Abstract
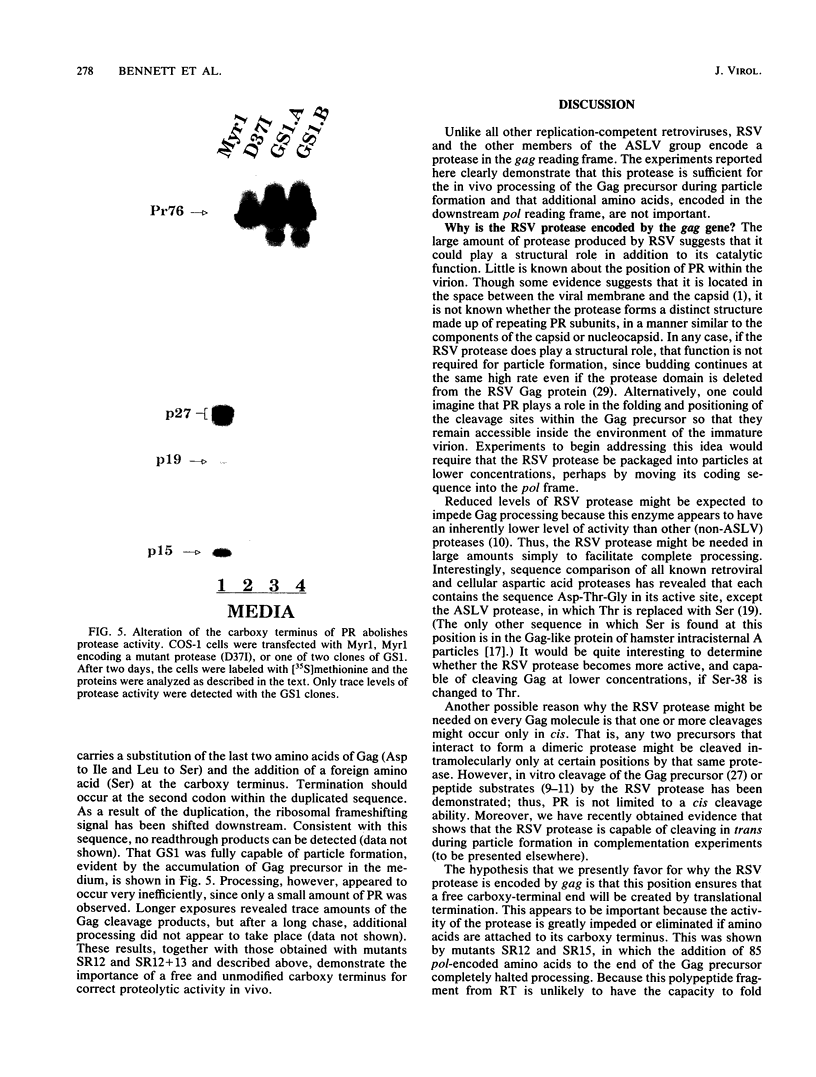
Rous sarcoma virus (RSV) and its relatives are unique in that they appear to encode their viral protease in the gag reading frame. As a result, this 124-amino-acid sequence is found at the carboxy terminus of each Gag precursor molecule and, upon ribosome frameshifting, embedded within each Gag-Pol molecule. However, rigorous proof has never been obtained for the activity of this 124-amino-acid Gag domain during virion assembly in vivo. If the active protease actually included amino acids encoded downstream in the pol reading frame, then the sequence organization would be more in line with those of other retroviruses. To examine this issue, mutations that disrupt the addition of amino acids by ribosome frameshifting were analyzed for their effects on particle assembly and Gag processing in a mammalian expression system (J. W. Wills, R. C. Craven, and J. A. Achacoso, J. Virol. 63:4331-4343, 1989). A 2-base substitution which created a nonsense mutation in the pol reading frame and was predicted to disrupt the hairpin structure of the ribosome frameshift signal had no effect on particle assembly or Gag processing, definitively showing that downstream amino acids are unnecessary. Mutations that fused the gag and pol reading frames to place 85 amino acids at the carboxy terminus of Gag hindered particle assembly and totally abolished the activity of the protease. A smaller fusion protein containing only the seven-amino-acid spacer peptide that links Gag and reverse transcriptase allowed particle formation but slowed processing. The reduced rate of processing exhibited by this mutant also revealed a previously unnoticed series of late maturation steps associated with the RSV capsid (CA) protein. Another mutant containing two substituted amino acids plus one additional amino acid at the carboxy terminus of protease nearly abolished processing. Together, these results demonstrate the importance of the carboxy terminus for proteolytic activity and suggest that this end must be unrestrained for optimal activity. If this hypothesis is correct, then the RSV protease may be encoded at the end of gag simply to ensure the production of a free carboxy terminus by translational termination.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolognesi D. P., Luftig R., Shaper J. H. Localization of RNA tumor virus polypeptides. I. Isolation of further virus substructures. Virology. 1973 Dec;56(2):549–564. doi: 10.1016/0042-6822(73)90057-3. [DOI] [PubMed] [Google Scholar]
- Felsenstein K. M., Goff S. P. Expression of the gag-pol fusion protein of Moloney murine leukemia virus without gag protein does not induce virion formation or proteolytic processing. J Virol. 1988 Jun;62(6):2179–2182. doi: 10.1128/jvi.62.6.2179-2182.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Billich S., Schulze T., Sukrow S., Moelling K. Partial purification and substrate analysis of bacterially expressed HIV protease by means of monoclonal antibody. EMBO J. 1988 Jun;7(6):1785–1791. doi: 10.1002/j.1460-2075.1988.tb03009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Townsley K., Varmus H. E., Majors J. Two efficient ribosomal frameshifting events are required for synthesis of mouse mammary tumor virus gag-related polyproteins. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4298–4302. doi: 10.1073/pnas.84.12.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh I., Ikawa Y., Yoshinaka Y. Retrovirus protease characterized as a dimeric aspartic proteinase. J Virol. 1989 May;63(5):2226–2232. doi: 10.1128/jvi.63.5.2226-2232.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotler M., Danho W., Katz R. A., Leis J., Skalka A. M. Avian retroviral protease and cellular aspartic proteases are distinguished by activities on peptide substrates. J Biol Chem. 1989 Feb 25;264(6):3428–3435. [PubMed] [Google Scholar]
- Kotler M., Katz R. A., Danho W., Leis J., Skalka A. M. Synthetic peptides as substrates and inhibitors of a retroviral protease. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4185–4189. doi: 10.1073/pnas.85.12.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotler M., Katz R. A., Skalka A. M. Activity of avian retroviral protease expressed in Escherichia coli. J Virol. 1988 Aug;62(8):2696–2700. doi: 10.1128/jvi.62.8.2696-2700.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lapatto R., Blundell T., Hemmings A., Overington J., Wilderspin A., Wood S., Merson J. R., Whittle P. J., Danley D. E., Geoghegan K. F. X-ray analysis of HIV-1 proteinase at 2.7 A resolution confirms structural homology among retroviral enzymes. Nature. 1989 Nov 16;342(6247):299–302. doi: 10.1038/342299a0. [DOI] [PubMed] [Google Scholar]
- Loeb D. D., Swanstrom R., Everitt L., Manchester M., Stamper S. E., Hutchison C. A., 3rd Complete mutagenesis of the HIV-1 protease. Nature. 1989 Aug 3;340(6232):397–400. doi: 10.1038/340397a0. [DOI] [PubMed] [Google Scholar]
- Miller M., Jaskólski M., Rao J. K., Leis J., Wlodawer A. Crystal structure of a retroviral protease proves relationship to aspartic protease family. Nature. 1989 Feb 9;337(6207):576–579. doi: 10.1038/337576a0. [DOI] [PubMed] [Google Scholar]
- Navia M. A., Fitzgerald P. M., McKeever B. M., Leu C. T., Heimbach J. C., Herber W. K., Sigal I. S., Darke P. L., Springer J. P. Three-dimensional structure of aspartyl protease from human immunodeficiency virus HIV-1. Nature. 1989 Feb 16;337(6208):615–620. doi: 10.1038/337615a0. [DOI] [PubMed] [Google Scholar]
- Ono M., Toh H., Miyata T., Awaya T. Nucleotide sequence of the Syrian hamster intracisternal A-particle gene: close evolutionary relationship of type A particle gene to types B and D oncovirus genes. J Virol. 1985 Aug;55(2):387–394. doi: 10.1128/jvi.55.2.387-394.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oroszlan S., Luftig R. B. Retroviral proteinases. Curr Top Microbiol Immunol. 1990;157:153–185. doi: 10.1007/978-3-642-75218-6_6. [DOI] [PubMed] [Google Scholar]
- Pearl L. H., Taylor W. R. A structural model for the retroviral proteases. Nature. 1987 Sep 24;329(6137):351–354. doi: 10.1038/329351a0. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Allen D. W., Niall H. D. Amino acid sequence of p15 from avian myeloblastosis virus complex. Biochemistry. 1981 Jun 23;20(13):3784–3791. doi: 10.1021/bi00516a018. [DOI] [PubMed] [Google Scholar]
- Schwartz D. E., Tizard R., Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983 Mar;32(3):853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- Skalka A. M. Retroviral proteases: first glimpses at the anatomy of a processing machine. Cell. 1989 Mar 24;56(6):911–913. doi: 10.1016/0092-8674(89)90621-1. [DOI] [PubMed] [Google Scholar]
- Smith A. J., Cho M. I., Hammarskjöld M. L., Rekosh D. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into viruslike particles. J Virol. 1990 Jun;64(6):2743–2750. doi: 10.1128/jvi.64.6.2743-2750.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart L., Schatz G., Vogt V. M. Properties of avian retrovirus particles defective in viral protease. J Virol. 1990 Oct;64(10):5076–5092. doi: 10.1128/jvi.64.10.5076-5092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt V. M., Wight A., Eisenman R. In vitro cleavage of avian retrovirus gag proteins by viral protease p15. Virology. 1979 Oct 15;98(1):154–167. doi: 10.1016/0042-6822(79)90534-8. [DOI] [PubMed] [Google Scholar]
- Weber I. T., Miller M., Jaskólski M., Leis J., Skalka A. M., Wlodawer A. Molecular modeling of the HIV-1 protease and its substrate binding site. Science. 1989 Feb 17;243(4893):928–931. doi: 10.1126/science.2537531. [DOI] [PubMed] [Google Scholar]
- Weldon R. A., Jr, Erdie C. R., Oliver M. G., Wills J. W. Incorporation of chimeric gag protein into retroviral particles. J Virol. 1990 Sep;64(9):4169–4179. doi: 10.1128/jvi.64.9.4169-4179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills J. W., Craven R. C., Achacoso J. A. Creation and expression of myristylated forms of Rous sarcoma virus gag protein in mammalian cells. J Virol. 1989 Oct;63(10):4331–4343. doi: 10.1128/jvi.63.10.4331-4343.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills J. W., Srinivas R. V., Hunter E. Mutations of the Rous sarcoma virus env gene that affect the transport and subcellular location of the glycoprotein products. J Cell Biol. 1984 Dec;99(6):2011–2023. doi: 10.1083/jcb.99.6.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodawer A., Miller M., Jaskólski M., Sathyanarayana B. K., Baldwin E., Weber I. T., Selk L. M., Clawson L., Schneider J., Kent S. B. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science. 1989 Aug 11;245(4918):616–621. doi: 10.1126/science.2548279. [DOI] [PubMed] [Google Scholar]