Abstract
Lambda phage vectors mediate gene transfer in cultured mammalian cells and in live mice, and in vivo phage-mediated gene expression is increased when mice are pre-immunized with bacteriophage lambda. We now show that, like eukaryotic viruses, bacteriophage vectors are subject to Fc receptor-mediated, antibody-dependent enhancement of infection in mammalian cells. Antibody-dependent enhancement of phage gene transfer required FcγRI, but not its associated γ chain, and was not supported by other FcγR family members (FcγRIIA, FcγRIIB and FcγRIII). Studies using chlorpromazine and latrunculin A revealed an important role for clathrin-mediated endocytosis (chlorpromazine) and actin filaments (latrunculin A) in antibody-enhanced phage gene transfer. This was confirmed by experiments using inhibitors of endosomal acidification (bafilomycin A1, monensin) and by immunocytochemical colocalization of internalized phage particles with early endosome-associated protein-1 (EAA1) . In contrast, microtubule-targeting agents (nocodazole, taxol) increased the efficiency of antibody-enhanced phage gene transfer. These results reveal an unexpected antibody-dependent, FcγRI-mediated enhancement of phage transduction in mammalian cells, and suggest new approaches to improve bacteriophage-mediated gene transfer.
Keywords: Bacteriophage lambda, antibody-dependent enhancement, Fc gamma receptor, gene transfer, virus vector, endocytosis, CD64, FcγRI, microtubule, actin
Introduction
Bacteriophage lambda is capable of transducing mammalian cells (Geier and Merril, 1972; Merril, Geier, and Petricciani, 1971), and bacteriophage lambda vectors that contain a mammalian gene expression cassette can express encoded genes in mammalian target cells, both in vitro (Eguchi et al., 2001; Zanghi et al., 2007) and in vivo (Clark and March, 2004; Lankes et al., 2007; March, Clark, and Jepson, 2004). The ability of lambda phage particles to transduce mammalian cells in vivo depends only partially on phagocytic uptake of phage, and is increased when mice are pre-immunized with bacteriophage lambda (Lankes et al., 2007).
Antibody-dependent enhancement (ADE) of virus infection is a paradoxical phenomenon in which virus specific antibodies fail to completely neutralize virus infectivity, and in stead permit the more efficient infection of susceptible host cells such as monocytes and macrophages (Takada and Kawaoka, 2003). This process can be mediated through cellular receptors specific for the Fc portion of IgG, and has been reported to occur in a wide range of mammalian viruses and virus infections, including dengue virus, HIV-1, influenza virus, measles virus, murine gamma herpesvirus 68, rabies virus and yellow fever virus (among others) (Gotoff et al., 1994; Guillon et al., 2002; Iankov et al., 2006; Littaua, Kurane, and Ennis, 1990; Peiris and Porterfield, 1979; Porterfield, 1981; Rosa et al., 2007; Schlesinger and Brandriss, 1981; Takeda, Sweet, and Ennis, 1990; Takeda, Tuazon, and Ennis, 1988; Tamura, Webster, and Ennis, 1991; Tamura, Webster, and Ennis, 1994; Wallace et al., 2003). ADE has also been reported to occur with mammalian virus vectors, and can lead to enhanced transduction of antigen-presenting cells by neutralized adenovirus-immune complexes, in a Fc receptor-dependent fashion (Leopold et al., 2006; Mercier et al., 2004).
In the present study, we sought to develop an in vitro model for antibody-dependent enhancement of mammalian cells by bacteriophage λ vectors, and to determine the underlying mechanism(s) involved in this process - including the role of cellular Fc gamma receptor (FcγR) receptors. FcγRs are expressed on a wide range of hematopoieitic cells (including B cells, macrophages, monocytes, natural killer cells, and neutrophils), and play an essential role in the recognition and elimination of immune complexes and immunoglobulin G (IgG)-opsonized pathogens (Daeron, 1997; Ravetch and Bolland, 2001). The FcγR family includes both high affinity receptors (such as FcγRI or CD64) and low affinity receptors such as FcγRIIA (CD32), and FcγRIII (CD16) (Ravetch and Bolland, 2001). Binding of immune complexes to these receptors results in their ligation and in the internalization of the bound complexes, either via endocytosis (in the case of small/soluble complexes) or phagocytosis (in the case of IgG-opsonized microorganisms) (Daeron, 1997; Ravetch and Bolland, 2001). FcγR crosslinking also results in the activation of cell signaling and kinase pathways that contribute to phagocytosis and endocytosis, and that mediate the downstream effects of receptor ligation (such as the degranulation, initiation of host inflammatory responses and cytokine production) (Huang et al., 2006; Ravetch and Bolland, 2001.
A number of FcγRs have been shown to contribute to antibody-dependent enhancement of virus infection, including FcγRIA (CD64) and FcγRIIA (CD32) in the case of dengue virus (Rodrigo et al., 2006; Schlesinger and Chapman, 1999), and FcγRIIA (CD32) in the case of adenovirus vectors (Leopold et al., 2006). Our experiments revealed that only FcγRIA, but not other FcγR family members (FcγRIIA, FcγRIIB and FcγRIIIA), was capable of supporting antibody-dependent enhancement of bacteriophage-mediated gene transfer; coexpression of the FcγR associated γ chain was not required for ADE. Finally, studies using pharmacologic inhibitors and immunocytochemical staining techniques revealed that clathrin-mediated endocytosis and actin filaments played a critical role in antibody-enhancement of phage gene transfer, while the microtubule network was found to play an inhibitory role in this process.
Results
Bacteriophage lambda vectors are capable of transducing mammalian cells in vivo, and the efficiency of this process is increased when mice are pre-immunized with lambda phage particles (Lankes et al., 2007). To investigate the underlying basis for this observation, we developed an in vitro model for the antibody-dependent enhancement of lambda phage-mediated gene expression. Our model comprised (i) a CV1 cell line stably transduced with human FcγRI and its associated γ-chain (FcγRI γ+ cells), plus (ii) phage-containing immune complexes that were incubated with these cells. To facilitate the quantitation of phage-mediated gene transfer, we took advantage of an available phage recombinant containing a luciferase-encoding mammalian expression cassette [λ(luc)]. These phage particles were then mixed with a rabbit antiserum directed against gpD, the major coat protein of bacteriophage λ, in order to generate phage-containing immune complexes (Figure 1).
Figure 1. GpD-specific antiserum enhances λ-phage mediated luciferase expression in FcγRI positive CV1 cells.
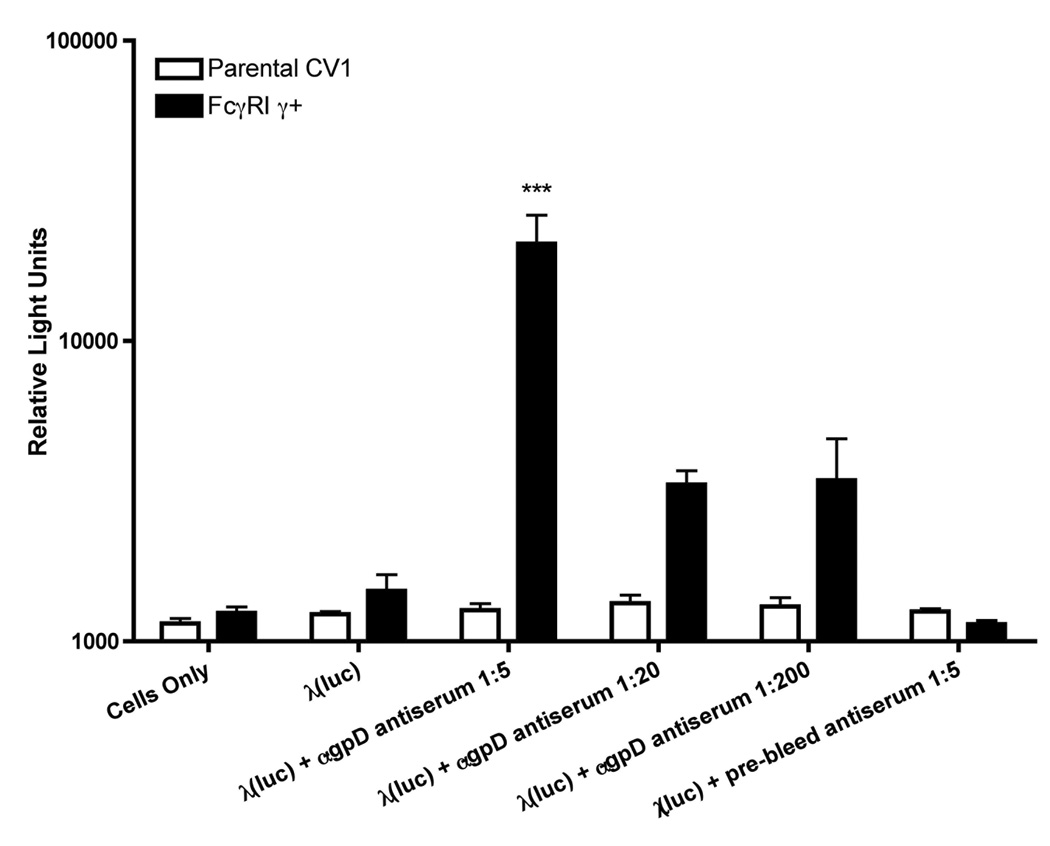
Luciferase encoding λ phage particles (λ luc) were pre-incubated with various dilutions of a rabbit antiserum raised against the major λ coat protein, gpD, or with a non-immune pre-bleed from the same animal. Immune complexes containing phage particles were then added to parental (white bars) or FcγRI γ+ expressing (black bars) CV1 cells, at a multiplicity of infection (MOI) of 106. Forty-eight hours later, cells were harvested, and luciferase activity was measured in cell lysates. The results shown represent mean values calculated from triplicate samples; error bars represent the standard deviation. The data are representative of 3 independent experiments that yielded similar results. The asterisks (***) denote a statistically significant difference from control cells/conditions (p value < 0.001, one-way ANOVA).
The gpD-specific antiserum was found to significantly enhance phage-mediated gene expression in FcγRI γ+ CV1 cells (which have previously been shown to express high levels of FcγRI on their surface; (Rodrigo et al., 2006)), but not in parental CV1 cells (which do not express FcγRI). This enhancement was greatest at a low dilution of the antiserum (corresponding to an estimated molar ratio close to 1 antibody molecule per gpD monomer on the phage capsid), and declined with serial dilution of the antibody (Figure 1). No enhancement was detected when phage was mixed with serum from the same rabbit, obtained prior to immunization with lamdba gpD protein (Figure 1; labeled as “prebleed”). In light of this result, subsequent experiments were performed using phage-containing immune complexes that were prepared using a serum dilution of 1:5.
To confirm that Fc receptor binding was required for the antibody-dependent enhancement of phage-mediated gene transfer in FcγRI γ+ CV1 cells, a follow-up experiment was performed in which the Fc-binding domain of the receptor was saturated with a FcγRI specific monoclonal antibody (10.1) whose binding site overlaps with the Fc-binding site on the receptor. Cultures treated with this blocking antibody showed a reduced level of luciferase reporter gene expression following incubation with phage-containing immune complexes, when compared with non-treated FcγRI γ+ CV1 cells or cells that were treated with an irrelevant, isotype-matched control antibody (Figure 2).
Figure 2. Antibody-dependent enhancement of λ-phage mediated gene expression in FcγRI-positive CV1 cells requires Fc receptor availability.

Luciferase encoding λ phage particles either added directly to target cells (λ luc), or were preincubated with a gpD-specific rabbit antiserum (λ luc + gpD) prior to addition to target cells at a MOI of 106. In some cases, the parental (white bars) or FcγRI γ+ expressing (black bars) CV1 cells were preincubated with 5 µg/ml of a FcγRI ligand-binding blocking antibody, 10.1 (αFcγRI), or IgG1 isotype-matched control antibody (IgG1 isotype) for 30 minutes prior to the addition of λ(luc) or λ(luc) + αgpD. Forty-eight hours later, cells were harvested, and luciferase activity was measured in cell lysates. The results shown represent mean values calculated from triplicate samples; error bars represent the standard deviation. The data are representative of 3 independent experiments that yielded similar results. The asterisks (***) denote a statistically significant difference from control cells/conditions (p value < 0.001, one-way ANOVA).
This result demonstrates that antibody-dependent enhancement of phage-mediated gene transfer required the presence of a Fcγ receptor on the surface of the target cells. Thus, the enhancement of gene transfer by phage-immune complexes cannot be attributed to the formation of a dense DNA-phage precipitate that is more efficiently deposited on cell surfaces by centrifugation, or non-specifically internalized by target cells.
We next wished to know whether Fcγ receptors other than FcγRI might also be capable to permitting antibody-dependent enhancement of phage-mediated gene transfer. In particular, although FcγRI binds IgG isotypes with nanomolar affinity, Fc-binding is not restricted to this receptor, and other receptors such as FcγRIIA have been implicated in antibody-dependent enhancement of susceptible cells by dengue virus (Rodrigo et al., 2006), and by adenovirus vectors (Leopold et al., 2006). Experiments were therefore performed to assess the ability of other FcγR family members to support antibody-dependent enhancement of phage-mediated gene transfer. To do this, COS-7 cells were transiently transfected with a panel of mammalian expression plasmids encoding a series of FcγR family members, in the presence or absence of the receptor-associated γ-chain. Surface expression of FcγRs on transiently transfected COS-7 cells was verified by flow cytometric analysis (Figure 3A); γ-chain expression was confirmed by immunoblot analysis of cell lysates using a γ-chain specific antibody (Figure 3B). Stably-transfected CV1 cells expressing FcγRI and the γ-chain subunit were used as a positive control for γ-chain expression (Figure 3B, lane 2). In the case of the γ chain-associated receptors, FcγRI and FcγRIII, cell surface expression in the transiently transfected COS-7 cells was detected even in the absence of a co-expressed γ-chain cDNA (Figure 3A). This is consistent with previous data showing that FcγRI can be expressed on the cell surface without an absolute requirement for the common γ chain (van Vugt et al., 1999).
Figure 3. FcγRs and associated γ chains are expressed in transiently transfected COS-7 cells.

Mammalian expression plasmid vectors encoding cDNAs corresponding to FcγRI, FcγRIIA, FcγRIIB, FcγRIII, or the common γ-chain subunit were transiently transfected into COS-7 cells using Lipofectamine-2000. Cells were collected 48-hours later and subjected to flow cytometric analysis for cell surface FcγR expression (A) or immunoblot analysis for γ chain expression (B). (A) Flow histograms are presented for COS-7 cells that were transiently transfected with expression plasmids encoding the indicated FcγR family members (CD16, CD32A, CD32B and CD64); in some cases, cells were also co-transfected with a plasmid encoding the common γ chain (CD16γ; CD64γ). In each panel, the shaded histogram represents receptor expression on non-transfected COS-7 cells, the dashed line denotes control staining with an irrelevant isotype-matched antibody, and the open histogram bounded by the thick solid line represents surface staining for the respective FcγR family member. (B) An immunoblot analysis of common γ chain expression in the transfected COS-7 cells is shown. Briefly, lysates were prepared from γ chain transfected cells, and equal quantities of cell protein were then electrophoretically separated under reducing conditions, on a 15% SDS polyacrylamide gel. Proteins were then transferred to nitrocellulose, and the membrane was probed with a γ chain-specific antibody. Lane 1 represents molecular weight markers (numbers denote the size of selected marker proteins in kilodaltons), while lanes 2–4 show γ-chain expression in lysates from various transfected cells. Shown are analyses of lysates from: the stably transfected FcγRI γ+ CV1 cell line (lane 2); COS-7 cells transiently transfected with the CD64 expression plasmid plus the γ chain expression plasmid (lane 3); or COS-7 cells transiently transfected with the CD16 expression plasmid plus the γ chain expression plasmid (lane 4). The expected molecular weight of the common γ-chain is ~10kD.
Having validated that the desired FcγRs were expressed on the surface of the transiently transfected COS-7 cells, and that cytoplasmic receptor-associated γ chain was also expressed, we performed gene transfer experiments using phage-containing immune complexes. As shown in Figure 4, the only Fc receptor that supported antibody-dependent enhancement of phage-mediated gene transfer was FcγRI. Expression of the receptor-associated γ-chain was not required for this effect. In addition, the other FcγR family members that were studied (FcγRIIA, FcγRIIB and FcγRIII [+/- the γ-chain]) all failed to support antibody-dependent enhancement of phage-mediated gene transfer (Figure 4).
Figure 4. Antibody-dependent enhancement of λ-phage mediated gene expression requires FcγRI, but not γ chain expression, and is not supported by other FcγR family members.
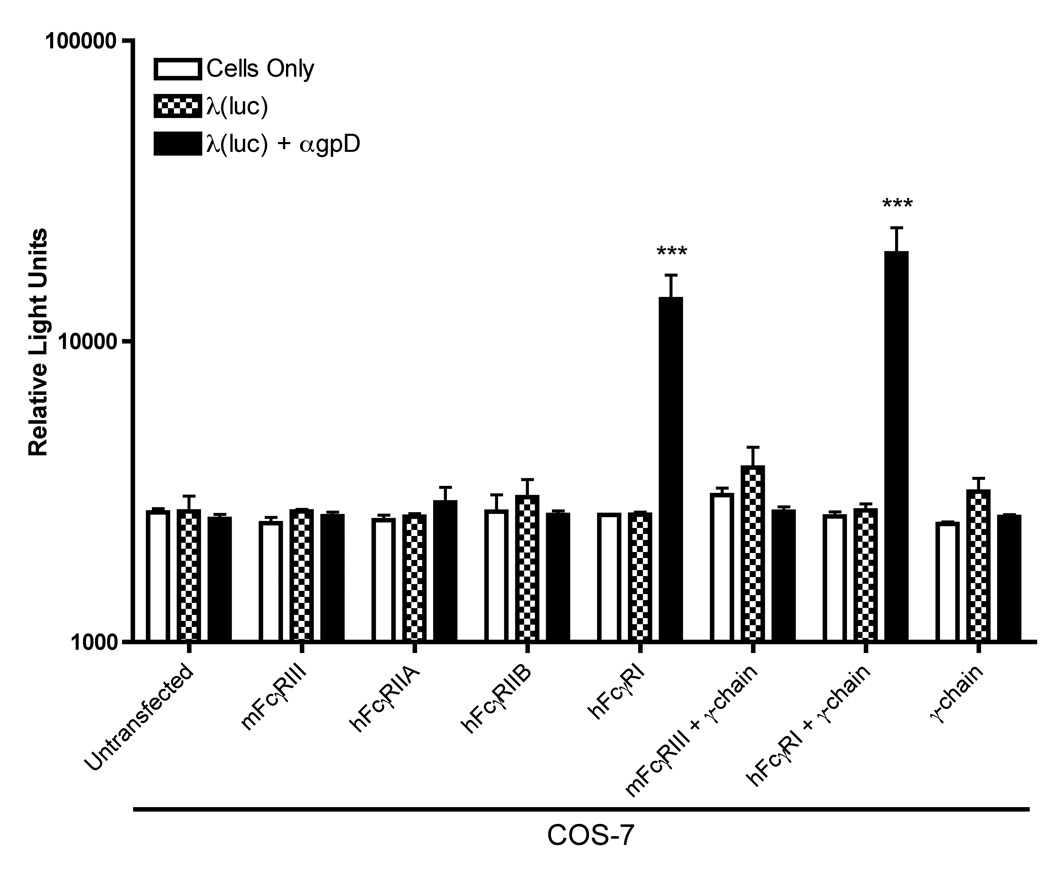
Luciferase encoding λ phage particles either added directly to target cells (λ luc), or were preincubated with a gpD-specific rabbit antiserum (λ luc + gpD) prior to addition to target cells at a MOI of 106. Phage were then added to COS-7 cells that had been transiently transfected with mammalian expression plasmids encoding the indicated FcγR family members (either alone, or in combination with the common γ chain). Forty-eight hours after the addition of phage, cells were harvested, and luciferase activity was measured in cell lysates. The data represent mean values calculated from triplicate samples; error bars represent the standard deviation. The data are representative of 3 independent experiments that yielded similar results. The asterisks (***) denote a statistically significant difference from control cells/conditions (p value < 0.001, one-way ANOVA).
To better understand the mechanisms involved in FcγRI-mediated, antibody-dependent enhancement of phage infectivity, we employed a panel of pharmacologic agents to dissect the intracellular mechanisms required for phage-mediated gene transfer. Drugs known to target clathrin-mediated endocytosis (chlorpromazine), actin polymerization (latrunculin A, cytochalasin D) and microtubules (nocodazole, taxol) were used in these experiments.
Chlorpromazine, an inhibitor of clathrin-coated pit formation at the plasma membrane, significantly decreased phage-mediated gene expression (Figure 5A), as did the actin polymerization inhibitors latrunculin A (Fig. 5A) and cytochalasin D (data not shown). In contrast, agents that targeted the microtubule network strongly augmented gene expression by phage-immune complexes. Both nocodazole (a microtubule-depolymerizing agent) and taxol (paclitaxel; a microtubule-stabilizing agent) augmented luciferase expression by roughly 10 to 50 fold, in comparison to non-treated cells (Figure 5B). These results suggest that the microtubule network plays an unexpected, inhibitory role in antibody-dependent enhancement of phage-mediated gene transfer.
Figure 5. Antibody-dependent enhancement of λ-phage mediated gene expression in FcγRI-positive cells is inhibited by chlorpromazine and latrunculin A, but augmented by nocodazole and taxol.

Luciferase encoding λ phage particles were preincubated with a gpD-specific rabbit antiserum (λ luc + gpD) prior to addition to target cells at a MOI of 106. Phage were then added to parental CV1 cells (open bars) or to CV1 cells stably expressing FcγRI together with the accessory γ-chain subunit (black bars; FcγR1 γ+). Cells were pretreated with (A) latrunculin A (2.5 µM), or chlorpromazine (5 µg/ml), or (B) nocodazole (5 µM) or taxol (1 µg/ml and 20 µg/ml), for 30 minutes at 37°C before the addition of bacteriophage. As controls, some cells were preincubated in the absence of any exogenous drug, or with DMSO alone. Forty-eight hours after the addition of phage, cells were harvested, and luciferase activity was measured in cell lysates. The data represent mean values calculated from triplicate samples; error bars represent the standard deviation. The data are representative of 3 independent experiments that yielded similar results. The asterisks (**, ***) denote a statistically significant difference from control cells/conditions (p value < 0.05[**] or p value < 0.001[***], one-way ANOVA).
The inhibitory effect of chlorpromazine on gene expression by phage-immune complexes (Fig. 5A), suggested that these complexes may enter cells via an endocytic pathway. We therefore conducted two sets of followup experiments to confirm this.
First, we performed transduction experiments in the presence of well-characterized pharmacologic agents that inhibit endosomal acidification via different mechanisms. To do this, we used bafilomycin A, which is an inhibitor of the vacuolar H+-ATPase (Bowman, Siebers, and Altendorf, 1988), and monensin, which is a proton ionophore (Berg et al., 1983; Harford et al., 1983; Mollenhauer, Morre, and Rowe, 1990). Both agents strongly inhibited gene expression by phage immune complexes (Fig. 6), without exerting overt cytotoxic effects (Table 1).
Figure 6. Antibody-dependent enhancement of λ-phage mediated gene expression in FcγRI-positive cells is prevented by inhibitors of endosomal acidification.

Luciferase encoding λ phage particles were preincubated with a gpD-specific rabbit antiserum (λ luc + gpD) prior to addition to target cells at a MOI of 106. Phage were then added to parental CV1 cells (open bars) or to CV1 cells stably expressing FcγRI together with the accessory γ-chain subunit (black bars; FcγR1 γ+). Cells were pretreated with medium alone (no drug), bafilomycin A1 (20 nM and 100 nM) or monensin (20 µM), for 30 minutes at 37°C before the addition of bacteriophage. Forty-eight hours after the addition of phage, cells were harvested, and luciferase activity was measured in cell lysates. The data represent mean values calculated from triplicate samples; error bars represent the standard deviation. The asterisks (***) denote a statistically significant difference from control cells (no drug) (p value < 0.001, one-way ANOVA).
Table 1.
Cell viability is unaffected by pharmacologic agents that modulate antibody-dependent enhancement of λ-phage mediated gene expression
| Drug | Parental CV1 | FcγRI γ+ |
|---|---|---|
| No treatment | 2.1 ±0.9 | 1.5 ±1.0 |
| Latrunculin A | 3.1 ±2.3 | 2.9 ±1.5 |
| Chlorpromazine | 3.2 ±1.4 | 2.7 ±1.0 |
| Chloroquine | 3.6 ±0.6 | 3.3 ±0.6 |
| Nocodazole | 5.0 ±1.0 | 2.6 ±0.6 |
| Taxol 1µg/ml | 2.0 ±0.2 | 1.6 ±0.6 |
| Taxol 20µg/ml | 3.7 ±0.5 | 3.1 ±0.5 |
| DMSO | 2.2 ±0.1 | 1.6 ±0.7 |
| Bafilomycin A1 (20 nM)* | 2.8 ±0.3 | 2.9 ±0.1 |
| Bafilomycin A1 (100 nM)* | 3.2 ±0.4 | 3.1 ±0.5 |
| Monensin* | 4.1 ±0.4 | 4.4 ±0.6 |
LEGEND. Parental CV1 and FcγRI γ+ cells were treated with the indicated drugs for 48 hours. Drug concentrations were as follows: latrunculin A (2.5 µM), chlorpromazine (5 µg/ml), chloroquine (50 µg/ml), nocodazole (5 µM), taxol (1 µg/ml and 20 µg/ml), bafilomycin A (20 nM and 100 nM) and monensin (20 µM). Cell viability was then measured by propidium iodide (PI) staining, followed by flow cytometric analysis; data shown represent the mean percentage of PI-positive cell, as calculated from three independent experiments (± standard deviation).
Toxicity of these compounds was assessed separately from that of the other compounds. In that separate analysis, PI staining of untreated cells was 1.9 ± 0.2% and 2.1 ± 0.5% for parental CV1 and FcγRI γ+ cells, respectively.
Second, we conducted immunocytochemical staining experiments to determine whether internalized phage particles were colocalized with endosomal marker proteins. To do this, cells were transduced with phage-immune complexes for 10 minutes, and then fixed, permeabilized and stained with antibodies specific for the major phage coat protein (gpD) or for the early endosome associated protein (EEA1) (Mu et al., 1995; Simonsen et al., 1998). The results of this analysis, shown in Fig. 7, reveal colocalization of phage and EEA1 (see yellow punctate dots in the "composite" panel). Thus, both sets of followup experiments confirmed that phage-immune complexes enter FcγRI-bearing cells via an endocytic pathway.
Figure 7. Internalized λ-phage immune complexes colocalize with EEA1 in FcγRI-positive cells.

Luciferase encoding λ phage particles were preincubated with a gpD-specific rabbit antiserum (λ luc + gpD) prior to addition to CV1 cells stably expressing FcγRI together with the accessory γ-chain subunit (FcγR1 γ+) at a MOI of 106. Cultures were incubated with phage-immune complexes at 4°C for one hour to allow binding, and then internalization was permitted by a brief (10 min) incubation at 37°C. Cells were then fixed, permeabilized and subjected to immunocytochemical staining. To do this, they were reacted with a polyclonal chicken antiserum directed against early endosome associated (EEA1) protein (Zymed Laboratories) and a polyclonal rabbit antiserum directed against the major phage capsid protein, gpD. Bound antibodies were then detected using an Alexa488 -conjugated goat anti-chicken IgG (Molecular Probes) and a Texas Red-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch). After washing, bound antibodies were visualized using a Nikon C1 visible light scanning confocal microscope. All images were taken using a 60x/1.4NA objective. A volume-rendered three-dimensional reconstruction of a series of optical sections taken at 0.3 µm steps in the Z dimension was calculated by the Nikon EZ-C1 analysis software. A representative 0.3 µm Z-stack confocal image, taken through the plane of the cell, is shown. The left panel shows immunostaining for EEA1 (green), the center panel shows staining for phage (red) and the right panel shows a composite image of the two. Note the abundant yellow staining, indicating colocalization of phage and EEA1.
Finally, we performed experiments to assess the efficiency of gene expression by phage-immune complexes. To do this, we took advantage of an available λ lysogen that contains a phage genome bearing a mammalian expression cassette encoding the enhanced GFP reporter gene [λ(GFP)] (Eguchi et al., 2001). λ(GFP) phage were produced and phage-immune complexes were used to transduce FcγRI-bearing CV1 cells. The results, shown in Figure 8A, reveal that about 1.5% of the cell population expressed GFP following exposure to λ(GFP) phage-immune complexes.
Figure 8. Efficiency of gene transfer by λ-phage immune complexes.
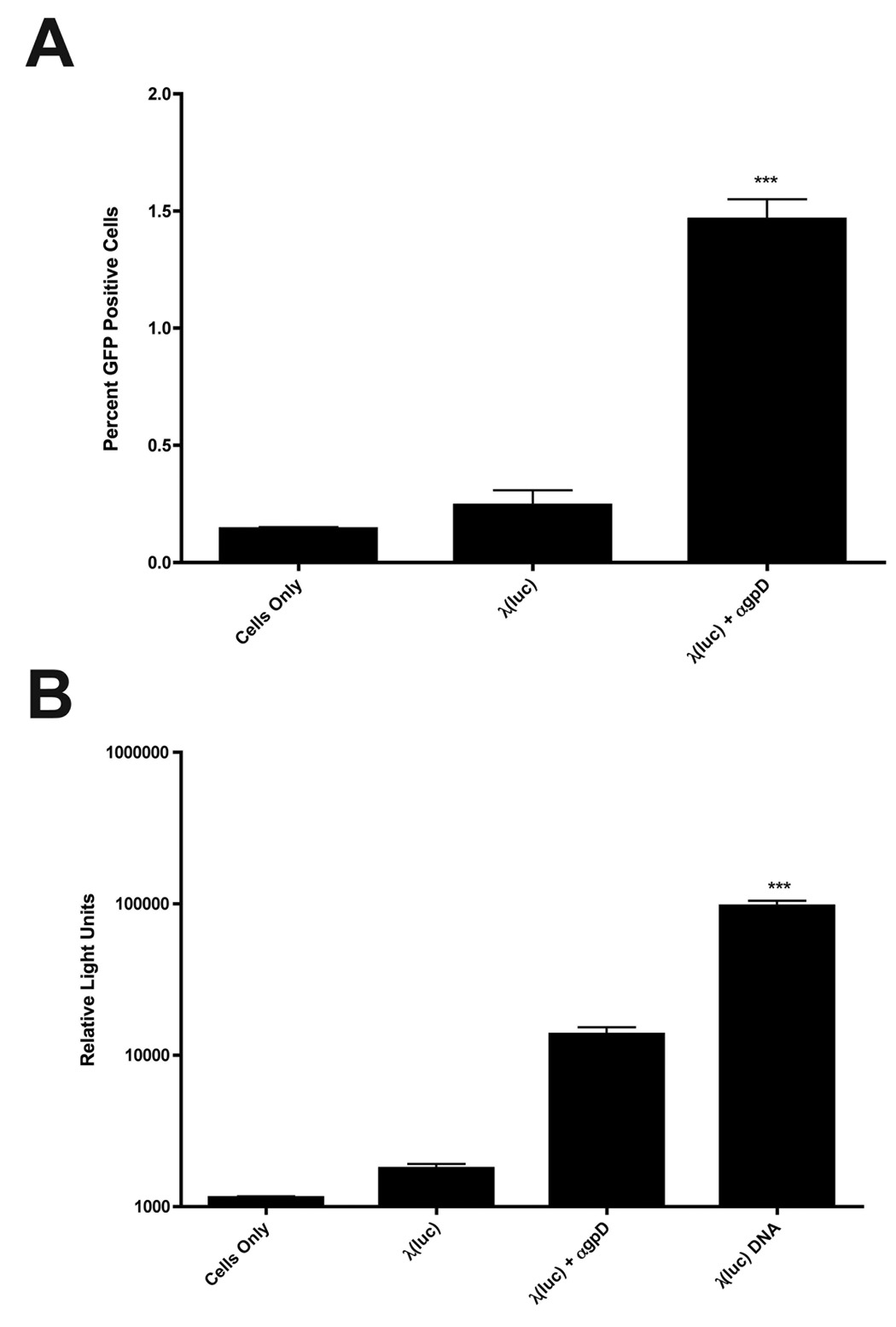
(A) CV1 cells stably expressing FcγRI and its associated • chain (FcγR1 γ+) were mixed with λ(GFP) particles or λ(GFP) immune complexes at a MOI of 106. 48 hours later, cells were harvested and the number of cells expressing GFP was quantitated by flow cytometry. (B) CV1 FcγR1 γ+ cells were mixed with λ(luc) particles or λ(luc) immune complexes at a MOI of 106, as described above. In parallel, CV1 FcγR1 γ+ cells were subjected to transient transfection with 500 ng of purified λ(luc) DNA using Lipofectamine2000. (A, B) The data represent mean values calculated from triplicate samples; error bars represent the standard deviation. The asterisks (***) denote a statistically significant difference from all other conditions (p value<0.001, one-way ANOVA).
In order to directly compare the efficiency of gene transfer by phage immune complexes to a standard DNA transfection methodology, we also performed an experiment in which FcγRI-bearing CV1 cells were either exposed to λ(luc) phage-immune complexes or transiently transfected with purified λ(luc) DNA using a cationic lipid (Lipofectamine2000). This experiment was conducted in 96-well plates, using our standard phage MOI (106) – thus, a total of 1010 phage particles were added into each well. To make the most direct possible comparison with phage-mediated gene transfer, cells were transiently transfected with an amount of purified λ(luc) DNA (500 ng) that corresponds to the DNA content of 1010 phage particles (Lankes et al., 2007). Measurement of luciferase activity in lysates of transduced or transfected cells revealed that the efficiency of gene transfer by phage-immune complexes was within an order of magnitude of that of lipofectamine-mediated DNA transfection (Fig. 8B).
Discussion
This study was designed to understand why prior immunization with lambda phage particles results in more efficient lambda phage-mediated gene expression in live mice (Lankes et al., 2007). We developed an in vitro model system to study this phenomenon, and demonstrated that antibody-dependent enhancement of phage can occur in susceptible mammalian cells, via a Fc receptor-mediated process.
In our experiments, expression of FcγRI alone – in the absence of its associated γ chain – was sufficient to render cells susceptible to enhanced transduction by phage-immune complexes. This suggests that gamma chain mediated signal transduction is not necessary for enhancement of gene transfer by phage-immune complexes. Prior studies have shown that cells which express FcγRI in the absence of the γ chain, or FcγRIIA with mutations in its cytoplasmic signaling domain, are unable to phagocytize large particles (Davis et al., 1995; Indik et al., 1991; Lowry et al., 1998; Odin et al., 1991; Rodrigo et al., 2006; Van den Herik-Oudijk et al., 1995), although they retain the ability to take up smaller immune complexes by endocytosis (Davis et al., 1995; Odin et al., 1991). Thus, the fact that the γ chain was not required for FcγRI-mediated enhancement of the infectivity of phage-containing immune complexes suggests that FcγRI-mediated phage uptake occurs via an endocytic mechanism, and not via phagocytosis.
This conclusion is supported by the fact that an inhibitor of clathrin-mediated endocytosis (chlorpromazine) blocked antibody-dependent enhancement of phage-mediated gene transfer. Our results with latrunculin A, nocodazole and taxol are also consistent with endocytic uptake of phage-immune complexes. Actin filaments, but not microtubules, are involved in clathrin-dependent endocytosis of ligands bound to the cell surface (Durrbach, Louvard, and Coudrier, 1996). In keeping with this, only latrunculin A (which affects actin polymerization) inhibited gene transfer by phage-immune complexes; in contrast, nocodazole or taxol (which target microtubules) augmented the gene transfer process.
Involvement of an endocytic pathway in uptake of phage-immune complexes was confirmed by additional studies using inhibitors of endosomal acidification (bafilomycin A and monensin), both of which inhibited antibody-dependent enhancement of phage-mediated gene transfer, and by direct observation of phage internalization using immunocytochemical staining. The latter studies revealed that, at 10 minutes following FcγR1-mediated internalization, phage particles were found be colocalized with the early endosome associated protein, EAA1.
The effects of nocodazole and taxol merit additional discussion. Both of these agents have been shown to enhance gene transfection in COS-7 cells mediated by cationic liposomes (Hasegawa, Hirashima, and Nakanishi, 2001). In addition, nocodazole and other microtubule-depolymerizing agents have been shown to enhance gene expression following transfection of vascular smooth muscle cells with cationic lipid-DNA complexes (Wang and MacDonald, 2004). Two mechanisms have been proposed to explain these results: (i) microtubules are required to transport liposome-DNA complexes to lysosomes in which DNA is degraded and (ii) the depolymerization of microtubules may lead to activation of NFκB and increased levels of gene transcription (Hasegawa, Hirashima, and Nakanishi, 2001; Wang and MacDonald, 2004).
Our results with taxol and nocodazole can best be explained by a role of microtubules in transporting internalized phage particles to a lysosomal compartment in which phage are degraded. This is because we obtained similar results with both taxol (a microtubule-stabilizing agent) and nocodazole (a microtubule-depolymerizing agent). Such a finding is inconsistent with a transcriptional effect on gene expression, since taxol is known to oppose the positive effects of microtubule-depolymerizing agents on NFκB activation (Rosette and Karin, 1995). It is, however, in good agreement with findings reported by Nakanishi and colleagues, who showed that both taxol and nocodazole interfered with microtubule-mediated transport of liposome-DNA complexes to lysosomes (Hasegawa, Hirashima, and Nakanishi, 2001).
The fact that phage-immune complex internalization proceeded via endocytosis rather than phagocytosis is consistent with recent studies on the Fc receptor-dependent uptake of adenovirus-immune complexes (Leopold et al., 2006). In contrast, dengue virus immune complexes have been shown to be internalized by phagocytosis as well (Rodrigo et al., 2006). The basis for this discordance is uncertain, but may reflect differences in the sizes and infectivities of the virus immune complexes (Almeida and Waterson, 1969; Parren and Burton, 2001).
More unexpected was the observation that phage-immune complexes led to efficient gene transduction in cells expressing FcγRI, but not in cells expressing FcγRIIA. FcγRIIA has been reported to support highly efficient uptake of dengue virus-immune complexes (Rodrigo et al., 2006), and one might therefore have expected it to also support the internalization of phage-immune complexes. Possible reasons for its failure to do so may again include differences in the sizes or infectivities of phage-immune complexes versus dengue virus-immune complexes. Whatever the underlying reason, this mechanistic difference in internalization of phage-versus dengue virus-immune complexes is consistent with the other fundamental differences in the uptake of these immune complexes (i.e., the fact that phage-immune complexes are internalized by an endocytic pathway, while dengue virus-immune complexes can enter cells by phagocytosis (Rodrigo et al., 2006)).
Figure 9 presents our working model for antibody-dependent enhancement of phage-mediated gene transfer. Our data are consistent with binding of phage-immune complexes to FcγRI, followed by clathrin-dependent endocytosis. The microtubule network then appears to play a role in transporting the internalizing phage particles to late endosomes or lysosomes, in which phage is degraded. This is consistent with the role of microtubules in transfering vesicular cargo from early endosomes to late endosomes (Gruenberg, Griffiths, and Howell, 1989). Some phage particles escape transport to this degradative compartment, and undergo uncoating and nuclear import – leading to expression of phage-encoded genes within the transduced mammalian target cell. Since gene expression is enhanced by 10 to 50 fold by disruption of the microtubule network, it appears likely that most of the internalized phage-immune complexes are ultimately destined for degradation, under normal circumstances.
Figure 9. Schematic model for antibody-dependent enhancement of λ-phage mediated luciferase expression in FcγRI-positive cells.
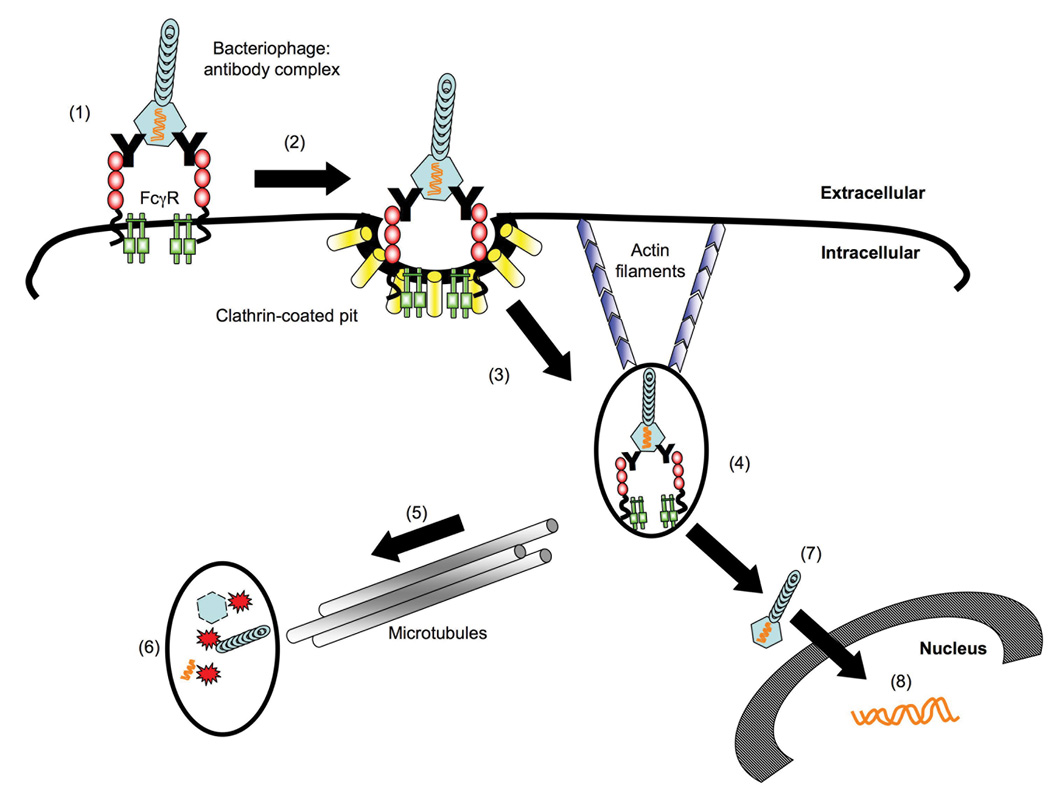
Bacteriophage λ:IgG complex binds and cross-links FcγRI (1), leading to localization of the bound complex to clathrin-coated pits (2). Clathrin-mediated endocytosis of phage-containing immune complexes then occurs, in an actin-dependent fashion (3); drugs that block clathrin-mediated endocytosis (chlorpromazine) or actin polymerization (latrunculin A) therefore inhibit phage-mediated gene transfer. The receptor-associated γ chain (shown in green in (1)) is not required for receptor-mediated internalization of phage-immune complexes. Bacteriophage λ is then trafficked to an early endosome/phagosome (4). Phagosome maturation occurs via the transport of early vesicles along microtubules (5) to late compartments containing degradative enzymes that break down the phage in late endosomes/phagolysosomes (6); drugs that target microtubules (nocodazole, taxol) therefore augment bacteriophage-mediated transduction. Some partially or fully uncoated phage particles/genomes are presumed to escape the endosome via an undefined mechanism (7), leading to nuclear import of phage DNA, and subsequent expression of phage-encoded genes (i.e., luciferase) (8).
The ability of phage-immune complexes to efficiently transduce FcγR-bearing mammalian cells has important implications for phage-mediated gene transfer (Eguchi et al., 2001; Hajitou et al., 2006; Ivanenkov, Felici, and Menon, 1999; Kassner et al., 1999; Larocca et al., 1999; Sathaliyawala et al., 2006; Zanghi et al., 2007), and for applications in which phage vectors are being used to elicit immune responses to encoded or displayed antigens (Chen et al., 2001; Clark and March, 2004; di Marzo Veronese et al., 1994; Irving, Pan, and Scott, 2001; Jepson and March, 2004; Minenkova et al., 1993; Sathaliyawala et al., 2006). For example, in vaccine applications, it may be beneficial to generate phage-immune complexes in vitro with the goal of targeting vector to FcγR-expressing antigen presenting cells (APC) in vivo. Similarly, it may be possible to generate high-titer anti-phage antibodies in vivo, and to then use these antibodies to facilitate vector targeting to FcγR-positive APC.
In conclusion, the results reported here show that, like mammalian viruses, bacteriophage vectors are subject to Fc receptor-mediated, antibody-dependent enhancement of infection. Our findings also show this process can be used to increase the efficiency of phage-mediated gene transfer to FcγR-positive mammalian cells, and provide new insights into the transduction of mammalian cells by bacteriophage vectors.
Materials and methods
Preparation of bacteriophage lambda lysogens and vector production
The λD1180(luc) lysogen (Dam15 del EcoRI-SacI cIts857 nin5 Sam100) was a kind gift from Dr. Mahito Nakanishi (Eguchi et al., 2001). This lysogen contains a firefly luciferase expression cassette under the transcriptional control of the cytomegalovirus immediate early promoter. Bacteriophage λD1180(luc) lysogens were prepared in Top10 (Invitrogen) Escherichia coli (E. coli) hosts and transformed with a wild-type gpD-containing plasmid, pTrc-gpD (Zanghi et al., 2005), to produce luciferase-expressing bacteriophage lambda [λ(luc)]. Induction of the lysogen and purification of the λ(luc) particles was performed as described (Zanghi et al., 2005). λ(luc) titers were measured by plaque forming units (PFU) on LE392 E. coli cells. In some experiments, λ(GFP) phage was used. In this case, phage was prepared from a λD1180(GFP) lysogen containing an enhanced GFP expression cassette under the transcriptional control of the cytomegalovirus immediate early promoter (Eguchi et al., 2001).
Polyclonal antiserum to gpD
Preparation of a polyclonal antiserum to the major coat protein of bacteriophage λ has been described (Zanghi et al., 2005). Briefly, purified gpD protein produced in a bacterial expression system was injected into rabbits (Sigma Genosys), and reactivity verified by immunoblot analysis against recombinant gpD as well as purified bacteriophage λ particles.
Cells lines, plasmids, and transfection reagents
CV-1 and COS-7 simian-derived kidney cells were obtained from ATCC and Sigma-Aldrich, respectively. A bi-cistronic expression plasmid encoding human FcγRI and γ-chain subunit (FcγRI γ+, clone N’x15) has been described (Rodrigo et al., 2006); preparation of the CV-1 cells stably transfected with this plasmid and their interaction with dengue virus immune complexes will be reported elsewhere (Rodrigo et al.). All cells lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum, and 100 U/ml penicillin, 100 µg/ml streptomycin, and 2mM L-glutamine. FcγRIγ+ cell cultures were maintained in culture media containing 100 µg/ml of hygromycin. Transient transfections were performed in 12-well tissue culture plates following the manufacturer’s protocol (Lipofectamine2000; Invitrogen, Carlsbad, CA). pORF9 expression vectors containing the human FcγRIA, human FcγRIIA, human FcγRIIB and murine FcγRIII open reading frames were purchased from InvivoGen (San Diego, CA). Three µg of plasmid DNA, alone or in combination with a γ chain expression plasmid (Rodrigo et al., 2006) (as indicated in the relevant Figure legends) was then mixed with transfection reagent and added to COS-7 monolayers. Forty-eight hours post-transfection, cells were harvested and then used either (i) to perform flow cytometric or immunoblot assays (for verification of FcγR and γ chain expression), or (ii) to conduct gene transduction studies using λ(luc) alone or phage-immune complexes.
Analysis of FcγR andγ-subunit expression in transfected COS-7 cells
Cell surface expression of FcγRs was analyzed by flow cytometry. FITC-conjugated monoclonal antibodies specific for human FcγRI (10.1), human FcλRIIA and FcγRIIB (3D3) or murine FcγRIII (2.4G2) were used, along with corresponding isotype-matched control antibodies (mouse IgG1 κ); all antibodies were purchased from BD Pharmingen (San Jose, CA). A minimum of 10,000 live/gated events were collected for each stained sample. Expression of the γ-chain subunit was verified by immunoblot using a rabbit polyclonal IgG antibody raised against the γ-subunit of FcεRI (Upstate Cell Signaling Solutions; Lake Placid, NY), followed by a horseradish peroxidase-linked, donkey α-rabbit Ig whole antibody (GE Healthcare Life Sciences).
Transduction of cells with λ(luc) or λ(luc)-containing immune complexes, and quantitation of luciferase activity in lysates from transduced cells
Cell lines transiently or stably expressing FcγRs (either alone or in combination with the receptor-associated γ-chain) were seeded at 1 × 104 cells/well in a flat-bottom 96-well tissue culture plate. Non-transfected CV1 and COS-7 cells were used as negative controls. After incubation overnight, culture medium was removed and replaced with serum-free medium 30 minutes prior to the addition of λ(luc) or λ(luc)-containing immune complexes. To generate λ(luc)-containing immune complexes, λ(luc) particles were mixed with specified dilutions of the gpD-specific rabbit antiserum (usually 1:5), incubated for 5 minutes at 37°C, and then added to tissue culture wells in a total of 50 µl infection volume.
Transduction of target cells was enhanced using centrifugal enhancement (“spinoculation”) (Forestell et al., 1996; Hudson, Misra, and Mosmann, 1976; O'Doherty, Swiggard, and Malim, 2000). Briefly, cell cultures were centrified at 1200 RPM (approx. 1000 g) for 10 minutes at 37°C, after addition of phage-immune complexes. Following centrifugation, cells were incubated for an additional 10 minutes at 37°C prior to the addition of 200 µl of serum-containing media, and incubated for 48 hours at 37°C. To measure luciferase expression, cells were washed with phosphate buffered saline (PBS) and lysed in passive lysis buffer (PLB) as instructed by the manufacturer (Promega). Protein concentration in the resulting cell lysates was determined by Bradford assay, and equal protein quantities were used in luciferase assays. Results for luciferase assays were reported in relative light units.
In Fc-receptor blocking studies, FcγRI γ+ cells were incubated with an α-FcγRI antibody (10.1) or an isotype-matched control antibody for 30 minutes on ice before the addition of phage-immune complexes; transduction proceeded as described above. In some experiments, pharmacologic agents were pre-incubated with the cells 30 minutes prior to the addition of λ(luc) or λ(luc)-immune complexes, and remained in the medium for the entire culture period. The drug concentrations used in the experiments were titrated on cells and optimal concentrations were selected, based on doses that did not induce cytotoxicity (Table 1).
Transduction of cells with λ(GFP) or λ(GFP)-containing immune complexes, and quantitation of GFP expression in transduced cells
In some experiments, cells were transduced with GFP-encoding phage [λ(GFP)]. Transduction was performed using the same methods described for λ(luc), and protein expression was quantitated 48 hours later, by flow cytometry using a FACScalibur (Becton-Dickinson).
Transient transfection of cells with purified λ(luc) DNA
In the experiment shown in Fig 8, 104 cells were seeded in 96-well plates and either transduced with phage-immune complexes at an MOI of 106, or transiently transfected with purified genomic DNA prepared from λ(luc) particles, using Lipofectamine2000 (Invitrogen). For transient tranfections, 500 ng of purified λ(luc) DNA was mixed with Lipofectamine2000 and incubated for 20 minutes prior to overlaying on CV1 monolayers (note that this amount of DNA is equivalent to that contained in 1010 phage particles; (Lankes et al., 2007)). Medium was removed 4 hours later and replaced with fresh serum-supplemented DMEM. Forty-eight hours post-transfection, cells were harvested and luciferase activity was assessed in cell lysates.
Immunocytochemical staining of cells transduced with λ(luc)-containing immune complexes
CV1 cells stably expressing Fc•RI together with the accessory γ-chain subunit (Fc•R1 γ+) were grown on glass cover slips. Cover slips were equilibrated on ice for 60 minutes prior to exposure to λ(luc)-containing immune complexes at a MOI of 106. Cultures were incubated on ice for an additional 60 minutes with phage-immune complexes prior to transferring cover slips to 37°C for 10 minutes. Cover slips were then washed with ice-cold PBS and fixed with 4% paraformaldehyde. Cells were next permeabilized with 0.1% Triton X-100 and reacted with a polyclonal chicken antiserum directed against early endosome associated (EEA1) protein (Zymed Laboratories, 1:100 dilution) and a polyclonal rabbit antiserum directed against the major phage capsid protein, gpD (1:100) for 60 minutes at room temperature. After 3 washes with PBS, performed at room temperature for 5 minutes, the bound primary antibodies were detected using an Alexa488-conjugated goat anti-chicken IgG (Molecular Probes, 1:100 dilution) and a Texas Red-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch, 1:100 dilution). After washing (as above), bound antibodies were visualized using a Nikon C1 visible light scanning confocal microscope. A volume-rendered three-dimensional reconstruction of a series of optical sections taken at 0.3 µm steps in the Z dimension was then calculated by the Nikon EZ-C1 analysis software, and recorded.
Data analysis and biostatistics
All figures show results that are representative of at least three independent experiments. Data values were calculated from experimental groups performed in triplicate, and the data are presented as the mean ± the standard deviation. Data were analyzed by one-way ANOVA using Prism 4 software (GraphPad, Inc), followed by Tukey’s Multiple Comparison post-test. A p value of less than 0.05 (p<0.05) was considered to be statistically significant.
Acknowledgments
The authors would like to thank Dr. Mahito Nakanishi and DNAVEC Corporation for providing λ phage vectors [λD1180 (Luc)], Drs. David Yule, Brian Ward and Tim Bushnell for assistance with microscopy, and Drs. John Frelinger and Heather Lankes for helpful discussions. This work was supported by NIH grants R01 DE14914 (to R.S.) and R21 AI058791 (to S.D.), and by a grant from the Pediatric Dengue Vaccine Initiative (TR03/04 to J.J.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almeida JD, Waterson AP. The morphology of virus-antibody interaction. Adv Virus Res. 1969;15:307–338. doi: 10.1016/S0065-3527(08)60878-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg T, Blomhoff R, Naess L, Tolleshaug H, Drevon CA. Monensin inhibits receptor-mediated endocytosis of asialoglycoproteins in rat hepatocytes. Exp Cell Res. 1983;148(2):319–330. doi: 10.1016/0014-4827(83)90156-8. [DOI] [PubMed] [Google Scholar]
- Bowman EJ, Siebers A, Altendorf K. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc Natl Acad Sci U S A. 1988;85(21):7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Scala G, Quinto I, Liu W, Chun TW, Justement JS, Cohen OJ, vanCott TC, Iwanicki M, Lewis MG, Greenhouse J, Barry T, Venzon D, Fauci AS. Protection of rhesus macaques against disease progression from pathogenic SHIV-89.6PD by vaccination with phage-displayed HIV-1 epitopes. Nat Med. 2001;7(11):1225–1231. doi: 10.1038/nm1101-1225. [DOI] [PubMed] [Google Scholar]
- Clark JR, March JB. Bacteriophage-mediated nucleic acid immunisation. FEMS Immunol Med Microbiol. 2004;40(1):21–26. doi: 10.1016/S0928-8244(03)00344-4. [DOI] [PubMed] [Google Scholar]
- Daeron M. Fc receptor biology. Annu Rev Immunol. 1997;15:203–234. doi: 10.1146/annurev.immunol.15.1.203. [DOI] [PubMed] [Google Scholar]
- Davis W, Harrison PT, Hutchinson MJ, Allen JM. Two distinct regions of FC gamma RI initiate separate signalling pathways involved in endocytosis and phagocytosis. Embo J. 1995;14(3):432–441. doi: 10.1002/j.1460-2075.1995.tb07019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Marzo Veronese F, Willis AE, Boyer-Thompson C, Appella E, Perham RN. Structural mimicry and enhanced immunogenicity of peptide epitopes displayed on filamentous bacteriophage. The V3 loop of HIV-1 gp120. J Mol Biol. 1994;243(2):167–172. doi: 10.1006/jmbi.1994.1643. [DOI] [PubMed] [Google Scholar]
- Durrbach A, Louvard D, Coudrier E. Actin filaments facilitate two steps of endocytosis. J Cell Sci. 1996;109(Pt 2):457–465. doi: 10.1242/jcs.109.2.457. [DOI] [PubMed] [Google Scholar]
- Eguchi A, Akuta T, Okuyama H, Senda T, Yokoi H, Inokuchi H, Fujita S, Hayakawa T, Takeda K, Hasegawa M, Nakanishi M. Protein transduction domain of HIV-1 Tat protein promotes efficient delivery of DNA into mammalian cells. J Biol Chem. 2001;276(28):26204–26210. doi: 10.1074/jbc.M010625200. [DOI] [PubMed] [Google Scholar]
- Forestell SP, Dando JS, Bohnlein E, Rigg RJ. Improved detection of replication-competent retrovirus. J Virol Methods. 1996;60(2):171–178. doi: 10.1016/0166-0934(96)02052-6. [DOI] [PubMed] [Google Scholar]
- Geier MR, Merril CR. Lambda phage transcription in human fibroblasts. Virology. 1972;47(3):638–643. doi: 10.1016/0042-6822(72)90553-3. [DOI] [PubMed] [Google Scholar]
- Gotoff R, Tamura M, Janus J, Thompson J, Wright P, Ennis FA. Primary influenza A virus infection induces cross-reactive antibodies that enhance uptake of virus into Fc receptor-bearing cells. J Infect Dis. 1994;169(1):200–203. doi: 10.1093/infdis/169.1.200. [DOI] [PubMed] [Google Scholar]
- Gruenberg J, Griffiths G, Howell KE. Characterization of the early endosome and putative endocytic carrier vesicles in vivo and with an assay of vesicle fusion in vitro. J Cell Biol. 1989;108(4):1301–1316. doi: 10.1083/jcb.108.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon C, Schutten M, Boers PH, Gruters RA, Osterhaus AD. Antibody-mediated enhancement of human immunodeficiency virus type 1 infectivity is determined by the structure of gp120 and depends on modulation of the gp120-CCR5 interaction. J Virol. 2002;76(6):2827–2834. doi: 10.1128/JVI.76.6.2827-2834.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajitou A, Trepel M, Lilley CE, Soghomonyan S, Alauddin MM, Marini FC, 3rd, Restel BH, Ozawa MG, Moya CA, Rangel R, Sun Y, Zaoui K, Schmidt M, von Kalle C, Weitzman MD, Gelovani JG, Pasqualini R, Arap W. A hybrid vector for ligand-directed tumor targeting and molecular imaging. Cell. 2006;125(2):385–398. doi: 10.1016/j.cell.2006.02.042. [DOI] [PubMed] [Google Scholar]
- Harford J, Wolkoff AW, Ashwell G, Klausner RD. Monensin inhibits intracellular dissociation of asialoglycoproteins from their receptor. J Cell Biol. 1983;96(6):1824–1828. doi: 10.1083/jcb.96.6.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa S, Hirashima N, Nakanishi M. Microtubule involvement in the intracellular dynamics for gene transfection mediated by cationic liposomes. Gene Ther. 2001;8(21):1669–1673. doi: 10.1038/sj.gt.3301573. [DOI] [PubMed] [Google Scholar]
- Huang ZY, Barreda DR, Worth RG, Indik ZK, Kim MK, Chien P, Schreiber AD. Differential kinase requirements in human and mouse Fc-gamma receptor phagocytosis and endocytosis. J Leukoc Biol. 2006;80(6):1553–1562. doi: 10.1189/jlb.0106019. [DOI] [PubMed] [Google Scholar]
- Hudson JB, Misra V, Mosmann TR. Cytomegalovirus infectivity: analysis of the phenomenon of centrifugal enhancement of infectivity. Virology. 1976;72(1):235–243. doi: 10.1016/0042-6822(76)90326-3. [DOI] [PubMed] [Google Scholar]
- Iankov ID, Pandey M, Harvey M, Griesmann GE, Federspiel MJ, Russell SJ. Immunoglobulin g antibody-mediated enhancement of measles virus infection can bypass the protective antiviral immune response. J Virol. 2006;80(17):8530–8540. doi: 10.1128/JVI.00593-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indik Z, Kelly C, Chien P, Levinson AI, Schreiber AD. Human Fc gamma RII, in the absence of other Fc gamma receptors, mediates a phagocytic signal. J Clin Invest. 1991;88(5):1766–1771. doi: 10.1172/JCI115496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving MB, Pan O, Scott JK. Random-peptide libraries and antigen-fragment libraries for epitope mapping and the development of vaccines and diagnostics. Curr Opin Chem Biol. 2001;5(3):314–324. doi: 10.1016/S1367-5931(00)00208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanenkov V, Felici F, Menon AG. Uptake and intracellular fate of phage display vectors in mammalian cells. Biochim Biophys Acta. 1999;1448(3):450–462. doi: 10.1016/s0167-4889(98)00162-1. [DOI] [PubMed] [Google Scholar]
- Jepson CD, March JB. Bacteriophage lambda is a highly stable DNA vaccine delivery vehicle. Vaccine. 2004;22(19):2413–2419. doi: 10.1016/j.vaccine.2003.11.065. [DOI] [PubMed] [Google Scholar]
- Kassner PD, Burg MA, Baird A, Larocca D. Genetic selection of phage engineered for receptor-mediated gene transfer to mammalian cells. Biochem Biophys Res Commun. 1999;264(3):921–928. doi: 10.1006/bbrc.1999.1603. [DOI] [PubMed] [Google Scholar]
- Lankes HA, Zanghi CN, Santos K, Capella C, Duke CM, Dewhurst S. In vivo gene delivery and expression by bacteriophage lambda vectors. J Appl Microbiol. 2007;102(5):1337–1349. doi: 10.1111/j.1365-2672.2006.03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocca D, Kassner PD, Witte A, Ladner RC, Pierce GF, Baird A. Gene transfer to mammalian cells using genetically targeted filamentous bacteriophage. Faseb J. 1999;13(6):727–734. doi: 10.1096/fasebj.13.6.727. [DOI] [PubMed] [Google Scholar]
- Leopold PL, Wendland RL, Vincent T, Crystal RG. Neutralized adenovirus-immune complexes can mediate effective gene transfer via an Fc receptor-dependent infection pathway. J Virol. 2006;80(20):10237–10247. doi: 10.1128/JVI.00512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littaua R, Kurane I, Ennis FA. Human IgG Fc receptor II mediates antibody-dependent enhancement of dengue virus infection. J Immunol. 1990;144(8):3183–3186. [PubMed] [Google Scholar]
- Lowry MB, Duchemin AM, Robinson JM, Anderson CL. Functional separation of pseudopod extension and particle internalization during Fc gamma receptor-mediated phagocytosis. J Exp Med. 1998;187(2):161–176. doi: 10.1084/jem.187.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March JB, Clark JR, Jepson CD. Genetic immunisation against hepatitis B using whole bacteriophage lambda particles. Vaccine. 2004;22(13–14):1666–1671. doi: 10.1016/j.vaccine.2003.10.047. [DOI] [PubMed] [Google Scholar]
- Mercier S, Rouard H, Delfau-Larue MH, Eloit M. Specific antibodies modulate the interactions of adenovirus type 5 with dendritic cells. Virology. 2004;322(2):308–317. doi: 10.1016/j.virol.2004.01.031. [DOI] [PubMed] [Google Scholar]
- Merril CR, Geier MR, Petricciani JC. Bacterial virus gene expression in human cells. Nature. 1971;233(5319):398–400. doi: 10.1038/233398a0. [DOI] [PubMed] [Google Scholar]
- Minenkova OO, Ilyichev AA, Kishchenko GP, Petrenko VA. Design of specific immunogens using filamentous phage as the carrier. Gene. 1993;128(1):85–88. doi: 10.1016/0378-1119(93)90157-x. [DOI] [PubMed] [Google Scholar]
- Mollenhauer HH, Morre DJ, Rowe LD. Alteration of intracellular traffic by monensin; mechanism, specificity and relationship to toxicity. Biochim Biophys Acta. 1990;1031(2):225–246. doi: 10.1016/0304-4157(90)90008-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu FT, Callaghan JM, Steele-Mortimer O, Stenmark H, Parton RG, Campbell PL, McCluskey J, Yeo JP, Tock EP, Toh BH. EEA1, an early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine "fingers" and contains a calmodulin-binding IQ motif. J Biol Chem. 1995;270(22):13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- O'Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74(21):10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odin JA, Edberg JC, Painter CJ, Kimberly RP, Unkeless JC. Regulation of phagocytosis and [Ca2+]i flux by distinct regions of an Fc receptor. Science. 1991;254(5039):1785–1788. doi: 10.1126/science.1837175. [DOI] [PubMed] [Google Scholar]
- Parren PW, Burton DR. The antiviral activity of antibodies in vitro and in vivo. Adv Immunol. 2001;77:195–262. doi: 10.1016/S0065-2776(01)77018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris JS, Porterfield JS. Antibody-mediated enhancement of Flavivirus replication in macrophage-like cell lines. Nature. 1979;282(5738):509–511. doi: 10.1038/282509a0. [DOI] [PubMed] [Google Scholar]
- Porterfield JS. Antibody-mediated enhancement of rabies virus. Nature. 1981;290(5807):542. doi: 10.1038/290542a0. [DOI] [PubMed] [Google Scholar]
- Ravetch JV, Bolland S. IgG Fc receptors. Annu Rev Immunol. 2001;19:275–290. doi: 10.1146/annurev.immunol.19.1.275. [DOI] [PubMed] [Google Scholar]
- Rodrigo WW, Jin X, Blackley SD, Rose RC, Schlesinger JJ. Differential enhancement of dengue virus immune complex infectivity mediated by signaling-competent and signaling-incompetent human Fcgamma RIA (CD64) or FcgammaRIIA (CD32) J Virol. 2006;80(20):10128–10138. doi: 10.1128/JVI.00792-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa GT, Gillet L, Smith CM, de Lima BD, Stevenson PG. IgG fc receptors provide an alternative infection route for murine gamma-herpesvirus-68. PLoS ONE. 2007;2:e560. doi: 10.1371/journal.pone.0000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosette C, Karin M. Cytoskeletal control of gene expression: depolymerization of microtubules activates NF-kappa B. J Cell Biol. 1995;128(6):1111–1119. doi: 10.1083/jcb.128.6.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathaliyawala T, Rao M, Maclean DM, Birx DL, Alving CR, Rao VB. Assembly of human immunodeficiency virus (HIV) antigens on bacteriophage T4: a novel in vitro approach to construct multicomponent HIV vaccines. J Virol. 2006;80(15):7688–7698. doi: 10.1128/JVI.00235-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger JJ, Brandriss MW. Growth of 17D yellow fever virus in a macrophage-like cell line, U937: role of Fc and viral receptors in antibody-mediated infection. J Immunol. 1981;127(2):659–665. [PubMed] [Google Scholar]
- Schlesinger JJ, Chapman SE. Influence of the human high-affinity IgG receptor FcgammaRI (CD64) on residual infectivity of neutralized dengue virus. Virology. 1999;260(1):84–88. doi: 10.1006/viro.1999.9816. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Lippe R, Christoforidis S, Gaullier JM, Brech A, Callaghan J, Toh BH, Murphy C, Zerial M, Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394(6692):494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- Takada A, Kawaoka Y. Antibody-dependent enhancement of viral infection: molecular mechanisms and in vivo implications. Rev Med Virol. 2003;13(6):387–398. doi: 10.1002/rmv.405. [DOI] [PubMed] [Google Scholar]
- Takeda A, Sweet RW, Ennis FA. Two receptors are required for antibody-dependent enhancement of human immunodeficiency virus type 1 infection: CD4 and Fc gamma R. J Virol. 1990;64(11):5605–5610. doi: 10.1128/jvi.64.11.5605-5610.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A, Tuazon CU, Ennis FA. Antibody-enhanced infection by HIV-1 via Fc receptor-mediated entry. Science. 1988;242(4878):580–583. doi: 10.1126/science.2972065. [DOI] [PubMed] [Google Scholar]
- Tamura M, Webster RG, Ennis FA. Antibodies to HA and NA augment uptake of influenza A viruses into cells via Fc receptor entry. Virology. 1991;182(1):211–219. doi: 10.1016/0042-6822(91)90664-w. [DOI] [PubMed] [Google Scholar]
- Tamura M, Webster RG, Ennis FA. Subtype cross-reactive, infection-enhancing antibody responses to influenza A viruses. J Virol. 1994;68(6):3499–3504. doi: 10.1128/jvi.68.6.3499-3504.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Herik-Oudijk IE, Capel PJ, van der Bruggen T, Van de Winkel JG. Identification of signaling motifs within human Fc gamma RIIa and Fc gamma RIIb isoforms. Blood. 1995;85(8):2202–2211. [PubMed] [Google Scholar]
- van Vugt MJ, Kleijmeer MJ, Keler T, Zeelenberg I, van Dijk MA, Leusen JH, Geuze HJ, van de Winkel JG. The FcgammaRIa (CD64) ligand binding chain triggers major histocompatibility complex class II antigen presentation independently of its associated FcR gamma-chain. Blood. 1999;94(2):808–817. [PubMed] [Google Scholar]
- Wallace MJ, Smith DW, Broom AK, Mackenzie JS, Hall RA, Shellam GR, McMinn PC. Antibody-dependent enhancement of Murray Valley encephalitis virus virulence in mice. J Gen Virol. 2003;84(Pt 7):1723–1728. doi: 10.1099/vir.0.18980-0. [DOI] [PubMed] [Google Scholar]
- Wang L, MacDonald RC. Effects of microtubule-depolymerizing agents on the transfection of cultured vascular smooth muscle cells: enhanced expression with free drug and especially with drug-gene lipoplexes. Mol Ther. 2004;9(5):729–737. doi: 10.1016/j.ymthe.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Zanghi CN, Lankes HA, Bradel-Tretheway B, Wegman J, Dewhurst S. A simple method for displaying recalcitrant proteins on the surface of bacteriophage lambda. Nucleic Acids Res. 2005;33(18):e160. doi: 10.1093/nar/gni158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanghi CN, Sapinoro R, Bradel-Tretheway B, Dewhurst S. A tractable method for simultaneous modifications to the head and tail of bacteriophage lambda and its application to enhancing phage-mediated gene delivery. Nucleic Acids Res. 2007;35(8):e59. doi: 10.1093/nar/gkm146. [DOI] [PMC free article] [PubMed] [Google Scholar]


