Abstract
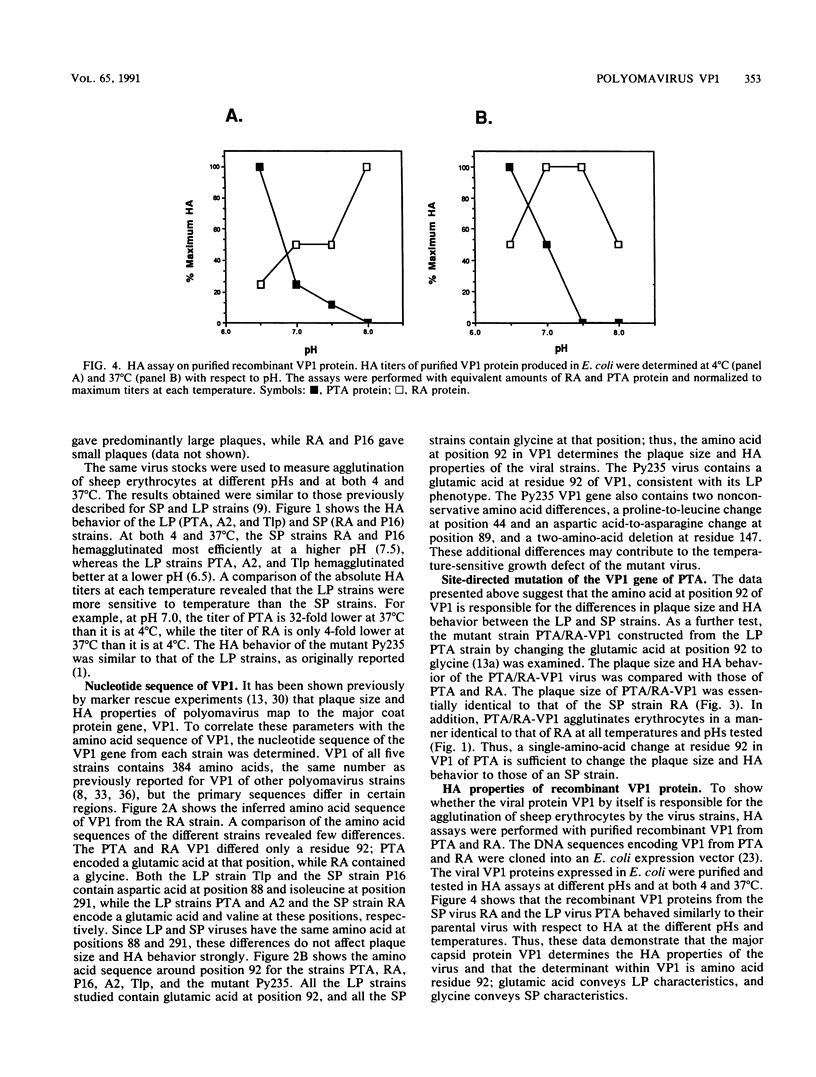
The plaque size and hemagglutination characteristics of five cloned wild-type strains of polyomavirus were determined. The strains fell into two groups, those with large or small plaques, each with distinctive hemagglutination behavior at different temperatures and pHs. The nucleotide sequence of VP1, the major capsid protein of the virus, was determined for each of the viral strains. The PTA (large-plaque) and RA (small-plaque) strains differed only at residue 92 of VP1, where there is a glutamic acid or glycine, respectively (R. Freund, A. Calderone, C. J. Dawe, and T. L. Benjamin, J. Virol. 65:335-341, 1991). The same amino acid difference in VP1 correlated with plaque size and hemagglutination properties of the other sequenced viruses. Mutagenesis converting amino acid 92 from glutamic acid to glycine converted the plaque size and hemagglutination behavior of the large-plaque PTA strain to that of a small-plaque strain. Furthermore, PTA and RA VP1 proteins produced in Escherichia coli behaved as their parental viruses did in hemagglutination assays. These results demonstrate that amino acid residue 92 of VP1 is involved in determining the plaque size and hemagglutination behavior of polyomavirus and strongly suggest that this region of the VP1 polypeptide interacts directly with cell receptors.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basilico C., DiMayorca G. Mutant of polyoma virus with impaired adsorption to BHK cells. J Virol. 1974 Apr;13(4):931–934. doi: 10.1128/jvi.13.4.931-934.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolen J. B., Consigli R. A. Differential adsorption of polyoma virions and capsids to mouse kidney cells and guinea pig erythrocytes. J Virol. 1979 Nov;32(2):679–683. doi: 10.1128/jvi.32.2.679-683.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRAWFORD L. V. The adsorption of polyoma virus. Virology. 1962 Oct;18:177–181. doi: 10.1016/0042-6822(62)90003-x. [DOI] [PubMed] [Google Scholar]
- Cahan L. D., Paulson J. C. Polyoma virus adsorbs to specific sialyloligosaccharide receptors on erythrocytes. Virology. 1980 Jun;103(2):505–509. doi: 10.1016/0042-6822(80)90208-1. [DOI] [PubMed] [Google Scholar]
- Cahan L. D., Singh R., Paulson J. C. Sialyloligosaccharide receptors of binding variants of polyoma virus. Virology. 1983 Oct 30;130(2):281–289. doi: 10.1016/0042-6822(83)90083-1. [DOI] [PubMed] [Google Scholar]
- Carmichael G. G., Schaffhausen B. S., Dorsky D. I., Oliver D. B., Benjamin T. L. Carboxy terminus of polyoma middle-sized tumor antigen is required for attachment to membranes, associated protein kinase activities, and cell transformation. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3579–3583. doi: 10.1073/pnas.79.11.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIAMOND L., CRAWFORD L. V. SOME CHARACTERISTICS OF LARGE-PLAQUE AND SMALL-PLAQUE LINES OF POLYOMA VIRUS. Virology. 1964 Feb;22:235–244. doi: 10.1016/0042-6822(64)90008-x. [DOI] [PubMed] [Google Scholar]
- Dawe C. J., Freund R., Mandel G., Ballmer-Hofer K., Talmage D. A., Benjamin T. L. Variations in polyoma virus genotype in relation to tumor induction in mice. Characterization of wild type strains with widely differing tumor profiles. Am J Pathol. 1987 May;127(2):243–261. [PMC free article] [PubMed] [Google Scholar]
- Deininger P., Esty A., LaPorte P., Friedmann T. Nucleotide sequence and genetic organization of the polyoma late region: features common to the polyoma early region and SV40. Cell. 1979 Nov;18(3):771–779. doi: 10.1016/0092-8674(79)90130-2. [DOI] [PubMed] [Google Scholar]
- Dubensky T. W., Freund R., Dawe C. J., Benjamin T. L. Polyomavirus replication in mice: influences of VP1 type and route of inoculation. J Virol. 1991 Jan;65(1):342–349. doi: 10.1128/jvi.65.1.342-349.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulbecco R., Vogt M. SIGNIFICANCE OF CONTINUED VIRUS PRODUCTION IN TISSUE CULTURES RENDERED NEOPLASTIC BY POLYOMA VIRUS. Proc Natl Acad Sci U S A. 1960 Dec;46(12):1617–1623. doi: 10.1073/pnas.46.12.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EDDY B. E., ROWE W. P., HARTLEY J. W., STEWART S. E., HUEBNER R. J. Hemagglutination with the SE polyoma virus. Virology. 1958 Aug;6(1):290–291. doi: 10.1016/0042-6822(58)90078-3. [DOI] [PubMed] [Google Scholar]
- EDDY B. E., STEWART S. E. Characteristics of the SE polyoma virus. Am J Public Health Nations Health. 1959 Nov;49:1486–1492. doi: 10.2105/ajph.49.11.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feunteun J., Sompayrac L., Fluck M., Benjamin T. Localization of gene functions in polyoma virus DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4169–4173. doi: 10.1073/pnas.73.11.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund R., Calderone A., Dawe C. J., Benjamin T. L. Polyomavirus tumor induction in mice: effects of polymorphisms of VP1 and large T antigen. J Virol. 1991 Jan;65(1):335–341. doi: 10.1128/jvi.65.1.335-341.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund R., Dawe C. J., Benjamin T. L. Duplication of noncoding sequences in polyomavirus specifically augments the development of thymic tumors in mice. J Virol. 1988 Oct;62(10):3896–3899. doi: 10.1128/jvi.62.10.3896-3899.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund R., Mandel G., Carmichael G. G., Barncastle J. P., Dawe C. J., Benjamin T. L. Polyomavirus tumor induction in mice: influences of viral coding and noncoding sequences on tumor profiles. J Virol. 1987 Jul;61(7):2232–2239. doi: 10.1128/jvi.61.7.2232-2239.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried H., Cahan L. D., Paulson J. C. Polyoma virus recognizes specific sialyligosaccharide receptors on host cells. Virology. 1981 Feb;109(1):188–192. doi: 10.1016/0042-6822(81)90485-2. [DOI] [PubMed] [Google Scholar]
- GOTLIEB-STEMATSKY T., LEVENTON S. Studies on the biological properties of two plaque variants isolated from SE polyoma virus. Br J Exp Pathol. 1960 Oct;41:507–519. [PMC free article] [PubMed] [Google Scholar]
- Garcea R. L., Salunke D. M., Caspar D. L. Site-directed mutation affecting polyomavirus capsid self-assembly in vitro. Nature. 1987 Sep 3;329(6134):86–87. doi: 10.1038/329086a0. [DOI] [PubMed] [Google Scholar]
- Goldman E., Benjamin T. L. Analysis of host range of nontransforming polyoma virus mutants. Virology. 1975 Aug;66(2):372–384. doi: 10.1016/0042-6822(75)90210-x. [DOI] [PubMed] [Google Scholar]
- HARTLEY J. W., ROWE W. P., CHANOCK R. M., ANDREWS B. E. Studies of mouse polyoma virus infection. IV. Evidence for mucoprotein erythrocyte receptors in polyoma virus hemagglutination. J Exp Med. 1959 Jul 1;110(1):81–91. doi: 10.1084/jem.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hong G. F. A systemic DNA sequencing strategy. J Mol Biol. 1982 Jul 5;158(3):539–549. doi: 10.1016/0022-2836(82)90213-3. [DOI] [PubMed] [Google Scholar]
- Leavitt A. D., Roberts T. M., Garcea R. L. Polyoma virus major capsid protein, VP1. Purification after high level expression in Escherichia coli. J Biol Chem. 1985 Oct 15;260(23):12803–12809. [PubMed] [Google Scholar]
- McCULLOCH E. A., HOWATSON A. F., SIMINOVITCH L., AXELRAD A. A., HAM A. W. A cytopathogenic agent from a mammary tumour in a C3H mouse that produces tumours in Swiss mice and hamsters. Nature. 1959 May 30;183(4674):1535–1536. doi: 10.1038/1831535a0. [DOI] [PubMed] [Google Scholar]
- McCutchan J. H., Pagano J. S. Enchancement of the infectivity of simian virus 40 deoxyribonucleic acid with diethylaminoethyl-dextran. J Natl Cancer Inst. 1968 Aug;41(2):351–357. [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Miller L. K., Fried M. Construction of the genetic map of the polyoma genome. J Virol. 1976 Jun;18(3):824–832. doi: 10.1128/jvi.18.3.824-832.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G. N., Paulson J. C., Daniels R. S., Skehel J. J., Wilson I. A., Wiley D. C. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983 Jul 7;304(5921):76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- Rogers G. N., Pritchett T. J., Lane J. L., Paulson J. C. Differential sensitivity of human, avian, and equine influenza A viruses to a glycoprotein inhibitor of infection: selection of receptor specific variants. Virology. 1983 Dec;131(2):394–408. doi: 10.1016/0042-6822(83)90507-x. [DOI] [PubMed] [Google Scholar]
- Rothwell V. M., Folk W. R. Comparison of the DNA sequence of the Crawford small-plaque variant of polyomavirus with those of polyomaviruses A2 and strain 3. J Virol. 1983 Nov;48(2):472–480. doi: 10.1128/jvi.48.2.472-480.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STANNERS C. P. STUDIES ON THE TRANSFORMATION OF HAMSTER EMBRYO CELLS IN CULTURE BY POLYOMA VIRUS. II. SELECTIVE TECHNIQUES FOR THE DETECTION OF TRANSFORMED CELLS. Virology. 1963 Nov;21:464–476. doi: 10.1016/0042-6822(63)90207-1. [DOI] [PubMed] [Google Scholar]
- Sahli R., McMaster G. K., Hirt B. DNA sequence comparison between two tissue-specific variants of the autonomous parvovirus, minute virus of mice. Nucleic Acids Res. 1985 May 24;13(10):3617–3633. doi: 10.1093/nar/13.10.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Weis W., Brown J. H., Cusack S., Paulson J. C., Skehel J. J., Wiley D. C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988 Jun 2;333(6172):426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- Wiley D. C., Wilson I. A., Skehel J. J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981 Jan 29;289(5796):373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]



