Abstract
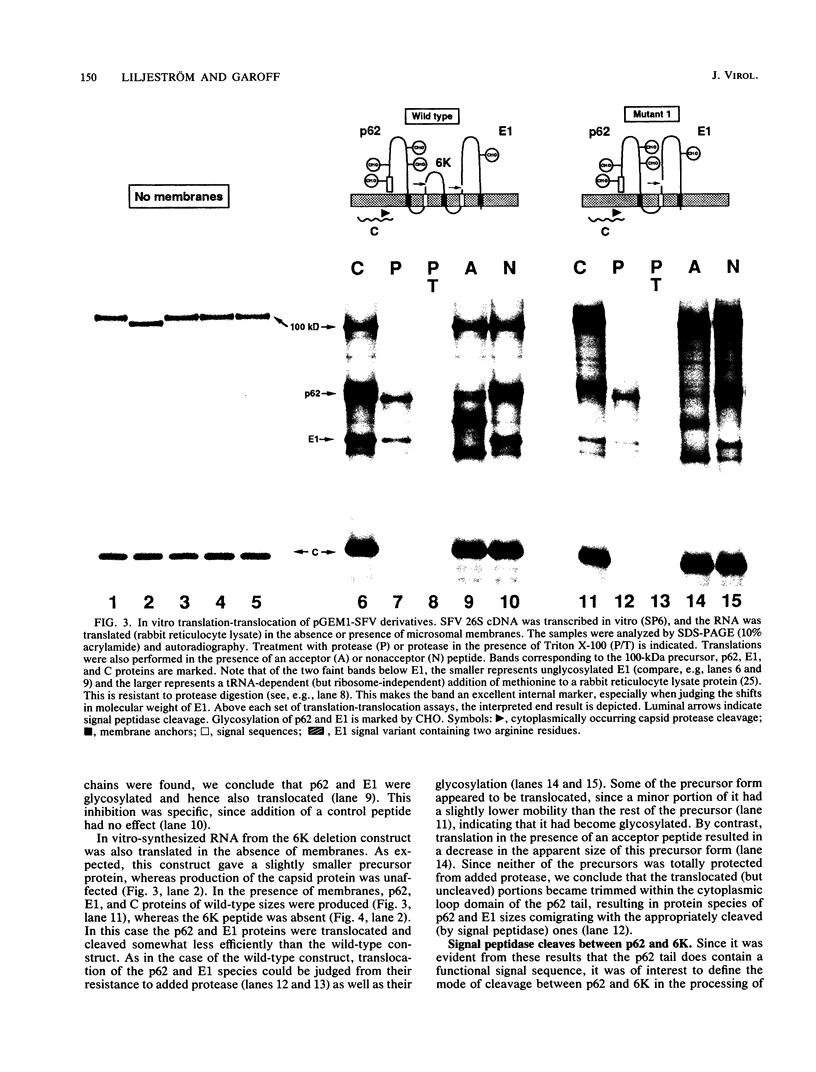
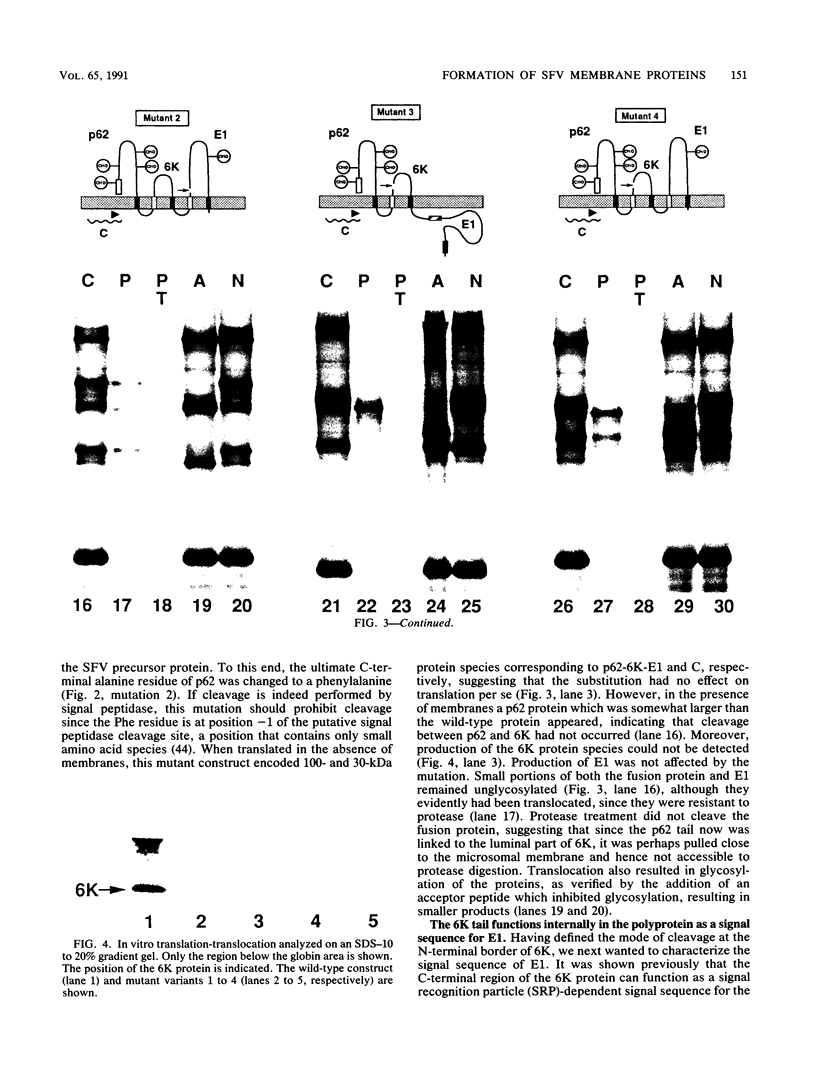
The proteolytic processes involved in the cotranslational production of the Semliki Forest virus proteins p62, 6K, and E1 from a common precursor polypeptide were analyzed by an in vitro translation-translocation assay. By studying the behavior of wild-type and mutant variants of the polyprotein, we show that the signal sequences responsible for membrane translocation of the 6K and E1 proteins reside in the C-terminal regions of p62 and 6K, respectively. We present evidence suggesting that the polyprotein is processed on the luminal side by signal peptidase at consensus cleavage sites immediately following the signal sequences. Our results also lead us to conclude that the 6K protein is a transmembrane polypeptide with its N terminus on the luminal side of the membrane (type I). Thus, the production of all three membrane proteins is directed by alternating signal and stop-transfer (anchor) sequences that function in translocation and cleavage of the virus precursor polyprotein. This also shows conclusively that internally located signal sequences can be cleaved by signal peptidase.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aliperti G., Schlesinger M. J. Evidence for an autoprotease activity of sindbis virus capsid protein. Virology. 1978 Oct 15;90(2):366–369. doi: 10.1016/0042-6822(78)90321-5. [DOI] [PubMed] [Google Scholar]
- Armstrong J., Niemann H., Smeekens S., Rottier P., Warren G. Sequence and topology of a model intracellular membrane protein, E1 glycoprotein, from a coronavirus. Nature. 1984 Apr 19;308(5961):751–752. doi: 10.1038/308751a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bonatti S., Migliaccio G., Blobel G., Walter P. Role of signal recognition particle in the membrane assembly of Sindbis viral glycoproteins. Eur J Biochem. 1984 May 2;140(3):499–502. doi: 10.1111/j.1432-1033.1984.tb08130.x. [DOI] [PubMed] [Google Scholar]
- Bos T. J., Davis A. R., Nayak D. P. NH2-terminal hydrophobic region of influenza virus neuraminidase provides the signal function in translocation. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2327–2331. doi: 10.1073/pnas.81.8.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain J. P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979 Sep 15;98(1):132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- Chang G. J., Trent D. W. Nucleotide sequence of the genome region encoding the 26S mRNA of eastern equine encephalomyelitis virus and the deduced amino acid sequence of the viral structural proteins. J Gen Virol. 1987 Aug;68(Pt 8):2129–2142. doi: 10.1099/0022-1317-68-8-2129. [DOI] [PubMed] [Google Scholar]
- Chung C. T., Miller R. H. A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res. 1988 Apr 25;16(8):3580–3580. doi: 10.1093/nar/16.8.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler D. F., Garoff H. Mutants of the membrane-binding region of Semliki Forest virus E2 protein. I. Cell surface transport and fusogenic activity. J Cell Biol. 1986 Mar;102(3):889–901. doi: 10.1083/jcb.102.3.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler D. F., Melancon P., Garoff H. Mutants of the membrane-binding region of Semliki Forest virus E2 protein. II. Topology and membrane binding. J Cell Biol. 1986 Mar;102(3):902–910. doi: 10.1083/jcb.102.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgarno L., Rice C. M., Strauss J. H. Ross River virus 26 s RNA: complete nucleotide sequence and deduced sequence of the encoded structural proteins. Virology. 1983 Aug;129(1):170–187. doi: 10.1016/0042-6822(83)90404-x. [DOI] [PubMed] [Google Scholar]
- Garoff H., Frischauf A. M., Simons K., Lehrach H., Delius H. Nucleotide sequence of cdna coding for Semliki Forest virus membrane glycoproteins. Nature. 1980 Nov 20;288(5788):236–241. doi: 10.1038/288236a0. [DOI] [PubMed] [Google Scholar]
- Garoff H., Huylebroeck D., Robinson A., Tillman U., Liljeström P. The signal sequence of the p62 protein of Semliki Forest virus is involved in initiation but not in completing chain translocation. J Cell Biol. 1990 Sep;111(3):867–876. doi: 10.1083/jcb.111.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garoff H., Kondor-Koch C., Pettersson R., Burke B. Expression of Semliki Forest virus proteins from cloned complementary DNA. II. The membrane-spanning glycoprotein E2 is transported to the cell surface without its normal cytoplasmic domain. J Cell Biol. 1983 Sep;97(3):652–658. doi: 10.1083/jcb.97.3.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garoff H., Kondor-Koch C., Riedel H. Structure and assembly of alphaviruses. Curr Top Microbiol Immunol. 1982;99:1–50. doi: 10.1007/978-3-642-68528-6_1. [DOI] [PubMed] [Google Scholar]
- Garoff H., Simons K., Dobberstein B. Assembly of the Semliki Forest virus membrane glycoproteins in the membrane of the endoplasmic reticulum in vitro. J Mol Biol. 1978 Oct 5;124(4):587–600. doi: 10.1016/0022-2836(78)90173-0. [DOI] [PubMed] [Google Scholar]
- Gierasch L. M. Signal sequences. Biochemistry. 1989 Feb 7;28(3):923–930. doi: 10.1021/bi00429a001. [DOI] [PubMed] [Google Scholar]
- Hahn C. S., Lustig S., Strauss E. G., Strauss J. H. Western equine encephalitis virus is a recombinant virus. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5997–6001. doi: 10.1073/pnas.85.16.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C. S., Strauss E. G., Strauss J. H. Sequence analysis of three Sindbis virus mutants temperature-sensitive in the capsid protein autoprotease. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4648–4652. doi: 10.1073/pnas.82.14.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C. S., Strauss J. H. Site-directed mutagenesis of the proposed catalytic amino acids of the Sindbis virus capsid protein autoprotease. J Virol. 1990 Jun;64(6):3069–3073. doi: 10.1128/jvi.64.6.3069-3073.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtley S. M., Helenius A. Protein oligomerization in the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:277–307. doi: 10.1146/annurev.cb.05.110189.001425. [DOI] [PubMed] [Google Scholar]
- Jackson R. J., Hunt T. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- Jou W. M., Verhoeyen M., Devos R., Saman E., Fang R., Huylebroeck D., Fiers W., Threlfall G., Barber C., Carey N. Complete structure of the hemagglutinin gene from the human influenza A/Victoria/3/75 (H3N2) strain as determined from cloned DNA. Cell. 1980 Mar;19(3):683–696. doi: 10.1016/s0092-8674(80)80045-6. [DOI] [PubMed] [Google Scholar]
- Kaderbhai M. A., Austen B. M. Dog pancreatic microsomal-membrane polypeptides analysed by two-dimensional gel electrophoresis. Biochem J. 1984 Jan 1;217(1):145–157. doi: 10.1042/bj2170145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkkinen N. Carboxyl-terminal sequence analysis of the four structural proteins of Semliki Forest virus. FEBS Lett. 1980 Jun 30;115(2):163–166. doi: 10.1016/0014-5793(80)81158-6. [DOI] [PubMed] [Google Scholar]
- Kinney R. M., Johnson B. J., Welch J. B., Tsuchiya K. R., Trent D. W. The full-length nucleotide sequences of the virulent Trinidad donkey strain of Venezuelan equine encephalitis virus and its attenuated vaccine derivative, strain TC-83. Virology. 1989 May;170(1):19–30. doi: 10.1016/0042-6822(89)90347-4. [DOI] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol. 1987;155:397–415. doi: 10.1016/0076-6879(87)55027-3. [DOI] [PubMed] [Google Scholar]
- Levinson R. S., Strauss J. H., Strauss E. G. Complete sequence of the genomic RNA of O'nyong-nyong virus and its use in the construction of alphavirus phylogenetic trees. Virology. 1990 Mar;175(1):110–123. doi: 10.1016/0042-6822(90)90191-s. [DOI] [PubMed] [Google Scholar]
- Machamer C. E., Rose J. K. A specific transmembrane domain of a coronavirus E1 glycoprotein is required for its retention in the Golgi region. J Cell Biol. 1987 Sep;105(3):1205–1214. doi: 10.1083/jcb.105.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markoff L., Lin B. C., Sveda M. M., Lai C. J. Glycosylation and surface expression of the influenza virus neuraminidase requires the N-terminal hydrophobic region. Mol Cell Biol. 1984 Jan;4(1):8–16. doi: 10.1128/mcb.4.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melancon P., Garoff H. Processing of the Semliki Forest virus structural polyprotein: role of the capsid protease. J Virol. 1987 May;61(5):1301–1309. doi: 10.1128/jvi.61.5.1301-1309.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melancon P., Garoff H. Reinitiation of translocation in the Semliki Forest virus structural polyprotein: identification of the signal for the E1 glycoprotein. EMBO J. 1986 Jul;5(7):1551–1560. doi: 10.1002/j.1460-2075.1986.tb04396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okun M. M., Eskridge E. M., Shields D. Truncations of a secretory protein define minimum lengths required for binding to signal recognition particle and translocation across the endoplasmic reticulum membrane. J Biol Chem. 1990 May 5;265(13):7478–7484. [PubMed] [Google Scholar]
- Rose J. K., Welch W. J., Sefton B. M., Esch F. S., Ling N. C. Vesicular stomatitis virus glycoprotein is anchored in the viral membrane by a hydrophobic domain near the COOH terminus. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3884–3888. doi: 10.1073/pnas.77.7.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss E. G., Rice C. M., Strauss J. H. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984 Feb;133(1):92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- Su T. Z., el-Gewely M. R. A multisite-directed mutagenesis using T7 DNA polymerase: application for reconstructing a mammalian gene. Gene. 1988 Sep 15;69(1):81–89. doi: 10.1016/0378-1119(88)90380-0. [DOI] [PubMed] [Google Scholar]
- Wiley D. C., Skehel J. J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Transcending the impenetrable: how proteins come to terms with membranes. Biochim Biophys Acta. 1988 Jun 9;947(2):307–333. doi: 10.1016/0304-4157(88)90013-5. [DOI] [PubMed] [Google Scholar]