Abstract
The specific-locus test (SLT) detects new mutants among mice heterozygous for seven recessive visible markers. Spontaneous mutations can be manifested not only as singleton whole-body mutants in controls (for which we report new data), but as mosaics—either visible (manifesting mottled coat color) in the scored generation (G2) or masked, among the wild-type parental generation (G1). Masked G1 mosaics reveal themselves by producing clusters of whole-body mutants in G2. We provide evidence that most, if not all, mosaics detected in the SLT (both radiation and control progenies) result from a single-strand spontaneous mutation subsequent to the last premeiotic mitosis and before the first postmeiotic one of a parental genome—the “perigametic interval.” Such events in the genomes of the G1 and G0 result, respectively, in visible and masked 50:50 mosaics. Per cell cycle, the spontaneous mutation rate in the perigametic interval is much higher than that in pregamete mitotic divisions. A clearly different locus spectrum further supports the hypothesis of different origin, and casts further doubt on the validity of the doubling-dose risk-estimation method. Because mosaics cannot have arisen in mitotic germ cells, and are not induced by radiation exposure in the perigametic interval, they should not be included in calculations of radiation-induced germ-line mutation rates. For per-generation calculations, inclusion of mosaics yields a spontaneous frequency 1.7 times that calculated from singletons alone for mutations contributed by males; including both sexes, the multiple is 2.2.
Keywords: mosaic mutants, cluster mutants, mutation frequencies, mutation spectra
Specific-locus mutations that are recovered in mosaic, rather than whole-body-mutant, mice, and that were first reported by us in 1964 (1, 2), can provide evidence concerning the time of occurrence and nature of spontaneous mutations. Here we present further evidence that most, if not all, mosaics that have been detected in the specific-locus test (SLT) resulted from a single-strand mutation (i.e., in one strand of the DNA helix) arising subsequent to the last premeiotic mitosis and before the first postmeiotic one (the “perigametic interval”) of a parental genome. Such mutations are manifested as either (i) visible mosaic mutants among the scored progeny (1–3) or (ii) analogous, masked, mosaics among the wild-type parents of the test cross (where the new recessive mutation is covered by a wild-type allele), which produce clusters of identical whole-body mutants among their offspring (2–5). We report on both types of mosaics detected in progenies, now totaling almost two million and over one million from irradiated and control males, respectively, and provide evidence that mosaics in both populations are of spontaneous origin. As expected from their analogous origin (in different generations), visible and masked mosaics are found with similar frequencies. Because, however, the visible mosaics are scored in much larger populations than the masked ones, and because of other complexities involving the latter, quantitative conclusions are more reliably based on the visible mosaics.
Large-scale mammalian germ-cell mutagenesis experiments have, for more than four decades, used the SLT (6), in which new recessive mutations at seven loci are recovered over a standard recessive allele. This method can detect a wide spectrum of induced and spontaneous DNA changes that can lead to either loss or gain of function. The SLT is well-suited for the identification of visibly mosaic mutants because five of the seven loci involve coat-color phenotypes. It is likewise suited for the detection of masked mosaics among the parents because the generally large sibships provide optimum conditions for recovering multiple mutants, and because these mutants can be readily identified by their phenotype as probably identical—a presumption that can be verified by subsequent genetic (and now also molecular) tests.
Singleton whole-body mutants are the type commonly reported for SLT studies. Before reporting and analyzing mosaic-type mutants, we bring up to date information on the frequency and spectrum of spontaneous singleton mutants, which will be compared with the mosaics.
The SLT System
The SLT for mutagenesis in male germ cells entails mating untreated females of the T tester stock, which is homozygous for seven recessive markers (genotype: a/a; b/b; p cch/p cch; d se/d se; s/s), with treated or control males that are homozygous for all the corresponding wild-type alleles (6). In almost all experiments, these wild-type males have been offspring of a cross of 101/Rl females by C3H/Rl males (G0 generation), and their strain designation is abbreviated H. We will refer to them as G1-generation males (Fig. 1). Among the (T × H)F1 offspring (which will be referred to as G2, or the “scored generation”), whole-body mutants are of the type m/m*, where m is used generically to designate any one of the standard recessive markers contributed by the T stock, and m* is any new mutation involving the same locus. [Because of the very close linkage between d and se (0.16 cM), the m/m* designation may include d se/Df(d se).]
Figure 1.
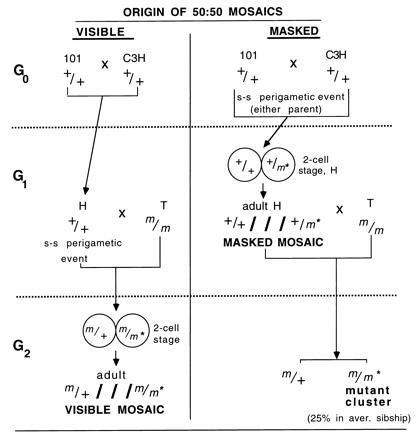
Mating scheme of the mouse SLT, and origin of the visible and masked mosaics. m designates any one of the seven marker alleles contributed by the T stock (a, b, p, cch, d, se, or s). m* designates a new spontaneous mutation at any one of the loci. + designates the corresponding wild-type allele. ///separates the two component genotypes of a mosaic. “s-s perigametic event” designates a single-strand mutation in the interval between the last premeiotic mitosis and the first postmeiotic one, an event that would produce a conceptus that is a 50:50 mosaic (see text). If the event occurs in an H mouse (in G1), it produces a scored-generation (G2) mosaic that is visible. An analogous event occurring in a 101 or C3H mouse (in G0) produces a G1 mosaic that is masked (because m* is covered by a dominant +) but reveals itself by producing clusters of mutants (m/m*) in the G2, the scored generation. (Note that mutant clusters can also be produced by rare H mice that are heterozygous for m*; for simplicity, these are omitted from the figure.) We provide evidence (see text) that most, if not all mosaics detected in the SLT are 50:50 mosaics.
Mosaic mutants in G2, the scored generation, are of the type m/+///m/m* (where///is used to separate the genotypes of the two component cell populations). Those mosaics in which m* involves one of the five loci that code for coat color (a, b, c, d, and p) are detectable by a mottled phenotype (“visible mosaics”). By contrast, among the H males (G1) that father the G2, any mosaics will be of genotype +/+///+/m* and, regardless of locus involved, will not be detectable by phenotype because each wild-type allele is fully dominant. These mice, which will be referred to as “masked mosaics,” are potentially detectable by clusters of mutants in their progenies. The crosses and types of mosaics are shown in Fig. 1.
The data on mosaics here reported come from all of our past specific-locus experiments in which males were exposed to radiations of a variety of doses, dose rates, and qualities (x-rays, gamma rays, neutrons from cyclotron, reactor, or atomic-bomb emissions, and radiations from internal emitters 3H2O and 239Pu citrate). Altogether, there were 61 experiments (not counting replicates separately) that screened the progenies of 26,167 irradiated males and yielded 1,815,704 scored offspring. Most of these radiation experiments were accompanied by concurrent controls, which screened the progenies of 10,882 unexposed males and yielded a total of 940,937 scored offspring, including 531,500 from our earliest experiments, already reported (7). Also included here are 99,817 offspring of 686 males from control groups that were concurrent with chemical mutagenesis experiments on males, bringing the total control offspring to 1,040,754.
Singleton Whole-Body Mutants
A Genetox Program report (8) summarized mutation frequencies for about 800,000 offspring of untreated males scored in SLTs at three laboratories. Observations on an additional ≈100,000 and ≈250,000 Oak Ridge National Laboratory and Neuherberg control offspring, respectively, were published subsequently (9, 10). This paper adds information on over 400,000 previously unreported Oak Ridge control offspring from experiments on males. Data for females were summarized earlier (11).
Singly occurring whole-body mutants will be referred to as singletons. The incidence of singletons among offspring of untreated males in the specific-locus cross is listed in Table 1 for our updated tabulations as well as for results reported by others. Table 1 excludes clusters of mutants, which are discussed below. The mutation rates per locus are quite similar for different laboratories, with completely overlapping 95% confidence limits (shown in parentheses). Summing all results for singletons, the average spontaneous mutation frequency for the seven specific loci is 6.6 [95% confidence limits (95% CL): 5.2, 8.3] × 10−6 per locus.
Table 1.
Singleton whole-body mutants in controls for specific-locus experiments
| Laboratory* | No. of mutants | No. of offspring | Mutation frequency/locus, × 10−6 |
|---|---|---|---|
| Harwell (4) | 9 | 157,421 | 8.2 (4.0, 15.2)† |
| Neuherberg (10) | 14 | 248,413 | 8.1 (4.7, 13.2) |
| Oak Ridge (12) | 0 | 38,448 | — |
| Oak Ridge‡ | 46 | 1,040,754 | 6.3 (4.7, 8.3) |
| Total | 69 | 1,485,036 | 6.6 (5.2, 8.3) |
This table excludes mutants recovered in clusters (i.e., progeny of invisible mosaics, see text).
Reference number is in parentheses. Stocks of males used at all laboratories have a common origin at Oak Ridge.
95% confidence limits in parentheses.
The spectrum among the loci for all 46 Oak Ridge control singleton mutants, as well as the viability/lethality and phenotype of homozygotes, are shown in Table 2, along with distributions reported for Harwell and Neuherberg mutants. Two kinds of d se mutants have been observed: (i) homozygous lethal and (ii) homozygous viable. The former are deletions involving both of these closely linked loci (13, 14). The latter give every indication of being homozygous for the standard alleles supplied by the T stock (13, 15), and we have suggested (13) that they represent cases of double nondisjunction, carrying two copies of chromosome 9 supplied by the T parent and none supplied by the H parent. They might, alternatively, represent (i) nondisjunction of only the T-stock chromosome 9, leading to initial Ts9 (d se/d se/++), with subsequent loss of the ++ chromosome from the fertilized egg, or (ii) somatic crossing-over proximal to d in an early cleavage, resulting in both d se/d se and ++/++ blastomeres, with only the former contributing to the embryo. Whichever of the alternative mechanisms accounts for the viable d se mutants, it is unlikely to be the explanation for the majority of the remaining mutants; of the 50 non-d se control mutants for which homozygous phenotypes are reported, 39 could clearly not have arisen by any of the hypothesized mechanisms (e.g., mutations to c, d +, + se, or p +).
Table 2.
Distribution among loci and homozygous viability of singleton whole-body spontaneous mutations
| Locus | Ref. 4 | Ref. 5 | This paper | Total |
|---|---|---|---|---|
| a | 0 | 0 | 0 | 0 |
| b | 2 | 5 (5v) | 8 (4v, 3xv, 1pl) | 15 |
| c | 0 | 2 (1v, 1nt) | 2 (1v, 1xv) | 4 |
| p | 1 | 3 (2v, 1pl) | 8 (6v, 1xpl, 1nt) | 12 |
| d | 2 | 1 (1v) | 12 (10 op, 2xv) | 15 |
| se | 0 | 0 | 1 (1v) | 1 |
| Df(d se) | 1 | 0 | 1 (1pl) | 2 |
| d se* | 1 | 0 | 5 (5v) | 6 |
| s | 2 | 0 | 9 (2v, 3jl, 4pl) | 11 |
v, Viable; x, phenotype intermediate between stock allele and wild-type; op, opisthtonic and juvenile lethal; jl, juvenile lethal; pl, prenatal lethal; nt, not tested.
See text for possible modes of origin.
Russell and colleagues (16–18) have reported the spectrum for 325 whole-body, nonclustered Oak Ridge mutants found among offspring derived from irradiated spermatogonia. These figures exclude neutron data as well as experiments involving 24h fractionation, which can alter the spectrum (19). In increasing order of frequency, the percentages of total mutants represented by Df(d se), se, a, c, d, p, b, and s mutants are 0.6, 1.5, 2.2, 9.8, 14.8, 15.4, 17.2, and 38.5, respectively. The corresponding percentages for the same loci among the 60 control mutations shown in Table 2 (omitting the viable d se mutations discussed above) are 3.0, 1.5, 0, 6.1, 22.7, 18.2, 22.7, and 16.7. In several respects, the spectra resemble each other in that a, se, and Df(d se) mutations are very infrequent, c mutations are the next lowest, and p, d, and b mutations are roughly similar amongst themselves. However, mutations involving s constitute almost 40% of all the radiation-induced mutations (almost as high as for p, d, and b combined), while fewer than 20% of control mutations involve s, a relative frequency lower than that for p, d, or b. Overall, the radiation-induced and control distributions differ significantly from each other (P < 0.02 by χ2).
The distributions also differ from each other with respect to the nature of mutations at individual loci. Since most of the analysis concerns historical data predating molecular analyses, comparisons can only be made regarding homozygous lethality/viability [subsequent findings have indicated that, for all loci, prenatal or neonatal lethality is associated with multilocus lesions (20, 21)]. Comparison of the control data in Table 2 with Oak Ridge reports on radiation-induced mutations for spermatogonia (16–18) illustrates this difference. For the b locus, for example, only 1 of 13 analyzed control mutations, but 14 of 28 radiation-induced mutations were homozygous lethal. At p, the corresponding ratios were 2 of 10 and 9 of 25. Fewer than half the control s-locus mutations were prenatally lethal, but there were 45 prenatal lethals among the 49 radiation-induced s-locus mutations. Other major differences in lesion size between spontaneous mutations and those radiation-induced in spermatogonial stem cells have been reported for the c locus and the d se region, with some of the results being based on molecular analyses (19).
A summary of spontaneous mouse mutations that arose in various standard inbred or mutant stocks at over a dozen loci indicates that many possible types of DNA alterations are represented, without any one mechanism appearing to be the predominant one (22). Analysis of a number of human spontaneous mutations suggests that the spectrum of events may vary with the locus (23). In the SLT, comparison of the same set of loci in offspring of control and irradiated fathers is, as discussed above, indicative of a different array of DNA alterations in the two populations. Moreover, because the nature of induced mutations (often established by molecular analysis) varies clearly with the mutagenic treatment, particularly with germ-cell stage (1, 19, 24), there can be no generalized similarity between induced and spontaneous mutations.
Visible Mosaic Mutants
For 33 of the 61 specific-locus experiments on males, our progeny data for irradiated and control groups were searched for any mice recorded as mottled, or possibly mottled, and 137 such records were found. For 78 of these (to be referred to as “undetermined”), genotype was not definitively determined, because (i) they were not mated (<15%), (ii) they were tested only for a dominant mode of inheritance, or (iii) (particularly in the case of females) they did not produce enough offspring by a mate capable of revealing the locus for which mosaicism is present. Of the 59 for which identification was possible, 21 were the result of genetic conditions other than mosaicism for mutation at one of the specific loci. These other conditions included X-linked mutations, X-autosome translocations (i.e., X-inactivation effects on translocated autosomal genes), autosomal dominants, and mottling alleles at one of the specific loci.
For the 38 genetically identified mosaics in the completely searched experiments, frequencies of occurrence (based on total observations shown in Table 3) were calculated under three scenarios: (i) the “minimum,” which assumes that none of the “undetermined” mottleds were mosaic; (ii) the “maximum,” which assumes that all were; and (iii) the “proportional,” which assumes that the ratio of specific-locus mosaics to other genetic conditions is the same in the undetermined as in the genetically tested group, i.e., 38/59. (Scenario ii is clearly an overestimate because several cases of “mottling” might have environmental causes, or be the result of mistaken phenotype assessment.) The frequencies under each of the three scenarios (Table 3) are similar for experimental and control populations. This is not surprising for irradiation of spermatogonia, because, even if single-strand lesions were being induced (which is unlikely), they would come to involve both chromatids after mitotic divisions. Calculations were made for a subset of the experimental population (about 56,000 offspring) that was clearly derived from postmitotic radiation. Minimum and proportional scenarios yielded frequencies of 1.8 × 10−5 and 5.4 × 10−5, respectively, i.e., very similar to those for the entire data set, namely, 2.1 × 10−5 and 4.8 × 10−5, respectively. One may therefore conclude that, as suggested earlier (1–3), mosaics in all irradiated as well as control groups are likely to be of spontaneous origin. Because visible mosaics are probably undetectable for two of the seven loci, se and s, the mutation frequency per locus under the proportional scenario is 4.8 × 10−5 × (null)/1;5, or 9.6 (95% CL: 7.7, 11.7) × 10−6. (Note that because of the inability to detect overt se and s mosaics, and the difficulty of verifying mosaicism at some of the other loci by breeding, a meaningful locus-spectrum cannot be derived.)
Table 3.
Frequencies of visible specific-locus mosaics
| Treatment | No. scored | Estimated frequency (×
10−5) under following scenario*
|
||
|---|---|---|---|---|
| Minimum | Maximum | Proportional | ||
| X | 1,116,558 | 1.7 | 7.3 | 4.6 |
| C | 725,852 | 2.6 | 4.8 | 4.4 |
| X + C | 1,842,410 | 2.1 | 6.3 | 4.8 |
X, father irradiated; C, father not irradiated.
See text.
Segregation ratios were determined for progenies of 50 mosaics (including 12 from experiments incompletely searched for mottleds). Of these, 32 (designated group 1) could be analyzed with a high degree of genetic confidence because the mutation was either to an allele different from that contributed by the T stock or had a linked, nonmutant marker; the remaining 18 mosaics were designated group 2 (designations as in ref. 2, which presented data for 35 of the 50 mosaics). The distribution for the proportion of mutant cells in the mosaic gonad (twice the frequency of segregation of the mutant allele) is shown in Fig. 2. The mean of frequencies is very close to 50%: for the 32 group-1 animals, mean = 47.0, and for all 50 visible mosaics analyzed, mean = 46.1.
Figure 2.
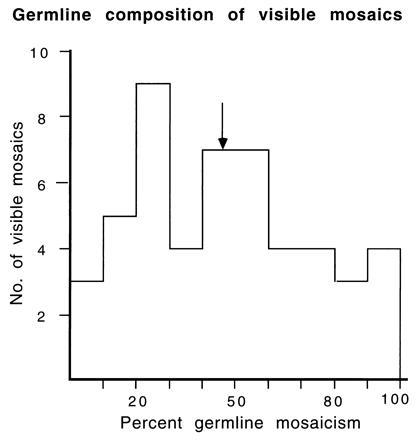
Distribution with respect to proportion of germ-line mosaicism for 50 visible specific-locus mosaics. The arrow represents the mean.
Because the mouse embryo proper develops not from the entire blastocyst but only from the inner cell mass, any individual mosaic derives from a small number of cells set aside, at random, from a much larger mixed cell pool—a process expected to lead to a broad distribution. Some time later in differentiation, another very small group of cells is set aside to form the germ line. The composition of the mixed cell pool from which these set-asides occur cannot be deduced from any one individual mosaic, but the distribution of the whole population of mosaics is informative. If, in each individual, one of the first two blastomeres was mutant (i.e., m/m* as opposed to m/+), the mean of the proportions characterizing individual mosaics would be 50%. The mean could be no higher than 50% (unless additional mutations to the same allele occurred in subsequent cell divisions). A mean lower than 50% would be expected if the population of mosaics included an admixture of individuals in which the original mutation had occurred at a later cleavage stage (as a result of which the mixed cell pool at the time of the set-asides would contain a lower proportion of mutant cells). Because the mean of the observed distribution was close to 50%, it may be concluded that the rate of the events that produce 50:50 mosaics must be very much higher than the mutation rate in subsequent cell divisions. [This conclusion may be drawn for ≈8 cell divisions because mosaicism can be detected when >1/200th of the fur is mutant (25).]
A 50:50 mosaic could result from one of two events: (i) a double-strand mutation at the 2-cell stage, or (ii) a single-strand mutation (i.e., in one strand of the DNA helix) occurring during or after meiosis (starting with premeiotic synthesis) in a parental germ cell, or in the genome contributed by that germ cell to the zygote, i.e., after the last pregametic mitosis and before the first postgametic one—an interval that will be referred to as the “perigametic interval.” Alternative i appears rather unlikely, because spontaneous mutations would probably more often be single- than double-strand. Further, if double-strand spontaneous mutations did occur in the 2-cell stage, roughly similar rates would be expected for the 4-cell, 8-cell, etc. stage, resulting in a mean for the distribution that was lower than the 50% observed. Within the perigametic interval (alternative ii), the zygote stage occupies only a few hours, while the prezygotic stages occupy weeks (in the male) or months (in the female). The likelihood of the event may, however, be governed not by length of time but by certain unknown critical states.
Masked Mosaics
To result in a visible 50:50 mosaic in the scored generation (G2) of an SLT (see preceding section), a spontaneous mutation would have to occur in the perigametic interval of the H (G1) genome. An analogous event occurring in the grandparental (G0) generation, i.e., in a C3H or 101 genome, would produce a 50:50 mosaic H male (Fig. 1). Such +/+///+/m* mosaicism would, however, not be obvious because the new recessive at one of the specific loci would be masked by the wild-type allele contributed by the other parent (101 or C3H). Because each H male in specific-locus experiments typically sires large sibships (the average sibship sizes for the 61 experiments were 86.5 and 69.4 for control and irradiated males, respectively; the overall size was 74.4), a +/+///+/m* mosaic male has a good chance of being identified by producing a cluster of mutants detected in the scored generation (Fig. 1).
While spontaneous mutation in a C3H or 101 perigametic genome (i.e., G0 generation) can result in a cluster in the progeny of either an irradiated or a control H male, irradiated H males may, in addition, produce treatment-induced (TI) clusters. The presumed origin of such clusters is radiation-induced killing of spermatogonia and subsequent multiplication of any mutant cell during mitotic expansion of the depleted spermatogonial population. Of 26,167 sibships from irradiated males, 44 contained small specific-locus clusters. Among these 44 (found in 20 experiments that involved treatments producing spermatogonial killing, as indicated by temporary sterile periods), there were 41 clusters of two, two clusters of three, and one cluster of five mutants.
In 39 of these 44 cases, the putative TI clusters constituted ≤3.7% of the experiment’s average sibship size; in the five cases for which the percentage was somewhat larger, the sterile period for the individual was quite long, e.g., in the male that yielded the single cluster of 5 (6.4% of his sibship), 44 weeks elapsed between exposure and conception of the first poststerile litter. Apart from the 44 presumed TI clusters, all other clusters in irradiated groups were assumed to be the result of spontaneous mosaicism of the H male. Additional evidence for this conclusion occurred in two cases in which one or more members of the cluster were derived from germ cells that had been in postspermatogonial stages at the time of irradiation—an event incompatible with TI clustering.
Clusters additional to the 44 identified as TI under the above criteria, are listed in Table 4. Two and four of these six mosaic-type clusters identified in Oak Ridge radiation experiments on males were found in control and irradiated groups, respectively; one each of these was published earlier (2, 3). Not included are two clusters (se and d) that were probably the result of heterozygosity of the respective H males (one irradiated and one control), which produced, respectively, 311/608 and 166/297 mutant/total progeny. While no reliable mean of individual germ-line proportions can be derived from only six mutants, there is no evidence that the distribution differs markedly from that of the much more meaningful one for the 50 visible mosaics (Fig. 2), which should be analogous in origin. For the six masked mosaics, the mean of the individual germ-line proportions that are mutant is 34%; if the two clusters assumed to result from heterozygosity were in fact produced by mosaics that, by chance, had a cell proportion at the extreme end of the binomial distribution, the mean for the eight is 52%.
Table 4.
Mutant clusters produced by masked mosaics
| Treatment | Allele | Offspring scored | Percentage of germ-line mosaic* |
|---|---|---|---|
| X | axv | 228 | 72.8 |
| X | dpl† | 787 | 50.6 |
| C | cv† | 402 | 42.8 |
| X | ajv | 325 | 17.8 |
| X | pv | 72 | 11.2 |
| C | ax | 391 | 8.7 |
Two additional clusters (d and se) were produced by what were probably heterozygotes (see text).
X, H male irradiated; C, H male nonirradiated; v, viable; x, phenotype intermediate between stock allele and wild-type; pl, prenatal lethal; j, jet (=a
).
Computed from double the frequency of segregating allele.
Altogether, 37,735 H males (26,167 irradiated, 11,568 control) had the opportunity to reveal masked mosaicism for any of seven loci. Because a mutation in either the C3H male or the 101 female parent of the H could have produced the six mosaics, the incidence must be divided by two to yield the mutation rate of 11.4 (95% CL: 4.9, 24.3) × 10−6 per locus (in the absence of other evidence, we are assuming equal likelihood in the two sexes and strains). Calculated for control and irradiated populations separately, the rates are 12.3 (95% CL: 2.2, 41.3) × 10−6 and 10.9 (95% CL: 3.7, 26.2) × 10−6, respectively; the fact that the point estimate for irradiated males is very similar to that for controls indicates that it is unlikely that many (if any) of the clusters assumed to be the result of masked mosaicism were instead TI clusters, or vice versa. Among H mice, masked mosaic males and females have a similar origin and could therefore be used jointly for calculating the frequency of cluster-generating mosaics. Preliminary indications are that, once the data compilation for female experiments is completed, the mutation rate for this event may change in a downward direction. Two clusters in controls (se and s) reported from Neuherberg (5) and one (b) from Harwell (4) cannot be included in these mutation-rate calculations because the number of H males screened was not stated.
It was notable that three of our six clusters involved the a locus (Table 4). A statistical comparison of the entire distribution for the altogether nine masked mosaics found at all three laboratories (three mutant at a and one mutant at each of the other loci) with that for the 66 control singletons (Table 2) shows a significant difference (P < 0.001 by χ2). [If the two presumed heterozygotes (mutant at se and d) were in fact mosaics, the difference becomes even greater.] This difference in spectrum lends further support to the hypothesis that the clustered mutations detectable in SLT sibships do not originate during the 40–80 mitotic divisions (26) that ultimately lead to formation of the H male’s sperm. It is intriguing to speculate on special spontaneous-mutation-producing mechanisms that might operate in the interval between the last premeiotic and first postmeiotic mitoses, but other organisms might be better suited to their elucidation than is the mouse. [Note that clusters of spontaneous sex-linked recessive lethals in Drosophila melanogaster constitute a smaller percentage of total progeny than do mouse clusters and have been concluded to result from a different mechanism, namely mosaicism of the testis of the cluster-producing male, due to mutational events in premeiotic germ cells (27).]
Mutation-Rate Comparisons and Applications
Table 5 lists spontaneous specific-locus mutation frequencies calculated from three sets of data: (i) whole-body singleton mutations among control offspring, (ii) visible mosaics in control and irradiated populations of the “scored generation” (G2), and (iii) masked mosaics among control and irradiated H males (G1) (Fig. 1). The 95% confidence limits of the three groups overlap, and the rates for the two types of mosaics (ii and iii) do not differ significantly from each other (P = 0.47, Fisher’s exact test); however, ii is significantly higher than i (P = 0.01) (iii is not significantly higher than i, P = 0.26). The means based on visible and masked mosaics are, respectively, 1.5 and 1.7 times that for whole-body singletons. Because of the wide confidence limits of the mean for masked mosaics, and because of other considerations discussed below, the latter multiple is a very uncertain one.
Table 5.
Comparison of spontaneous mutation frequencies
| Frequency based on | Frequency per locus, ×10−6 | Source |
|---|---|---|
| Whole-body mutants | 6.6 (5.2, 8.3) | Table 1 |
| Visible mosaics | 9.6 (7.7, 11.7) | Table 3,* |
| Masked mosaics | 11.4 (4.9, 24.3) | 6/37,735 × 1/7 × 1/2† |
Numbers in parenthesis are 95% confidence limits.
Proportional scenario.
See text.
Singleton whole-body control mutants can have arisen in any of several mitoses in the cell lineage ancestral to G1 gametes, whereas we have concluded that each of the two analogous types of mosaic detected by us arose from events occurring after the last pregametic mitosis of a parental genome and prior to the first postgametic one. Per cell cycle, the latter spontaneous events must, therefore, be much more frequent than are mutational events occurring during any pregamete mitotic division.
The spontaneous mutations that give rise to detectable mosaics (either masked or visible) cannot have arisen in mitotic germ cells, and, therefore, these mosaics should be excluded from control frequencies used for calculating mutagen-induced spermatogonial mutation rates. We have provided evidence to show that the mosaics detected in the SLT result from single-strand mutations occurring during the perigametic interval of the the respective parental genome (G1 masked mosaics, from mutation in a G0 genome; G2 visible mosaics, from mutation in the G1 genome). Radiation appears not to induce this type of mutation, and, therefore, mosaics (masked or visible) should not be included in calculations of radiation-induced mutation rates for any spermatogenic stage. The mutagen N-ethyl-N-nitrosourea has given clear evidence of inducing single-strand mutations in the perigametic interval, and some other chemicals may also do so (28, 29).
Singleton whole-body spontaneous mutants could trace to mutations in the cell lineage ancestral to parental germ cells (see above), or might have one of two other origins: (i) as m/m*///m/+ mosaics in which, by chance, only the m/m* blastomeres contributed to the embryo, or (ii) as offspring of a +/m*///+/+ masked 50:50 mosaic that produced too small a sibship to reveal a cluster. The category of control singletons therefore includes some mutations additional to those arising in germ cells. Despite these complexities, the control mutation frequency of 6.6 (95% CL: 5.2, 8.3) × 10−6 for singleton whole-body mutants (Tables 1 and 5) is probably the best approximation available for calculations of mutation rates induced in any spermatogenic stage by radiations or other mutagens that do not give evidence of inducing single-strand mutations in the perigametic interval.
To calculate the frequency of spontaneous mutations arising in males per generation, the best estimate is derived from G2 of the SLT; i.e., visible mosaics would be included with singleton whole-body mutants (Fig. 3). Based on the mutation rates listed in Table 5, and allowing for the fact that the mosaics contribute only half as many new mutations to the next generation (in this case, G3) than do the whole-body mutants, the per-generation rate is 1.7 times that calculated from singletons alone (i.e., [6.6 + {½ × 9.6}]/6.6).
Figure 3.
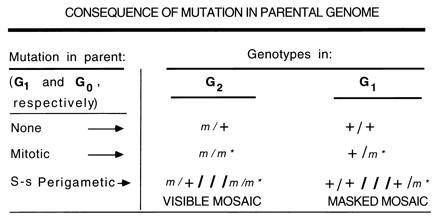
Consequence of either mitotic or single-strand perigametic mutation in a parental genome. The G2 and G1 genotypes resulting from mutations in the G1 and G0 genomes, respectively, are shown.
Analogously (see Fig. 3), but less reliably, one might calculate a per-generation rate from mutations detectable in G1 (rather than G2) by adding masked mosaics (weighted by ½) to heterozygotes (also masked). There are several drawbacks to this alternative: (i) heterozygotes might have arisen not in one of the parents of the H mouse, but in some prior generation of either the 101 or C3H strain (examination of prior stock records can decrease, but not eliminate, the possibility that this might have been the case); (ii) a putative heterozygote might, in fact, be a 50:50 mosaic with a cell proportion at the extreme end of the binomial distribution; (iii) unless frequency calculations are restricted to untreated males, the cluster that identified a putative mosaic male might, in fact, be a TI cluster; (iv) the confidence limits on both putative heterozygotes and putative masked mosaics are very wide, the number of cases being very much smaller than those for the respectively analogous whole-body mutants and visible mosaics in the G2. A calculation based on the present data for putative heterozygotes and putative masked mosaics (both of which could arise in either 101 or C3H) yields a per-generation rate of 9.4 × 10−6 per locus (namely, ½ [2/37,735 + {½ × 6/37,735}] × (null)/1;7). This is 1.4 times the rate calculated from singleton whole-body mutations in the “scored generation,” roughly similar to—but considerably more uncertain than—the ratio of 1.7 calculated above from G2 data. (Illustrating some of the problems, the multiple becomes 1.2 if the presumed heterozygotes were in fact mosaics; it becomes 1.7, if two of the presumed TI clusters were in fact mosaic clusters.) Because of these numerous uncertainties, we recommend that the per-generation rate not be calculated on the basis of G1 data (i.e., cluster-producing masked mosaics and heterozygotes).
The per-generation spontaneous mutation rate has been used in the past for the “doubling-dose method” to estimate the relative genetic risks from radiation-induced and spontaneous mutations. The validity of the doubling-dose method has long been controversial, notably because of the evidence of qualitative differences between induced and spontaneous mutations (30). We have now presented even further evidence for qualitative differences; thus, the mutations of perigametic origin differ from spontaneous singletons which, in turn, differ from induced mutations. Additionally, regardless of the nature of the mutations (which, in itself, undoubtedly affects phenotype), it has become evident that an appreciable fraction of new spontaneous (but not radiation-induced) mutations in any one generation will result in mosaics which, overall, will have less extreme phenotypes than do whole-body mutants. Whether and how to use the mosaic mutants in calculating the doubling dose is not discussed here. For those wishing to know the total per-generation spontaneous mutation rate in the mouse, regardless of mutation type, we have presented data indicating that for mutations contributed by males, inclusion of mosaics yields a per-generation frequency 1.7 times that calculated from singletons alone. Adding the female contribution of singletons (1.6 × 10−6; ref. 11) and mosaics (the latter rate assumed same as for males) produces a multiple of 2.2 of the combined singleton frequencies.
Acknowledgments
We are deeply grateful to Prof. Dean R. Parker, Dr. E. M. Rinchik, and Dr. D. K. Johnson for their critical reading of the manuscript and their valuable suggestions; and to P. R. Hunsicker, E. M. Kelly, and the numerous technicians of past years, who helped generate the data on which this paper is based. This research was jointly sponsored by the Office of Health and Environmental Research, U.S. Department of Energy under contract DE-AC05-96OR22464 with Lockheed Martin Energy Research Corp, and by the National Institute of Environmental Health Sciences under IAG No. 1-Y01-ES-50318-00.
Footnotes
Abbreviations: SLT, specific-locus test; TI, treatment induced; 95% CL, 95% confidence limits.
References
- 1.Russell W L. In: Genetics Today. Geerts S J, editor. Vol. 2. Oxford: Pergamon; 1964. pp. 257–264. [Google Scholar]
- 2.Russell L B. In: The Role of Chromosomes in Development. Locke M, editor. New York: Academic; 1964. pp. 153–181. [Google Scholar]
- 3.Russell L B. Genetics. 1979;91:141–147. doi: 10.1093/genetics/91.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Searle A G. Adv Radiat Biol. 1974;4:131–207. [Google Scholar]
- 5.Ehling U H, Neuhäuser-Klaus A. In: Problems of Threshold in Chemical Mutagenesis. Tazima J, Kondo S, Kuroda Y, editors. Tokyo: Nissan Science Foundation; 1984. pp. 15–35. [Google Scholar]
- 6.Russell W L. Cold Spring Harbor Symp Quant Biol. 1951;16:327–336. doi: 10.1101/sqb.1951.016.01.024. [DOI] [PubMed] [Google Scholar]
- 7.Russell W L. Proc Natl Acad Sci USA. 1962;48:1724–1727. doi: 10.1073/pnas.48.10.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell L B, Selby P B, von Halle E, Sheridan W, Valcovic L. Mutat Res. 1981;86:329–354. doi: 10.1016/0165-1110(81)90010-5. [DOI] [PubMed] [Google Scholar]
- 9.Russell L B, Hunsicker P R, Shelby M D. Mutat Res. 1992;282:151–158. doi: 10.1016/0165-7992(92)90089-z. [DOI] [PubMed] [Google Scholar]
- 10.Ehling U H, Neuhäuser-Klaus A. Mutat Res. 1995;328:73–82. doi: 10.1016/0027-5107(94)00198-e. [DOI] [PubMed] [Google Scholar]
- 11.Russell L B, Russell W L. Mutat Res. 1992;296:107–127. doi: 10.1016/0165-1110(92)90035-8. [DOI] [PubMed] [Google Scholar]
- 12.Selby P B. Mutat Res. 1973;18:63–75. doi: 10.1016/0027-5107(73)90021-3. [DOI] [PubMed] [Google Scholar]
- 13.Russell L B. Mutat Res. 1971;11:107–123. doi: 10.1016/0027-5107(71)90036-4. [DOI] [PubMed] [Google Scholar]
- 14.Rinchik E M, Russell L B, Copeland N G, Jenkins N A. Genetics. 1986;112:321–342. doi: 10.1093/genetics/112.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehling U H, Neuhäuser-Klaus A. Mutat Res. 1994;307:229–236. doi: 10.1016/0027-5107(94)90296-8. [DOI] [PubMed] [Google Scholar]
- 16.Russell W L, Russell L B. Radiat Res Suppl. 1959;1:296–305. [Google Scholar]
- 17.Russell, W. L., Cumming, R. B., Kelly, E. M. & Phipps, E. L. (1979) Behavior of Tritium in the Environment, IAEA-SM-232/85 (Int. Atomic Energy Agency, Vienna), pp. 489–497.
- 18.Russell W L, Kelly E M. Proc Natl Acad Sci USA. 1982;79:539–541. doi: 10.1073/pnas.79.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russell L B, Rinchik E M. Mutat Res. 1993;288:187–195. doi: 10.1016/0027-5107(93)90084-s. [DOI] [PubMed] [Google Scholar]
- 20.Rinchik E M, Russell L B. In: Genome Analysis. Davies K, Tilghman S, editors. Vol. 1. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. pp. 121–158. [Google Scholar]
- 21.O’Brien T P, Metallinos D L, Chen H, Shin M K, Tilghman S M. Genetics. 1996;143:447–461. doi: 10.1093/genetics/143.1.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Favor J. Mutat Res. 1994;304:107–118. doi: 10.1016/0027-5107(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 23.Mohrenweiser H. Mutat Res. 1994;304:119–137. doi: 10.1016/0027-5107(94)90322-0. [DOI] [PubMed] [Google Scholar]
- 24.Russell L B, Russell W L, Rinchik E M, Hunsicker P R. Banbury Rep. 1990;34:271–289. [Google Scholar]
- 25.Russell L B, Major M H. Genetics. 1957;42:161–175. doi: 10.1093/genetics/42.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyon M F. Mutat Res. 1981;87:323–345. doi: 10.1016/0165-1110(81)90018-x. [DOI] [PubMed] [Google Scholar]
- 27.Woodruff R C, Thompson J N., Jr J Evol Biol. 1992;5:457–464. [Google Scholar]
- 28.Russell L B, Bangham J W, Stelzner K F, Hunsicker P R. Proc Natl Acad Sci USA. 1988;85:9167–9170. doi: 10.1073/pnas.85.23.9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Favor J, Neuhäuser-Klaus A. Annu Rev Genet. 1994;28:27–47. doi: 10.1146/annurev.ge.28.120194.000331. [DOI] [PubMed] [Google Scholar]
- 30.Russell W L. Banbury Rep. 1990;34:385–395. [Google Scholar]


