Abstract
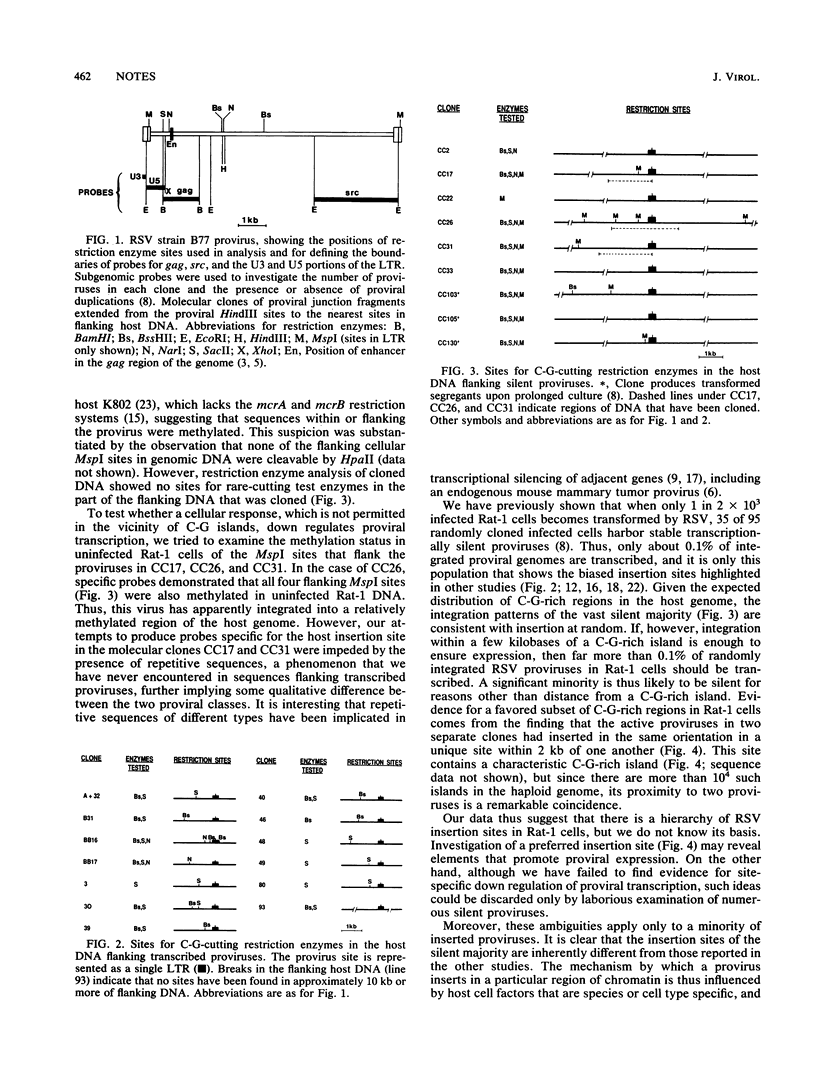
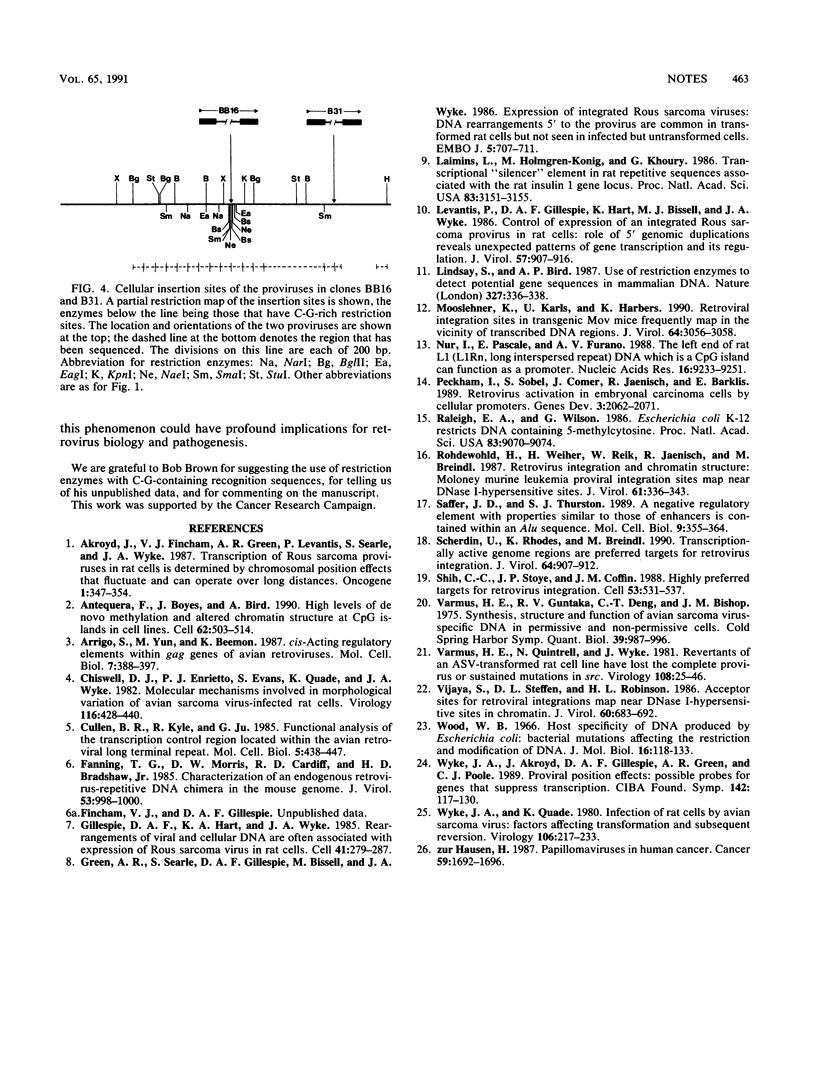
Transcribed Rous sarcoma proviruses in Rat-1 DNA tend to integrate closely 3' to C-G-rich restriction enzyme sites, and 2 of 13 such proviruses were found to have inserted at the same locus. However, most integrated proviruses were transcriptionally silent insertions at sites indistinguishable from random. We conclude that Rous sarcoma proviruses in rat cells usually fail to integrate in a favorable site for transcription, in contrast to studies on proviral integration in permissive hosts.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akroyd J., Fincham V. J., Green A. R., Levantis P., Searle S., Wyke J. A. Transcription of Rous sarcoma proviruses in rat cells is determined by chromosomal position effects that fluctuate and can operate over long distances. Oncogene. 1987;1(4):347–354. [PubMed] [Google Scholar]
- Antequera F., Boyes J., Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990 Aug 10;62(3):503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- Arrigo S., Yun M., Beemon K. cis-acting regulatory elements within gag genes of avian retroviruses. Mol Cell Biol. 1987 Jan;7(1):388–397. doi: 10.1128/mcb.7.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiswell D. J., Enrietto P. J., Evans S., Quade K., Wyke J. A. Molecular mechanisms involved in morphological variation of avian sarcoma virus-infected rat cells. Virology. 1982 Jan 30;116(2):428–440. doi: 10.1016/0042-6822(82)90137-4. [DOI] [PubMed] [Google Scholar]
- Fanning T. G., Morris D. W., Cardiff R. D., Bradshaw H. D., Jr Characterization of an endogenous retrovirus-repetitive DNA chimera in the mouse genome. J Virol. 1985 Mar;53(3):998–1000. doi: 10.1128/jvi.53.3.998-1000.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green A. R., Searle S., Gillespie D. A., Bissell M., Wyke J. A. Expression of integrated Rous sarcoma viruses: DNA rearrangements 5' to the provirus are common in transformed rat cells but not seen in infected but untransformed cells. EMBO J. 1986 Apr;5(4):707–711. doi: 10.1002/j.1460-2075.1986.tb04271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L., Holmgren-König M., Khoury G. Transcriptional "silencer" element in rat repetitive sequences associated with the rat insulin 1 gene locus. Proc Natl Acad Sci U S A. 1986 May;83(10):3151–3155. doi: 10.1073/pnas.83.10.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levantis P., Gillespie D. A., Hart K., Bissell M. J., Wyke J. A. Control of expression of an integrated Rous sarcoma provirus in rat cells: role of 5' genomic duplications reveals unexpected patterns of gene transcription and its regulation. J Virol. 1986 Mar;57(3):907–916. doi: 10.1128/jvi.57.3.907-916.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay S., Bird A. P. Use of restriction enzymes to detect potential gene sequences in mammalian DNA. 1987 May 28-Jun 3Nature. 327(6120):336–338. doi: 10.1038/327336a0. [DOI] [PubMed] [Google Scholar]
- Miller A. D., Law M. F., Verma I. M. Generation of helper-free amphotropic retroviruses that transduce a dominant-acting, methotrexate-resistant dihydrofolate reductase gene. Mol Cell Biol. 1985 Mar;5(3):431–437. doi: 10.1128/mcb.5.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooslehner K., Karls U., Harbers K. Retroviral integration sites in transgenic Mov mice frequently map in the vicinity of transcribed DNA regions. J Virol. 1990 Jun;64(6):3056–3058. doi: 10.1128/jvi.64.6.3056-3058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nur I., Pascale E., Furano A. V. The left end of rat L1 (L1Rn, long interspersed repeated) DNA which is a CpG island can function as a promoter. Nucleic Acids Res. 1988 Oct 11;16(19):9233–9251. doi: 10.1093/nar/16.19.9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckham I., Sobel S., Comer J., Jaenisch R., Barklis E. Retrovirus activation in embryonal carcinoma cells by cellular promoters. Genes Dev. 1989 Dec;3(12B):2062–2071. doi: 10.1101/gad.3.12b.2062. [DOI] [PubMed] [Google Scholar]
- Raleigh E. A., Wilson G. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9070–9074. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohdewohld H., Weiher H., Reik W., Jaenisch R., Breindl M. Retrovirus integration and chromatin structure: Moloney murine leukemia proviral integration sites map near DNase I-hypersensitive sites. J Virol. 1987 Feb;61(2):336–343. doi: 10.1128/jvi.61.2.336-343.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffer J. D., Thurston S. J. A negative regulatory element with properties similar to those of enhancers is contained within an Alu sequence. Mol Cell Biol. 1989 Feb;9(2):355–364. doi: 10.1128/mcb.9.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherdin U., Rhodes K., Breindl M. Transcriptionally active genome regions are preferred targets for retrovirus integration. J Virol. 1990 Feb;64(2):907–912. doi: 10.1128/jvi.64.2.907-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih C. C., Stoye J. P., Coffin J. M. Highly preferred targets for retrovirus integration. Cell. 1988 May 20;53(4):531–537. doi: 10.1016/0092-8674(88)90569-7. [DOI] [PubMed] [Google Scholar]
- Varmus H. E., Guntaka R. V., Deng C. T., Bishop J. M. Synthesis, structure and function of avian sarcoma virus-specific DNA in permissive and nonpermissive cells. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):987–996. doi: 10.1101/sqb.1974.039.01.113. [DOI] [PubMed] [Google Scholar]
- Vijaya S., Steffen D. L., Robinson H. L. Acceptor sites for retroviral integrations map near DNase I-hypersensitive sites in chromatin. J Virol. 1986 Nov;60(2):683–692. doi: 10.1128/jvi.60.2.683-692.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]
- Wyke J. A., Akroyd J., Gillespie D. A., Green A. R., Poole C. Proviral position effects: possible probes for genes that suppress transcription. Ciba Found Symp. 1989;142:117–130. doi: 10.1002/9780470513750.ch8. [DOI] [PubMed] [Google Scholar]
- Wyke J. A., Quade K. Infection of rat cells by avian sarcoma virus: factors affecting transformation and subsequent reversion. Virology. 1980 Oct 30;106(2):217–233. doi: 10.1016/0042-6822(80)90246-9. [DOI] [PubMed] [Google Scholar]
- zur Hausen H. Papillomaviruses in human cancer. Cancer. 1987 May 15;59(10):1692–1696. doi: 10.1002/1097-0142(19870515)59:10<1692::aid-cncr2820591003>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]


