Abstract
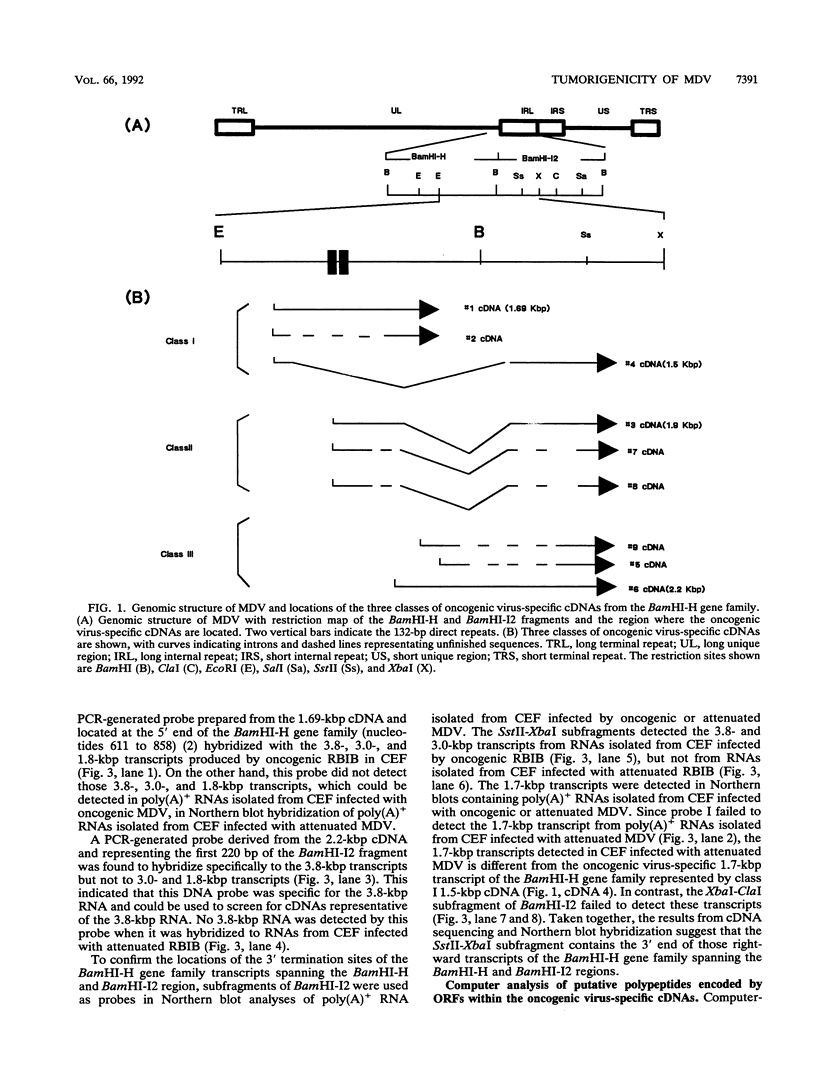
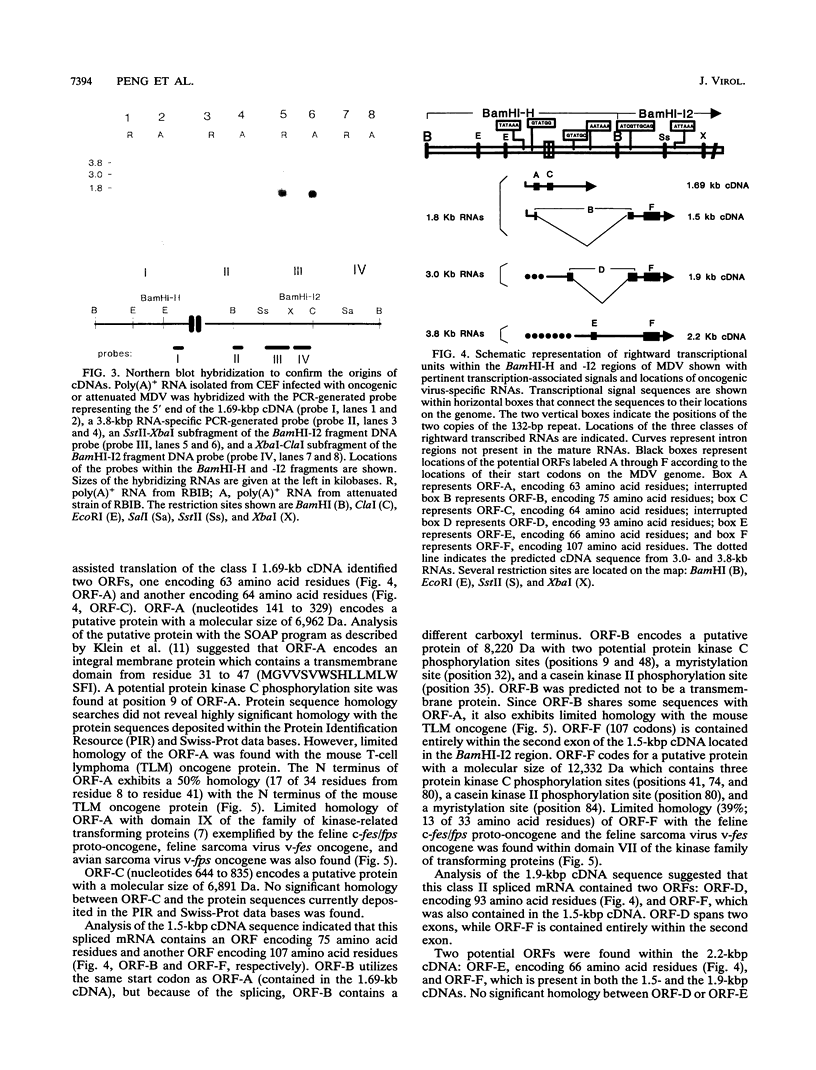
It has been reported that loss of the tumorigenic potential of attenuated Marek's disease virus (MDV) is strongly associated with amplification of the 132-bp repeat sequences found within the BamHI-D and BamHI-H fragments contained within the long terminal repeat and the long internal repeat, respectively. The expansion of this region results in loss of transcripts that are 3.8, 3.0, and 1.8 kbp long that are produced by tumorigenic strains of MDV. This evidence suggests that production of one or more of these three RNAs is strongly associated with the tumorigenic potential of the virus. In this study, we have cloned and sequenced 1.69-, 1.5-, 1.9-, and 2.2-kbp cDNAs from the BamHI-H gene family RNAs associated with tumorigenicity. The 1.69- and 2.2-kbp cDNAs are derived from nonspliced transcripts, whereas the 1.5- and 1.9-kbp cDNAs are from single spliced mRNAs spanning the BamHI-H and BamHI-I2 fragments of MDV DNA. Sequence analysis has shown two potential open reading frames in each of the cDNAs. The putative 63-amino-acid protein encoded by the first open reading frame in the 1.69-kbp cDNA and a putative 75-amino-acid protein encoded by the first open reading frame in the 1.5-kbp cDNA showed limited homology with the mouse T-cell lymphoma oncogene and the fes/fps family of kinase-related transforming proteins.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradley G., Hayashi M., Lancz G., Tanaka A., Nonoyama M. Structure of the Marek's disease virus BamHI-H gene family: genes of putative importance for tumor induction. J Virol. 1989 Jun;63(6):2534–2542. doi: 10.1128/jvi.63.6.2534-2542.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley G., Lancz G., Tanaka A., Nonoyama M. Loss of Marek's disease virus tumorigenicity is associated with truncation of RNAs transcribed within BamHI-H. J Virol. 1989 Oct;63(10):4129–4135. doi: 10.1128/jvi.63.10.4129-4135.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. B., Velicer L. F. Multiple bidirectional initiations and terminations of transcription in the Marek's disease virus long repeat regions. J Virol. 1991 May;65(5):2445–2451. doi: 10.1128/jvi.65.5.2445-2451.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fukuchi K., Tanaka A., Schierman L. W., Witter R. L., Nonoyama M. The structure of Marek disease virus DNA: the presence of unique expansion in nonpathogenic viral DNA. Proc Natl Acad Sci U S A. 1985 Feb;82(3):751–754. doi: 10.1073/pnas.82.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks S. K., Quinn A. M., Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988 Jul 1;241(4861):42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Iwata A., Ueda S., Ishihama A., Hirai K. Sequence determination of cDNA clones of transcripts from the tumor-associated region of the Marek's disease virus genome. Virology. 1992 Apr;187(2):805–808. doi: 10.1016/0042-6822(92)90483-6. [DOI] [PubMed] [Google Scholar]
- Kawamura M., Hayashi M., Furuichi T., Nonoyama M., Isogai E., Namioka S. The inhibitory effects of oligonucleotides, complementary to Marek's disease virus mRNA transcribed from the BamHI-H region, on the proliferation of transformed lymphoblastoid cells, MDCC-MSB1. J Gen Virol. 1991 May;72(Pt 5):1105–1111. doi: 10.1099/0022-1317-72-5-1105. [DOI] [PubMed] [Google Scholar]
- Klein P., Kanehisa M., DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985 May 28;815(3):468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- Lane M. A., Sainten A., Doherty K. M., Cooper G. M. Isolation and characterization of a stage-specific transforming gene, Tlym-I, from T-cell lymphomas. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2227–2231. doi: 10.1073/pnas.81.7.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M. A., Tobin M. B. Genomic sequence of the mouse oncogene tlm. Nucleic Acids Res. 1990 Jun 11;18(11):3410–3410. doi: 10.1093/nar/18.11.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maotani K., Kanamori A., Ikuta K., Ueda S., Kato S., Hirai K. Amplification of a tandem direct repeat within inverted repeats of Marek's disease virus DNA during serial in vitro passage. J Virol. 1986 May;58(2):657–660. doi: 10.1128/jvi.58.2.657-660.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maray T., Malkinson M., Becker Y. RNA transcripts of Marek's disease virus (MDV) serotype-1 in infected and transformed cells. Virus Genes. 1988 Oct;2(1):49–68. doi: 10.1007/BF00569736. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schat K. A., Buckmaster A., Ross L. J. Partial transcription map of Marek's disease herpesvirus in lytically infected cells and lymphoblastoid cell lines. Int J Cancer. 1989 Jul 15;44(1):101–109. doi: 10.1002/ijc.2910440119. [DOI] [PubMed] [Google Scholar]
- Silva R. F., Witter R. L. Genomic expansion of Marek's disease virus DNA is associated with serial in vitro passage. J Virol. 1985 Jun;54(3):690–696. doi: 10.1128/jvi.54.3.690-696.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S., Tanaka A., Nonoyama M. Transcription of the Marek's disease virus genome in a nonproductive chicken lymphoblastoid cell line. Virology. 1979 Feb;93(1):127–133. doi: 10.1016/0042-6822(79)90281-2. [DOI] [PubMed] [Google Scholar]
- Sugaya K., Bradley G., Nonoyama M., Tanaka A. Latent transcripts of Marek's disease virus are clustered in the short and long repeat regions. J Virol. 1990 Dec;64(12):5773–5782. doi: 10.1128/jvi.64.12.5773-5782.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]