Abstract
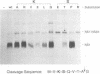
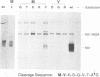
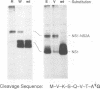
We have previously shown that proper processing of dengue type 4 virus NS1 from the NS1-NS2A region of the viral polyprotein requires a hydrophobic N-terminal signal and the downstream NS2A. Results from deletion analysis indicate that a minimum length of eight amino acids at the C terminus of NS1 is required for cleavage at the NS1-NS2A junction. Comparison of this eight-amino-acid sequence with the corresponding sequences of other flaviviruses suggests a consensus cleavage sequence of Met/Leu-Val-Xaa-Ser-Xaa-Val-Xaa-Ala. Site-directed mutagenesis was performed to construct mutants of NS1-NS2A that contained a single amino acid substitution at different positions of the consensus cleavage sequence or at the immediate downstream position. Three to eight different substitutions were made at each position. A total of 50 NS1-NS2A mutants were analyzed for their cleavage efficiency relative to that of the wild-type dengue type 4 virus sequence. As predicted, nearly all substitutions at positions P1, P3, P5, P7, and P8, occupied by conserved amino acids, yielded low levels of cleavage, with the exception that Pro or Ala substituting for Ser (P5) was tolerated. Substitutions of an amino acid at the remaining positions occupied by nonconserved amino acids generally yielded high levels of cleavage. However, some substitutions at nonconserved positions were not tolerated. For example, substitution of Gly or Glu for Gln (P4) and substitution of Val or Glu for Lys (P6) each yielded a low level of cleavage. Overall, these data support the proposed cleavage sequence motif deduced by comparison of sequences among the flaviviruses. This study also showed that in addition to the eight-amino-acid sequence, the amino acid immediately following the NS1-NS2A cleavage site plays a role in cleavage.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bazan J. F., Fletterick R. J. Detection of a trypsin-like serine protease domain in flaviviruses and pestiviruses. Virology. 1989 Aug;171(2):637–639. doi: 10.1016/0042-6822(89)90639-9. [DOI] [PubMed] [Google Scholar]
- Blundell T. L., Lapatto R., Wilderspin A. F., Hemmings A. M., Hobart P. M., Danley D. E., Whittle P. J. The 3-D structure of HIV-1 proteinase and the design of antiviral agents for the treatment of AIDS. Trends Biochem Sci. 1990 Nov;15(11):425–430. doi: 10.1016/0968-0004(90)90280-o. [DOI] [PubMed] [Google Scholar]
- Bonner W. M. Use of fluorography for sensitive isotope detection in polyacrylamide gel electrophoresis and related techniques. Methods Enzymol. 1983;96:215–222. doi: 10.1016/s0076-6879(83)96019-6. [DOI] [PubMed] [Google Scholar]
- Cahour A., Falgout B., Lai C. J. Cleavage of the dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions is mediated by viral protease NS2B-NS3, whereas NS4A/NS4B may be processed by a cellular protease. J Virol. 1992 Mar;66(3):1535–1542. doi: 10.1128/jvi.66.3.1535-1542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S., Brechling K., Moss B. Vaccinia virus expression vector: coexpression of beta-galactosidase provides visual screening of recombinant virus plaques. Mol Cell Biol. 1985 Dec;5(12):3403–3409. doi: 10.1128/mcb.5.12.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T. J., Grakoui A., Rice C. M. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol. 1991 Nov;65(11):6042–6050. doi: 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T. J., McCourt D. W., Rice C. M. Production of yellow fever virus proteins in infected cells: identification of discrete polyprotein species and analysis of cleavage kinetics using region-specific polyclonal antisera. Virology. 1990 Jul;177(1):159–174. doi: 10.1016/0042-6822(90)90470-c. [DOI] [PubMed] [Google Scholar]
- Chambers T. J., Weir R. C., Grakoui A., McCourt D. W., Bazan J. F., Fletterick R. J., Rice C. M. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8898–8902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coia G., Parker M. D., Speight G., Byrne M. E., Westaway E. G. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J Gen Virol. 1988 Jan;69(Pt 1):1–21. doi: 10.1099/0022-1317-69-1-1. [DOI] [PubMed] [Google Scholar]
- Deubel V., Kinney R. M., Trent D. W. Nucleotide sequence and deduced amino acid sequence of the nonstructural proteins of dengue type 2 virus, Jamaica genotype: comparative analysis of the full-length genome. Virology. 1988 Jul;165(1):234–244. doi: 10.1016/0042-6822(88)90677-0. [DOI] [PubMed] [Google Scholar]
- Elroy-Stein O., Fuerst T. R., Moss B. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5' sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6126–6130. doi: 10.1073/pnas.86.16.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgout B., Chanock R., Lai C. J. Proper processing of dengue virus nonstructural glycoprotein NS1 requires the N-terminal hydrophobic signal sequence and the downstream nonstructural protein NS2a. J Virol. 1989 May;63(5):1852–1860. doi: 10.1128/jvi.63.5.1852-1860.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgout B., Pethel M., Zhang Y. M., Lai C. J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991 May;65(5):2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Donchenko A. P., Koonin E. V., Blinov V. M. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res. 1989 May 25;17(10):3889–3897. doi: 10.1093/nar/17.10.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn Y. S., Galler R., Hunkapiller T., Dalrymple J. M., Strauss J. H., Strauss E. G. Nucleotide sequence of dengue 2 RNA and comparison of the encoded proteins with those of other flaviviruses. Virology. 1988 Jan;162(1):167–180. doi: 10.1016/0042-6822(88)90406-0. [DOI] [PubMed] [Google Scholar]
- Henchal E. A., Putnak J. R. The dengue viruses. Clin Microbiol Rev. 1990 Oct;3(4):376–396. doi: 10.1128/cmr.3.4.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Lai C. J. Cleavage of dengue virus NS1-NS2A requires an octapeptide sequence at the C terminus of NS1. J Virol. 1990 Sep;64(9):4573–4577. doi: 10.1128/jvi.64.9.4573-4577.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie K., Mohan P. M., Sasaguri Y., Putnak R., Padmanabhan R. Sequence analysis of cloned dengue virus type 2 genome (New Guinea-C strain). Gene. 1989 Feb 20;75(2):197–211. doi: 10.1016/0378-1119(89)90266-7. [DOI] [PubMed] [Google Scholar]
- Kräusslich H. G., Wimmer E. Viral proteinases. Annu Rev Biochem. 1988;57:701–754. doi: 10.1146/annurev.bi.57.070188.003413. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Zhao B. T., Hori H., Bray M. Infectious RNA transcribed from stably cloned full-length cDNA of dengue type 4 virus. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5139–5143. doi: 10.1073/pnas.88.12.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackow E., Makino Y., Zhao B. T., Zhang Y. M., Markoff L., Buckler-White A., Guiler M., Chanock R., Lai C. J. The nucleotide sequence of dengue type 4 virus: analysis of genes coding for nonstructural proteins. Virology. 1987 Aug;159(2):217–228. doi: 10.1016/0042-6822(87)90458-2. [DOI] [PubMed] [Google Scholar]
- Mandl C. W., Heinz F. X., Kunz C. Sequence of the structural proteins of tick-borne encephalitis virus (western subtype) and comparative analysis with other flaviviruses. Virology. 1988 Sep;166(1):197–205. doi: 10.1016/0042-6822(88)90161-4. [DOI] [PubMed] [Google Scholar]
- Mandl C. W., Heinz F. X., Stöckl E., Kunz C. Genome sequence of tick-borne encephalitis virus (Western subtype) and comparative analysis of nonstructural proteins with other flaviviruses. Virology. 1989 Nov;173(1):291–301. doi: 10.1016/0042-6822(89)90246-8. [DOI] [PubMed] [Google Scholar]
- Markoff L. In vitro processing of dengue virus structural proteins: cleavage of the pre-membrane protein. J Virol. 1989 Aug;63(8):3345–3352. doi: 10.1128/jvi.63.8.3345-3352.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P. W. Maturation of Japanese encephalitis virus glycoproteins produced by infected mammalian and mosquito cells. Virology. 1989 Apr;169(2):354–364. doi: 10.1016/0042-6822(89)90161-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B., Elroy-Stein O., Mizukami T., Alexander W. A., Fuerst T. R. Product review. New mammalian expression vectors. Nature. 1990 Nov 1;348(6296):91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- Osatomi K., Sumiyoshi H. Complete nucleotide sequence of dengue type 3 virus genome RNA. Virology. 1990 Jun;176(2):643–647. doi: 10.1016/0042-6822(90)90037-r. [DOI] [PubMed] [Google Scholar]
- Parks T. D., Dougherty W. G. Substrate recognition by the NIa proteinase of two potyviruses involves multiple domains: characterization using genetically engineered hybrid proteinase molecules. Virology. 1991 May;182(1):17–27. doi: 10.1016/0042-6822(91)90643-p. [DOI] [PubMed] [Google Scholar]
- Pletnev A. G., Yamshchikov V. F., Blinov V. M. Nucleotide sequence of the genome and complete amino acid sequence of the polyprotein of tick-borne encephalitis virus. Virology. 1990 Jan;174(1):250–263. doi: 10.1016/0042-6822(90)90073-z. [DOI] [PubMed] [Google Scholar]
- Preugschat F., Strauss J. H. Processing of nonstructural proteins NS4A and NS4B of dengue 2 virus in vitro and in vivo. Virology. 1991 Dec;185(2):689–697. doi: 10.1016/0042-6822(91)90540-r. [DOI] [PubMed] [Google Scholar]
- Preugschat F., Yao C. W., Strauss J. H. In vitro processing of dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J Virol. 1990 Sep;64(9):4364–4374. doi: 10.1128/jvi.64.9.4364-4374.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Lenches E. M., Eddy S. R., Shin S. J., Sheets R. L., Strauss J. H. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985 Aug 23;229(4715):726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- Ruiz-Linares A., Cahour A., Després P., Girard M., Bouloy M. Processing of yellow fever virus polyprotein: role of cellular proteases in maturation of the structural proteins. J Virol. 1989 Oct;63(10):4199–4209. doi: 10.1128/jvi.63.10.4199-4209.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speight G., Coia G., Parker M. D., Westaway E. G. Gene mapping and positive identification of the non-structural proteins NS2A, NS2B, NS3, NS4B and NS5 of the flavivirus Kunjin and their cleavage sites. J Gen Virol. 1988 Jan;69(Pt 1):23–34. doi: 10.1099/0022-1317-69-1-23. [DOI] [PubMed] [Google Scholar]
- Strauss J. H., Strauss E. G. Evolution of RNA viruses. Annu Rev Microbiol. 1988;42:657–683. doi: 10.1146/annurev.mi.42.100188.003301. [DOI] [PubMed] [Google Scholar]
- Sumiyoshi H., Mori C., Fuke I., Morita K., Kuhara S., Kondou J., Kikuchi Y., Nagamatu H., Igarashi A. Complete nucleotide sequence of the Japanese encephalitis virus genome RNA. Virology. 1987 Dec;161(2):497–510. doi: 10.1016/0042-6822(87)90144-9. [DOI] [PubMed] [Google Scholar]
- Tritch R. J., Cheng Y. E., Yin F. H., Erickson-Viitanen S. Mutagenesis of protease cleavage sites in the human immunodeficiency virus type 1 gag polyprotein. J Virol. 1991 Feb;65(2):922–930. doi: 10.1128/jvi.65.2.922-930.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengler G., Czaya G., Färber P. M., Hegemann J. H. In vitro synthesis of West Nile virus proteins indicates that the amino-terminal segment of the NS3 protein contains the active centre of the protease which cleaves the viral polyprotein after multiple basic amino acids. J Gen Virol. 1991 Apr;72(Pt 4):851–858. doi: 10.1099/0022-1317-72-4-851. [DOI] [PubMed] [Google Scholar]
- Zhao B. T., Prince G., Horswood R., Eckels K., Summers P., Chanock R., Lai C. J. Expression of dengue virus structural proteins and nonstructural protein NS1 by a recombinant vaccinia virus. J Virol. 1987 Dec;61(12):4019–4022. doi: 10.1128/jvi.61.12.4019-4022.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Mackow E., Buckler-White A., Markoff L., Chanock R. M., Lai C. J., Makino Y. Cloning full-length dengue type 4 viral DNA sequences: analysis of genes coding for structural proteins. Virology. 1986 Nov;155(1):77–88. doi: 10.1016/0042-6822(86)90169-8. [DOI] [PubMed] [Google Scholar]