Abstract
Recent studies have suggested that the retention of selectable marker cassettes (like PGK–Neo, in which a hybrid gene consisting of the phosphoglycerate kinase I promoter drives the neomycin phosphotransferase gene) in targeted loci can cause unexpected phenotypes in “knockout” mice due to disruption of expression of neighboring genes within a locus. We have studied targeted mutations in two multigene clusters, the granzyme B locus and the β-like globin gene cluster. The insertion of PGK–Neo into the granzyme B gene, the most 5′ gene in the granzyme B gene cluster, severely reduced the normal expression of multiple genes within the locus, even at distances greater than 100 kb from the mutation. Similarly, the insertion of a PGK–Neo cassette into the β-globin locus control region (LCR) abrogates the expression of multiple globin genes downstream from the cassette. In contrast, a targeted mutation of the promyelocyte-specific cathepsin G gene (which lies just 3′ to the granzyme genes in the same cluster) had minimal effects on upstream granzyme gene expression. Although the mechanism of these long distance effects are unknown, the expression of PGK–Neo can be “captured” by the regulatory domain into which it is inserted. These results suggest that the PGK–Neo cassette can interact productively with locus control regions and thereby disrupt normal interactions between local and long-distance regulatory regions within a tissue-specific domain.
Keywords:
PGK–Neo (a hybrid gene consisting of the phosphoglycerate kinase I promoter driving the neomycin phosphotransferase gene) is a widely used cassette employed as a selectable marker for homologous recombination in embryonic stem cells. Recent studies have suggested that targeted mutations that retain the PGK–Neo cassette may yield unexpected phenotypes due to the altered expression of neighboring genes (1); the mechanism is unknown. One example of this phenomenon has recently been described using targeted mutations of the β-globin locus control region (LCR). The human β-globin LCR consists of a series of erythroid-specific DNase I hypersentive sites located upstream from the human β-globin gene cluster (2–4). To define the functions of individual LCR elements, homologous recombination techniques have been used to make several specific mutations within the human or murine β-globin LCRs (5–8). Remarkably, the insertion of PGK–Neo abrogrates the expression of multiple globin genes downstream from the cassette. Deletion of the selectable marker cassette results in restoration of LCR activity, suggesting that the cassette disrupts normal interactions between the LCR and downstream regulatory elements. Other examples of this phenomenon have recently been suggested in mice bearing targeted mutations of the myogenic basic helix–loop–helix gene MRF4, the Igκ light chain intronic enhancer, and individual genes in the Hox gene clusters (9–18). These examples underscore the unpredictable phenotypes that can be caused by retained PGK–Neo cassettes.
Recent studies have identified a cluster of closely linked hematopoietic serine protease genes (granzymes) that are tightly regulated and expressed specifically in activated cytotoxic lymphocytes (19–23). Our laboratory has shown that cytotoxic T lymphocytes (CTLs), natural killer (NK) cells, and lymphokine-activated killer (LAK) cells require one of these enzymes, granzyme B, for the rapid induction of apoptosis in allogeneic target cells (24). The granzyme B loci of humans and mice are found in syntenic regions of chromosome 14 (25–28). The human locus is composed of four functional genes: granzymes B and H, cathepsin G, and mast cell chymase. Granzyme B is expressed exclusively in activated CTLs and NK and LAK cells, while granzyme H is expressed preferentially in NK and LAK cells (27). Cathepsin G is expressed in promyelocytes (29), and chymase is expressed preferentially in mast cells (30). Granzyme B is located at the 5′ end of the human cluster, followed by granzyme H, cathepsin G, and mast cell chymase. Previous studies have suggested that mouse granzymes B–G and cathepsin G are also tightly clustered (26, 28, 31).
In this report, we show that insertion of PGK–Neo into the murine granzyme B gene abrogates the expression of several granzyme genes within the granzyme B gene cluster. In addition, we have learned that PGK–Neo can be governed by regulatory elements in the domains into which it is inserted. These observations have important implications for the interpretation of phenotypes in knockout animals where the selectable marker is retained within the targeted locus.
MATERIALS AND METHODS
Isolation and Characterization of P1 and BAC Clones.
PCR primers for granzymes B, D, and E were used to screen a murine P1 library (Genome Systems, St. Louis). A 0.4-kb EcoRI–HindIII genomic DNA fragment derived from the 5′ flanking region of murine cathepsin G was used to screen a murine BAC library (Genome Systems). To screen for the presence of each gene on the overlapping clones, the following strategies were used: granzyme B was screened by Southern blot analysis using a 0.38-kb BamHI–EcoRI 3′ flanking fragment, and granzyme C was screened using a 0.5-kb PCR-derived genomic DNA fragment (position −1177 to −1677 of the 5′ flanking region of granzyme C). Granzymes D, E, F, and G and MMCP-2 were screened by PCR using exon-specific primers derived from sequences in exon 3–4 and exon 4–5 splice junctions (all primer sequences are available upon request from the authors). All PCR products were subcloned into pCR2.1 (Invitrogen) and subjected to DNA sequence analysis. Several EcoRI, HindIII, and PstI fragments spanning the entire genomic sequences of granzymes D, E, and G were also isolated from the P1 clones, subcloned into pUC 19 vectors, and sequenced to determine the genomic organization of each gene. DNA from each P1 and BAC clone was digested or double-digested using rare-cutting restriction endonucleases BssHII, XhoI, SmaI, SpeI, SstI, NruI, and NgoAIV. DNA fragments were separated on 1% agarose gels using a CHEF DRII apparatus (Bio-Rad), transferred onto Hybond membranes (Amersham), hybridized with probes specific for granzymes B, C, D, and F, cathepsin G, or MMCP-2 as described above, and washed using stringencies as described (27).
Splenocyte Activations and RNA Analysis.
Total splenocytes were activated in a one-way mixed lymphocyte reaction (MLR) as described (24). Generation of LAK cells was also performed as described (32).
Total cellular RNA was prepared and analyzed by S1 nuclease protection as described (27). The gene-specific probes for murine granzyme A (24), murine granzyme B (24), murine cathepsin G (28), and murine β2-microglobulin (24) have been previously described. Murine granzymes C, F, and G and MMCP-2 were generated by PCR using exon-specific primers that amplified genomic fragments containing the intron 3–exon 4 splice junctions. Granzyme D- and E-specific probes were generated using primers that amplified a genomic fragment containing the intron 4–exon 5 splice junction. The protected probe fragment sizes are indicated under the probes in Fig. 1. The probe for PGK–Neo was prepared from a 0.7-kb PCR product using the primer set: 5′-CCGGGTAGGGGAGGCGCT-3′ (derived from PGK sequence) and 5′-GCGCAAGGAACGCCCGTCGTG-3′ (derived from Neo sequence), using pMC1Neo (33) as the template.
Figure 1.

Mouse granzyme B cluster. The human granzyme B cluster, located on chromosome 14q11.2, has been described elsewhere (27). (A) Overlapping murine P1 and BAC clones were characterized by rare-cutting restriction analysis, and the relative position of each granzyme gene was determined by Southern blot analysis using specific probes for granzymes B, C, D, and F, CG, and MMCP-2. The relative position of granzymes E and G on overlapping clones was determined by PCR using exon-specific primers. The sizes of the P1 and BAC clones were determined by CHEF gel analysis. Slashes between genes indicate approximate distances. (B) The genomic organization of the granzymes and cathepsin G. Coding sequences are designated as solid boxes and introns are designated as open boxes. Specific probes for S1 nuclease protection assays and the lengths (in nucleotides) of the fragments protected by correctly spliced mRNAs are shown for each gene. Asterisks designate a newly defined gene structure.
RESULTS
Structure of the Murine Granzyme B Locus.
In mice, seven granzymes (A–G) have been described to date. The granzyme A gene resides on chromosome 13 (31), and the granzyme B cluster is found on chromosome 14 (26). We screened P1 and BAC libraries with primers specific for granzymes D, E, and B and cathepsin G and obtained several clones. We next used exon-specific primers and PCR techniques to define the specific granzyme genes found on each P1 or BAC clone. Each PCR product was subcloned and sequenced to confirm its identity. Granzymes D, E, and G (to our knowledge, whose gene structure has not been reported) were sequenced completely to determine their genomic organizations (Fig. 1B). Three overlapping P1 clones (P1-86, P1-38, and P1-39) with inserts averaging 80–90 kb long were shown to contain granzymes B–G (Fig. 1A). To determine the order of the granzyme genes within the cluster, DNA prepared from each P1 clone was digested or double-digested with various combinations of rare-cutting endonucleases (BssHII, XhoI, SmaI, SpeI, SstI, NruI, and NgoAIV), separated on CHEF gels, and transferred to nylon membranes. The blots were hybridized sequentially with probes specific for granzymes B, C, F, and D (data not shown). We determined that granzyme B is located at the 5′ end of the cluster, followed by granzymes C, F, G, D, and E. Granzyme C lies approximately 23 kb downstream from granzyme B and 25 kb upstream from granzyme F. Granzymes F and D colocalized on the same 75-kb NruI fragment (data not shown). We also determined that BAC clones 50 and 51 contain cathepsin G and and one of the chymase genes, MMCP-2. Cathepsin G lies 30–50 kb downstream from granzyme E and mapped within 80 kb of MMCP-2 on the same NgoAIV fragment (data not shown). MMCP-1, -4, and -5, which are known to reside in the chymase gene cluster on chromosome 14 (34), presumably lie further downstream. The transcriptional orientation of each gene in the cluster has not yet been determined; all genes in the human cluster are in the same orientation (Fig. 1).
Granzyme Gene Expression in Cytotoxic Lymphocytes.
We examined granzyme expression in activated splenocytes generated by MLR and in LAK cells. Using a series of specific S1 probes designed to discriminate among the highly related granzyme transcripts, we examined MLR RNA samples for the expression of granzymes in the cluster. Granzyme A and B mRNA levels were most abundant in MLR cells harvested on day 5 (Fig. 2A). A small amount of granzyme C mRNA was detected in MLR RNA, but no mRNA from granzymes D, E, F, and G was detected (Fig. 2A). This was confirmed with RNA derived from MLR on days 2–5, using the same set of probes (data not shown).
Figure 2.
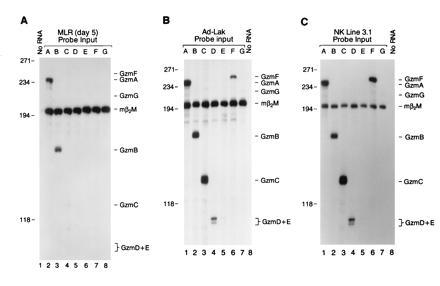
Expression of granzymes A–G in activated lymphocytes. (A) Twenty micrograms of total cellular RNA derived from a day 5 MLR culture was hybridized with a panel of specific probes for granzymes A–G (see Fig. 1). The designated granzyme probe and β2-microglobulin probe were cohybridized with each RNA sample, and S1 analysis was performed. Note that mRNAs encoding granzymes A and B are most abundant, with small amounts of granzyme C detected on day 5. Similar results were obtained using RNA obtained from day 2–5 MLR (data not shown). (B) LAK cells were generated by culturing splenocytes in the presence of high-dose IL-2 for 10 days. Twenty micrograms of total RNA was hybridized with specific probes for granzymes A–G (see Fig. 1). (C) Total RNA derived from the NK tumor cell line NK 3.1 (CD3−, NK1.1+, maintained in recombinant human IL-2 at 500 units/ml), was characterized using specific probes for granzymes A–G. Note that LAK cells and NK 3.1 cells both contain abundant levels of granzyme A, B, C, D, and F mRNAs.
We next examined granzyme expression in LAK cells derived from splenocytes cultured in the presence of high-dose interleukin 2 (IL-2) (1000 units/ml) for 8–10 days. In contrast to CTLs obtained by MLR, LAK cells express substantial amounts of granzymes C, D, and F mRNA, in addition to mRNA for granzymes A and B (Fig. 2B); low level expression of granzymes E and G was also consistently detected. Antibody-mediated depletion of T cells from LAK cultures [using anti-CD8, anti-T-cell receptor (TcR) αβ, and anti-TcR γδ] had no effect on the results (data not shown). In addition, we have also developed NK-like cell lines (CD3−, NK1.1+) from splenic tumors harvested from transgenic mice expressing simian virus 40 (SV40) large tumor (T) antigen driven by the human granzyme H promoter (D.M.M. and T.J.L., unpublished results). mRNA derived from several of these cell lines also contains abundant granzyme A, B, C, D, and F mRNAs, similar to that observed for LAK cells (an analysis of one cell line, NK 3.1, is shown in Fig. 2C).
Granzyme Expression in Granzyme B −/− and Cathepsin G −/− LAK Cells.
We next examined granzyme gene expression in LAK cells derived from mice containing targeted mutations in either granzyme B or cathepsin G. The granzyme B mutation was generated by replacing exon 1 with a standard PGK–Neo cassette (24). This mutation removes a 350-bp AvrII fragment starting within the 5′ untranslated region (22 bp upstream from the initiation codon) and extending into intron 1. PGK–Neo is transcribed in the same direction as the granzyme B gene. The cathepsin G mutation consists of a 393-bp PstI–BglII deletion that removes the 3′ end of exon 3, all of intron 3, and the 5′ end of exon 4; the same PGK–Neo cassette was inserted in the opposite transcriptional orientation from cathepsin G. RNA and protein analyses have confirmed that this mutation is null for cathepsin G (D.M.M. and T.J.L., unpublished results). The proliferative potential of splenocytes in response to high-dose IL-2 was not affected by either mutation (data not shown). We assayed granzyme A mRNA levels (since this gene is located on chromosome 13) as an indicator of adequate lymphocyte activation. Remarkably, insertion of the PGK–Neo cassette into the granzyme B gene caused a nearly complete loss of granzyme C expression, a severe reduction in granzyme F expression, and a clearcut reduction in granzyme D and G mRNA levels in LAK cells (Fig. 3). Expression of granzyme E (the most distal gene in the cluster, located ≈150 kb downstream from granzyme B) is minimal in LAK cells and is not significantly changed by the granzyme B mutation. Insertion of PGK–Neo into the cathepsin G gene did not alter granzyme B, C, E, F, and G gene expression in LAK cells. The CG mutation reduces granzyme D mRNA levels by approximately 30%, perhaps due to the proximity of granzyme D and cathepsin G in the cluster. The granzyme B mutation did not alter cathepsin G expression in the bone marrow (ref. 24 and Fig. 4). We determined that the reduction in granzyme C, F, D, and G expression was not due to gross alterations in these genes, since Southern blot analyses revealed that all genes were present and nonrearranged (data not shown).
Figure 3.
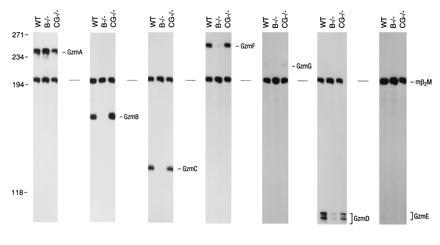
Expression of granzymes in LAK cells derived from wild-type (WT), granzyme B −/−, and cathepsin G −/− mice. LAK cell total RNA (20 μg), generated from WT, B −/−, and cathepsin G −/− mice was analyzed for the expression of granzymes A–G using S1 nuclease protection assays. Equivalent levels of granzyme A mRNA in all three samples indicates that all samples were similarly activated. Note that levels of granzymes C, F, and D mRNAs are substantially reduced in LAK cells derived from granzyme B −/− mice. These experiments were performed three times, yielding identical results. Autoradiograms for granzyme A, B, C, F, and D panels were exposed overnight. Granzyme G and E panels were exposed for 72 h.
Figure 4.
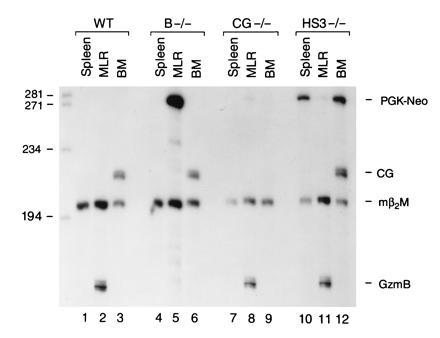
Expression of PGK–Neo in wild-type (WT), granzyme B −/−, cathepsin G −/−, and HS-3 −/− mice. RNA (15 μg) derived from resting spleen, MLR, and bone marrow from each animal was analyzed using S1 nuclease protection assays; each mRNA sample was cohybridized with probes for granzyme B, cathepsin G, PGK–Neo, and β2-microglobulin. Note that abundant, correctly initiated PGK–Neo mRNA is detected in MLR lymphocytes (or LAK cells, data not shown) only when the cassette is located within the granzyme B gene (lane 5 versus lanes 8 and 11). PGK–Neo is detected in the spleen and bone marrow of mice containing the cassette in the murine β-globin LCR (lanes 10 and 12); both of these organs contain erythroid precursors in adult mice. PGK–Neo mRNA is not detected in the marrow of the mice containing the cathepsin G −/− mutation. The granzyme B mutation does not affect expression of cathepsin G in marrow (lane 6), as reported (24). These experiments were repeated four times with identical results.
PGK–Neo Expression in Granzyme B −/−, Cathepsin G −/−, and 5′ HS-3 −/− Mice.
We next wanted to determine whether PGK–Neo expression was affected by its position in the granzyme B cluster. We designed a PGK–Neo probe such that correctly initiated PGK–Neo mRNA protects a probe fragment of 281 nt from S1 digestion. This probe, and probes for granzyme B, cathepsin G, and β2-microglobulin, were hybridized simultaneously with test RNAs and subjected to S1 analysis (Fig. 4). Samples derived from wild-type mice contain no detectable PGK–Neo mRNA (Fig. 4, lanes 1–3). “Resting” splenocytes derived from granzyme B −/− mice have no detectable granzyme B or PGK–Neo mRNA (lane 4), as expected. Remarkably, abundant PGK–Neo mRNA is detected in granzyme B −/− MLR RNA (Fig. 4, lane 5). This transcript is correctly initiated from the PGK promoter, since “readthrough” transcripts from the granzyme B promoter would yield a protected probe fragment of 770 nt; no readthrough transcripts of this size were detected (data not shown). A small amount of PGK–Neo mRNA is detected in CG −/− and HS-3 −/− MLR RNA samples (lanes 8 and 11), indicating that PGK–Neo is normally expressed at low levels in activated T cells. As expected, no cathepsin G mRNA was detected in the bone marrow of cathepsin G −/− mice (lane 9); PGK–Neo mRNA was not detected in the bone marrow of these animals either, indicating that PGK–Neo is not “activated” by cathepsin G regulatory sequences. In contrast, substantial amounts of correctly initiated PGK–Neo mRNA are detected in the resting spleen and bone marrow of HS-3 −/− mice (lanes 10 and 12); these organs contain a significant population of erythroid precursors. These results suggest that PGK–Neo expression is regulated by the β-globin LCR in erythroid cells.
DISCUSSION
In these studies, we have established that the PGK–Neo cassette can severely affect the expression of several genes within a multigene cluster. This cassette, when present in the LCR of the β-globin gene cluster, causes downregulation of the embryonic and adult globin genes downstream from the mutation. In this report, we have learned that a PGK–Neo cassette located in the granzyme B gene disrupts the expression of multiple granzyme genes downstream from granzyme B in LAK cells. However, a PGK–Neo cassette located in the cathepsin G gene (just downstream from the granzyme genes) caused minimal alterations in the expression of the upstream granzymes, suggesting that it lies within a separate regulatory domain within this multigene cluster. These results are supported by the observation that the PGK–Neo cassette becomes highly inducible in activated cytotoxic lymphocytes when it is located within the granzyme B gene, but it is not when it is located within the cathepsin G gene or the mouse β-globin LCR. Conversely, PGK–Neo is expressed at high levels in the spleen and bone marrow of mice containing the cassette in the β-globin LCR, suggesting that PGK–Neo has been captured by this erythroid regulatory element. Thus, these results have implications for how PGK–Neo cassettes can potentially alter the expression of multiple genes within a locus and suggests a mechanism for how that disregulation might occur.
Insertion of a PGK–Neo cassette into the granzyme B gene, the most 5′ gene in the granzyme cluster, caused a severe reduction of granzyme C, D, F, and G expression in LAK cells derived from granzyme B −/− splenocytes; the granzyme D gene lies >100 kb from the mutation. However, the granzyme B mutation had no effect on the expression of cathepsin G, a promyelocyte-specific gene that lies 30–50 kb downstream from the minimally expressed granzyme E gene. Conversely, the presence of PGK–Neo in the cathepsin G gene had minimal effects on the expression of the upstream granzyme genes in LAK cells. These results imply that the active granzyme domain may be regulated by a common regulatory element and that the insertion of PGK–Neo disrupts a normal interaction between this putative element and the downstream genes (Fig. 5). Since cathepsin G is expressed predominantly in myeloid precursors, this gene may lie in a separate domain within the locus and may have separate elements that regulate its expression.
Figure 5.

Model of competition between the PGK–Neo cassette and the individual granzyme promoters for a productive interaction with a putative LCR. Insertion of PGK–Neo may disrupt normal interactions between the putative LCR and the granzyme genes, leading to activation of the PGK–Neo gene and inactivation of multiple genes within the locus. This disruption is position-specific, since insertion of PGK–Neo into the cathepsin G gene has minimal effects on granzyme gene expression.
We demonstrate here that PGK–Neo is strongly induced in the activated lymphocytes of granzyme B −/− mice. These results suggest that the PGK–Neo cassette is captured by the regulatory elements of the granzyme B gene or domain, causing PGK–Neo to adopt the regulatory behavior of the granzyme B gene itself. These results are reminiscent of studies in transgenic mice that revealed that the human β-globin LCR can direct erythroid-specific expression of the Thy-1 promoter or the thymidine kinase promoter (35); similarly, the human CD2 LCR can redirect human β-globin gene expression to T lymphocytes in transgenic animals (36). The capture of PGK–Neo expression by regulatory elements that lie within targeted domains provides a tentative explanation for “neighborhood” effects; the PGK–Neo cassette may act as a “sink” for LCR activity (5, 6).
Since multiple granzymes are coordinately expressed in LAK cells and in clonal murine NK cell lines (Fig. 2 B and C), the putative LCR-like element of this domain may be capable of interacting with multiple genes simultaneously. Similarly, the two human γ-globin genes can apparently interact simultaneously with the human β-globin LCR (37). However, the presence of PGK–Neo at the 5′ end of the granzyme B gene cluster virtually eliminates all downstream interactions in LAK cells, which suggests that the putative LCR preferentially interacts with PGK–Neo, as it does for the β-globin LCR mutations (7, 8).
These results have led us to reassess the phenotype observed in granzyme B −/− mice. CTLs derived from these mice have a profound defect in their ability to rapidly induce apoptosis in allogeneic target cells. Since the CTLs derived from MLRs predominantly express granzymes A and B, we strongly suspect that the defect in the induction of apoptosis is due to the lack of granzyme B itself. Granzyme B −/− LAK cells and NK cells have a similar, severe reduction in their ability to rapidly induce apoptosis in susceptible targets. However, since LAK cells clearly express granzymes B, C, D, and F and since all of these genes are disregulated by the mutation, we do not know whether the cytotoxic defect observed is due to the loss of granzyme B or the loss of other granzymes in the cluster. To discriminate between these possibilities, we are creating mice with the same granzyme B mutation, but with the PGK–Neo cassette removed; we are also inserting the PGK–Neo cassette between granzymes B and C to determine whether all of the downstream genes can be disrupted by this mutation.
These results demonstrate the potential of a PGK–Neo cassette to mitigate the expression of several genes within a multigene locus, making interpretation of specific phenotypes potentially difficult to ascertain. This data, coupled with recent data generated with the murine β-globin LCR mutations, the Igκ intronic enhancer knockout, and mutations in the MRF4 and Hox genes, strongly suggest that deletion of the PGK–Neo cassette by site-specific recombinases may be very important for the accurate interpretation of targeted mutations located within multigene loci.
Acknowledgments
We thank Jeff Milbrandt and Mark Groudine for helpful suggestions and advice. Robin Wesselschmidt and Pam Goda provided expert animal care and technical assistance. We thank Dr. Wayne Yokoyama and Indira Mehta for T-cell-depleted LAK RNA. Nancy Reidelberger provided expert assistance with preparation of the manuscript. This work was supported by National Institutes of Health Grants CA49712, DK49786, and DK38682 and the Washington University–Monsanto Agreement (T.J.L.).
Footnotes
Abbreviations: LCR, locus control region; CTL, cytotoxic T lymphocyte; NK, natural killer; LAK, lymphocyte-activated killer; MLR, mixed lymphocyte reaction; IL-2, interleukin 2.
Data deposition: The sequences reported in this paper have been deposited in the GenBank data base (accession nos. U266472–U66474).
References
- 1.Olson E N, Arnold H-H, Rigby P W J, Wold B J. Cell. 1996;84:1–4. doi: 10.1016/s0092-8674(00)81073-9. [DOI] [PubMed] [Google Scholar]
- 2.Tuan D, Solomon W, Li Q, London I. Proc Natl Acad Sci USA. 1985;82:6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forrester W C, Thompson C, Elder J T, Groudine M. Proc Natl Acad Sci USA. 1986;83:1359–1363. doi: 10.1073/pnas.83.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhar V, Nandi A, Schildkraut C L, Skoultchi A. Mol Cell Biol. 1990;10:4324–4333. doi: 10.1128/mcb.10.8.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim C, Epner E, Forrester W, Groudine M. Genes Dev. 1992;6:928–938. doi: 10.1101/gad.6.6.928. [DOI] [PubMed] [Google Scholar]
- 6.Fiering S, Kim C, Epner E, Groudine M. Proc Natl Acad Sci USA. 1993;90:8469–68473. doi: 10.1073/pnas.90.18.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin D I K, Enver T, Ley T J, Groudine M. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 8.Hug B A, Wesselschmidt R L, Fiering S, Bender M A, Epner E, Groudine M, Ley T J. Mol Cell Biol. 1996;16:2906–2912. doi: 10.1128/mcb.16.6.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun T, Bober E, Rudnicki M A, Jaenisch R, Arnold H-H. Development (Cambridge, UK) 1994;120:3083–3092. doi: 10.1242/dev.120.11.3083. [DOI] [PubMed] [Google Scholar]
- 10.Braun T, Arnold H-H. EMBO J. 1995;14:1176–1186. doi: 10.1002/j.1460-2075.1995.tb07101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patapoutian A, Kyo Yoon J, Miner J H, Wang S, Stark K, Wold B. Development (Cambridge, UK) 1995;121:3347–3358. doi: 10.1242/dev.121.10.3347. [DOI] [PubMed] [Google Scholar]
- 12.Zhang W, Behringer R R, Olson E N. Genes Dev. 1995;9:1388–1399. doi: 10.1101/gad.9.11.1388. [DOI] [PubMed] [Google Scholar]
- 13.Jeannotte L, Lemieux M, Charron J, Poirier F, Robertson E J. Genes Dev. 1993;7:2085–2096. doi: 10.1101/gad.7.11.2085. [DOI] [PubMed] [Google Scholar]
- 14.Ramirez-Solis R, Zheng H, Whiting J, Krumlauf R, Bradley A. Cell. 1993;73:279–294. doi: 10.1016/0092-8674(93)90229-j. [DOI] [PubMed] [Google Scholar]
- 15.Kostic D, Capecchi M R. Mech Dev. 1994;46:231–247. doi: 10.1016/0925-4773(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 16.Horan G S B, Wu K, Wolgemuth D J, Behringer R R. Proc Natl Acad Sci USA. 1994;91:12644–12648. doi: 10.1073/pnas.91.26.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horan G S B, Nagy Kovacs E, Behringer R R, Featherstone M S. Dev Biol. 1995;169:359–372. doi: 10.1006/dbio.1995.1150. [DOI] [PubMed] [Google Scholar]
- 18.Xu Y, Davidson L, Alt F W, Baltimore D. Immunity. 1996;4:377–385. doi: 10.1016/s1074-7613(00)80251-4. [DOI] [PubMed] [Google Scholar]
- 19.Masson D, Tschopp J. Cell. 1987;49:679–685. doi: 10.1016/0092-8674(87)90544-7. [DOI] [PubMed] [Google Scholar]
- 20.Jenne D, Rey C, Haefliger J-A, Qiao B-Y, Groscurth P, Tschopp J. Proc Natl Acad Sci USA. 1988;85:4814–4818. doi: 10.1073/pnas.85.13.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenne D, Rey C, Masson D, Stanley K K, Herz J, Plaetinck G, Tschopp J. J Immunol. 1988;140:318–323. [PubMed] [Google Scholar]
- 22.Jenne D, Masson D, Zimmer M, Haefliger J-A, Li W-H, Tschopp J. Biochemistry. 1989;28:7953–7961. doi: 10.1021/bi00445a060. [DOI] [PubMed] [Google Scholar]
- 23.Bleackley R C, Lobe C G, Duggan B, Ehrman N, Fregeau C, Meier M, Letellier M, Havele C, Shaw J, Paetkau V. Immunol Rev. 1988;103:5–19. doi: 10.1111/j.1600-065x.1988.tb00746.x. [DOI] [PubMed] [Google Scholar]
- 24.Heusel J W, Wesselschmidt R L, Shresta S, Russell J H, Ley T J. Cell. 1994;76:977–987. doi: 10.1016/0092-8674(94)90376-x. [DOI] [PubMed] [Google Scholar]
- 25.Hohn P A, Popescu N C, Hanson R D, Salverson G, Ley T J. J Biol Chem. 1989;264:13412–13419. [PubMed] [Google Scholar]
- 26.Crosby J L, Bleackley R C, Nadeau J H. Genomics. 1990;6:252–259. doi: 10.1016/0888-7543(90)90564-b. [DOI] [PubMed] [Google Scholar]
- 27.Heusel J W, Hanson R D, Silverman G A, Ley T J. J Biol Chem. 1991;266:6152–6158. [PubMed] [Google Scholar]
- 28.Heusel J W, Scarpati E M, Jenkins N A, Gilbert D J, Copeland N G, Shapiro SD, Ley T J. Blood. 1993;81:1614–1623. [PubMed] [Google Scholar]
- 29.Hanson R D, Connolly N L, Burnett D, Campbell E J, Senior R M, Ley T J. J Biol Chem. 1990;265:1524–1530. [PubMed] [Google Scholar]
- 30.Caughey G A, Schaumberg T H, Zerwede E H, Butterfield J H, Hanson R D, Silverman G A, Ley T J. Genomics. 1993;15:614–620. doi: 10.1006/geno.1993.1115. [DOI] [PubMed] [Google Scholar]
- 31.Mattei M G, Harper K, Brunet J F, Denizot F, Mattei J F, Goldstein P, Giraud F. Cytogenet Cell Genet. 1987;46:657–658. [Google Scholar]
- 32.Shresta S, MacIvor D M, Heusel J W, Russell J H, Ley T J. Proc Natl Acad Sci USA. 1995;92:5679–5683. doi: 10.1073/pnas.92.12.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soriano T, Montgomery C, Geske R, Bradley A. Cell. 1991;64:639–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 34.Gurish M F, Nadeau J H, Johnson K R, McNeil H P, Grattan K M, Austen K F, Stevens R L. J Biol Chem. 1993;268:11372–11379. [PubMed] [Google Scholar]
- 35.Blom van Assendelft G, Hanscombe O, Grosveld F, Greaves D R. Cell. 1989;56:969–977. doi: 10.1016/0092-8674(89)90630-2. [DOI] [PubMed] [Google Scholar]
- 36.Greaves D R, Wilson F D, Lang G, Kioussis D. Cell. 1989;56:979–986. doi: 10.1016/0092-8674(89)90631-4. [DOI] [PubMed] [Google Scholar]
- 37.Bresnick E H, Felsenfeld G. Proc Natl Acad Sci USA. 1994;91:1314–1317. doi: 10.1073/pnas.91.4.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]


