Wells AL, Wyckoff JB, Sahai E, Singer RH, Segall JE, Condeelis JS. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 2004;64:8585–8594. doi: 10.1158/0008-5472.CAN-04-1136. [DOI] [PubMed] [Google Scholar]
- 17.Kobel M, Weidensdorfer D, Reinke C, Lederer M, Schmitt WD, Zeng K, Thomssen C, Hauptmann S, Huttelmaier S. Expression of the RNA-binding protein IMP1 correlates with poor prognosis in ovarian carcinoma. Oncogene. 2007;26:7584–7589. doi: 10.1038/sj.onc.1210563. [DOI] [PubMed] [Google Scholar]
- 18.Dimitriadis E, Trangas T, Milatos S, Foukas PG, Gioulbasanis I, Courtis N, Nielsen FC, Pandis N, Dafni U, Bardi G, et al. Expression of oncofetal RNA-binding protein CRD-BP/IMP1 predicts clinical outcome in colon cancer. Int J Cancer. 2007;121:486–494. doi: 10.1002/ijc.22716. [DOI] [PubMed] [Google Scholar]
- 19.Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Mol Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- •20.Vikesaa J, Hansen TV, Jonson L, Borup R, Wewer UM, Christiansen J, Nielsen FC. RNA-binding IMPs promote cell adhesion and invadopodia formation. Embo J. 2006;25:1456–1468. doi: 10.1038/sj.emboj.7601039. [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript presents data on loss of function experiments on the ZBP1 ortholog IMP1. Loss of IMP1 function resulted in defects in cell adhesion, cell spreading, and invadopod formation.
- 21.de Hoog CL, Foster LJ, Mann M. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell. 2004;117:649–662. doi: 10.1016/s0092-8674(04)00456-8. [DOI] [PubMed] [Google Scholar]
- ••22.Lapidus K, Wyckoff J, Mouneimne G, Lorenz M, Soon L, Condeelis JS, Singer RH. ZBP1 enhances cell polarity and reduces chemotaxis. J Cell Sci. 2007;120:3173–3178. doi: 10.1242/jcs.000638. [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript demonstrated that expression of ZBP1 in carcinoma cells that normally lack this protein prevented metastatic progression in living animals.
- 23.Hansen TV, Hammer NA, Nielsen J, Madsen M, Dalbaeck C, Wewer UM, Christiansen J, Nielsen FC. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol. 2004;24:4448–4464. doi: 10.1128/MCB.24.10.4448-4464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •24.Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for beta-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat Neurosci. 2006;9:1265–1273. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]; The authors show that BDNF stimulates local translation to induce attractive growth cone turning. Using antisense oligos that target the 3′UTR of β-actin mRNA and block the putative mRNA binding proteins, the authors find that this local stimulation by BDNF also induces local interaction between β-actin mRNA and Vg1RBP (a Xenopus ZBP1 homolog).
- •25.Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show that netrin-1 stimulates movement of the β-actin mRNA binding protein, Vg1RBP, into Xenopus retinal axon filopodia. This coincides with an asymmetric increase in β-actin mRNA translation associated with netrin mediated growth cone turning.
- ••26.Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, Kendall M, Smith DS, Bassell GJ, Twiss JL. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol. 2007;178:965–980. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that specific neurotrophins result in differential recruitment of mRNAs to sites of signalling, showing that there is specificity to the response of individual mRNAs to individual signals in regenerating axons. This suggests that RNP mediated regulation is an extremely flexible and can provide a wide range of regulatory possibilities.
- 27.Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eom T, Antar LN, Singer RH, Bassell GJ. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J Neurosci. 2003;23:10433–10444. doi: 10.1523/JNEUROSCI.23-32-10433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 30.Martin KC, Zukin RS. RNA trafficking and local protein synthesis in dendrites: an overview. J Neurosci. 2006;26:7131–7134. doi: 10.1523/JNEUROSCI.1801-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dahm R, Kiebler M, Macchi P. RNA localisation in the nervous system. Semin Cell Dev Biol. 2007;18:216–223. doi: 10.1016/j.semcdb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 32.Wu CW, Zeng F, Eberwine J. mRNA transport to and translation in neuronal dendrites. Anal Bioanal Chem. 2007;387:59–62. doi: 10.1007/s00216-006-0916-1. [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto M, Setou M, Inokuchi K. Transcriptome analysis reveals the population of dendritic RNAs and their redistribution by neural activity. Neurosci Res. 2007;57:411–423. doi: 10.1016/j.neures.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 34.Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci. 2006;26:13390–13399. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moccia R, Chen D, Lyles V, Kapuya E, E Y, Kalachikov S, Spahn CM, Frank J, Kandel ER, Barad M, et al. An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J Neurosci. 2003;23:9409–9417. doi: 10.1523/JNEUROSCI.23-28-09409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile x mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J Neurosci. 2004;24:2648–2655. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steward O, Worley PF. Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron. 2001;30:227–240. doi: 10.1016/s0896-6273(01)00275-6. [DOI] [PubMed] [Google Scholar]
- 38.Wells DG, Dong X, Quinlan EM, Huang YS, Bear MF, Richter JD, Fallon JR. A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons. J Neurosci. 2001;21:9541–9548. doi: 10.1523/JNEUROSCI.21-24-09541.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grooms SY, Noh KM, Regis R, Bassell GJ, Bryan MK, Carroll RC, Zukin RS. Activity bidirectionally regulates AMPA receptor mRNA abundance in dendrites of hippocampal neurons. J Neurosci. 2006;26:8339–8351. doi: 10.1523/JNEUROSCI.0472-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 41.Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 42.Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Bassell GJ, Kosik KS. Translocation of RNA granules in living neurons. J Neurosci. 1996;16:7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shan J, Munro TP, Barbarese E, Carson JH, Smith R. A molecular mechanism for mRNA trafficking in neuronal dendrites. J Neurosci. 2003;23:8859–8866. doi: 10.1523/JNEUROSCI.23-26-08859.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 45.Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 46.Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••47.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]; This work provides an example of the function of miRNA mediated translational repression in dendrites, showing that miRNA targeting occurs to dendritic spines and provides a specific target that is silenced in stimulation sensitive manner due to the mRNA-miRNA interaction.
- •48.Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]; In Drosophila, the miRNA pathway suppresses the dendritic expression of CaMKII and mRNA transport pathway factors, and synaptic stimulation de-represses these mRNAs through degrading the miRNA factors.
- •49.Costa-Mattioli M, Gobert D, Harding H, Herdy B, Azzi M, Bruno M, Bidinosti M, Ben Mamou C, Marcinkiewicz E, Yoshida M, et al. Translational control of hippocampal synaptic plasticity and memory by the eIF2alpha kinase GCN2. Nature. 2005;436:1166–1173. doi: 10.1038/nature03897. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper finds that an eIF2α kinase is important for memory, since GCN2-/- mice show a much lower threshold for LTP in hippocampal neurons.
- ••50.Costa-Mattioli M, Gobert D, Stern E, Gamache K, Colina R, Cuello C, Sossin W, Kaufman R, Pelletier J, Rosenblum K, et al. eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell. 2007;129:195–206. doi: 10.1016/j.cell.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper follows Costa-Mattioli et al. 2005 confirming that the phosphorylation state of eIF2α is responsible for the effect of GCN2 knockout on learning, with a central role in regulating plasticity. Together these papers represent an elegant use of mouse genetic models to define at the molecular level the role translation plays in learning and memory.
- •51.Banko JL, Hou L, Poulin F, Sonenberg N, Klann E. Regulation of eukaryotic initiation factor 4E by converging signaling pathways during metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2006;26:2167–2173. doi: 10.1523/JNEUROSCI.5196-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that metabotropic glutamate receptor signaling signals the cap-dependent translation machinery through eIF4E, the cap binding protein and its regulators.
- 52.Kanhema T, Dagestad G, Panja D, Tiron A, Messaoudi E, Havik B, Ying SW, Nairn AC, Sonenberg N, Bramham CR. Dual regulation of translation initiation and peptide chain elongation during BDNF-induced LTP in vivo: evidence for compartment-specific translation control. J Neurochem. 2006;99:1328–1337. doi: 10.1111/j.1471-4159.2006.04158.x. [DOI] [PubMed] [Google Scholar]
- •53.Sutton MA, Taylor AM, Ito HT, Pham A, Schuman EM. Postsynaptic decoding of neural activity: eEF2 as a biochemical sensor coupling miniature synaptic transmission to local protein synthesis. Neuron. 2007;55:648–661. doi: 10.1016/j.neuron.2007.07.030. [DOI] [PubMed] [Google Scholar]; This paper investigates how the phosphorylation state of eEF2, the translational elongation factor, is sensitive to synaptic activity in hippocampal neurons and how this is related to translational capacity in neurons.
- 54.Jordan BA, Fernholz BD, Khatri L, Ziff EB. Activity-dependent AIDA-1 nuclear signaling regulates nucleolar numbers and protein synthesis in neurons. Nat Neurosci. 2007;10:427–435. doi: 10.1038/nn1867. [DOI] [PubMed] [Google Scholar]