Abstract
Background
The genome sequence of the sea-ice bacterium Psychromonas ingrahamii 37, which grows exponentially at -12C, may reveal features that help to explain how this extreme psychrophile is able to grow at such low temperatures. Determination of the whole genome sequence allows comparison with genes of other psychrophiles and mesophiles.
Results
Correspondence analysis of the composition of all P. ingrahamii proteins showed that (1) there are 6 classes of proteins, at least one more than other bacteria, (2) integral inner membrane proteins are not sharply separated from bulk proteins suggesting that, overall, they may have a lower hydrophobic character, and (3) there is strong opposition between asparagine and the oxygen-sensitive amino acids methionine, arginine, cysteine and histidine and (4) one of the previously unseen clusters of proteins has a high proportion of "orphan" hypothetical proteins, raising the possibility these are cold-specific proteins.
Based on annotation of proteins by sequence similarity, (1) P. ingrahamii has a large number (61) of regulators of cyclic GDP, suggesting that this bacterium produces an extracellular polysaccharide that may help sequester water or lower the freezing point in the vicinity of the cell. (2) P. ingrahamii has genes for production of the osmolyte, betaine choline, which may balance the osmotic pressure as sea ice freezes. (3) P. ingrahamii has a large number (11) of three-subunit TRAP systems that may play an important role in the transport of nutrients into the cell at low temperatures. (4) Chaperones and stress proteins may play a critical role in transforming nascent polypeptides into 3-dimensional configurations that permit low temperature growth. (5) Metabolic properties of P. ingrahamii were deduced. Finally, a few small sets of proteins of unknown function which may play a role in psychrophily have been singled out as worthy of future study.
Conclusion
The results of this genomic analysis provide a springboard for further investigations into mechanisms of psychrophily. Focus on the role of asparagine excess in proteins, targeted phenotypic characterizations and gene expression investigations are needed to ascertain if and how the organism regulates various proteins in response to growth at lower temperatures.
Background
Well over half of the earth's surface is cold: deep oceans, mountains, polar regions. Likewise, Earth's solar system contains many planets and planetary bodies that are also cold. The cold environments on Earth are teeming with life [1] offering hope that other cold environments in our solar system such as Mars and Jupiter's moon, Europa, may harbor life [2]. For this reason it is surprising that so little is know about the lifestyle, particularly of microbial psychrophiles at low temperatures.
Psychrophiles have been studied primarily to understand biological mechanisms of adaptation to extreme conditions. Microbial physiologists have long been interested in psychrophiles as they employ mechanisms allowing them to maintain life processes at temperatures where rates of reactions and molecular properties present challenges. In reaching for an understanding of how life processes work at extremes of temperature, most of the focus to date has been on the properties of enzymes of extremophiles (reviewed by [3,4]). No single consistent answer has emerged to account for adaptation to temperature extremes. To date, no single type of modification is uniformly found in the enzymes of psychrophiles; instead numerous small and subtle differences appear to account for their increased flexibility thereby enabling them to function at low temperatures.
Recently whole genome sequences have been determined for psychrophiles Colwellia psychrerythraea 34 H [5], Idiomarina loihiensis L2TR [6], and Pseudoalteromonas haloplanktis TAC125 [7]. We now add the genomic sequence of the extreme species, Psychromonas ingrahamii 37 which grows at even colder temperatures. Availability of complete genome sequences provides the opportunity to search all of the proteins of the organisms for similarities and differences that might have bearing on the ability of the organism to grow at low temperatures.
The extreme psychrophile, Psychromonas ingrahamii was isolated from sea ice from the Arctic. It grows exponentially with a doubling time of 240 hours at -12°C and may well grow at even lower temperatures [8]. These temperatures do not necessarily solidify salt water or cytoplasm into ice. Liquid water has been shown to exist at grain contacts as low as -20C [2].
Results and Discussion
The P. ingrahamii genome
The single, circular chromosome of 4.56 Mb constituting the genome of P. ingrahamii 37 was sequenced as a set of contigs by the DOE Joint Genome Institute Production Genomics Facility, 2800 Mitchell Drive, Walnut Creek, CA 94598, and finished at DOE Joint Genome Institute, Bioscience Division, Los Alamos National Laboratory, Los Alamos, NM 87545. It was annotated at Oak Ridge National Laboratory, Oak Ridge, TN 37831 and deposited as GenBank file CP000510.1. Altogether 3708 genes were identified, 3545 of which were proteins of 83 residues or longer. A second round of annotation is described below.
Properties of proteins
Size
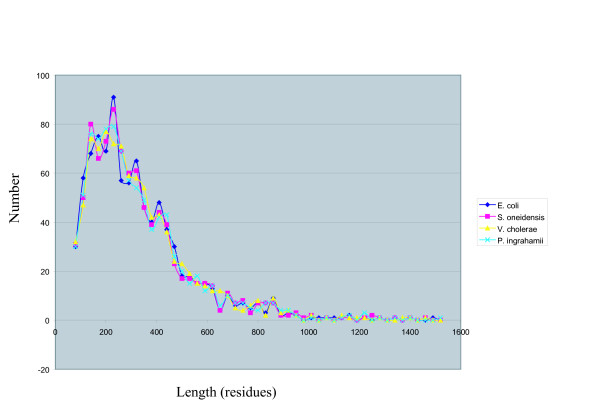
One can ask whether the sizes of proteins of a psychrophile differ from those of a mesophile. The sequences of all proteins of P. ingrahamii were compared to sequences of all proteins of three other bacteria: Shewanella oneidensis MR-1, Vibrio cholerae and Escherichia coli K-12 MG1655. 916 protein sequences were conserved among all four bacteria. The great majority of the conserved proteins were enzymes. The distribution of lengths of the 916 orthologous proteins were compared, revealing that the distribution was about the same for the comparable proteins in all four bacteria (Figure 1). Ideas that proteins that are required to function at low temperatures would be either shorter or longer than those of mesophiles are not borne out.
Figure 1.
Distribution of lengths of 916 orthologous proteins in four bacteria. Distribution of lengths of proteins as numbers of amino acid residues, ranging from 83 to 1501, in increments of 30. Black diamond = E. coli, red square = S. oneidensis, yellow triangle = V. cholerae, blue cross = P. ingrahamii.
Amino acid composition
Amino acid composition of an organism's proteins is affected by nucleotide composition of the DNA. The GC content of P. ingrahamii DNA was determined experimentally to be 40% [8], verified by the composition of the total genome nucleotide sequence (40.1%). Overall amino acid content of the encoded proteins compared to those of V. cholerae, S. oneidensis and E. coli is shown in Table 1 (P. Sharp, personal comunication). The amino acids isoleucine, asparagine, lysine, phenylalanine and tyrosine all are present in higher percentage in P. ingrahamii.
Table 1.
Amino acid composition (%) of total proteins of 4 bacteria
| Organisms* | S | L | R | P | T | V | A | G | I | F |
| Esccol | 5.83 | 10.65 | 5.54 | 4.43 | 5.41 | 7.1 | 9.49 | 7.37 | 6 | 3.9 |
| Shewone | 6.47 | 10.97 | 4.64 | 4.06 | 5.37 | 6.76 | 9.42 | 6.79 | 6.03 | 3.97 |
| Vibcho | 6.33 | 10.9 | 4.95 | 4.02 | 5.19 | 7.07 | 9.15 | 6.68 | 6.04 | 4.08 |
| Psying | 4.82 | 10.99 | 4.08 | 3.71 | 5.49 | 6.57 | 8.49 | 6.68 | 7.53 | 4.47 |
| Y | C | H | Q | N | K | D | E | W | M | |
| Esccol | 2.85 | 1.17 | 2.27 | 4.43 | 3.95 | 4.41 | 5.14 | 5.75 | 1.53 | 2.8 |
| Shewone | 3.05 | 1.09 | 2.33 | 4.93 | 4.12 | 5.14 | 5.29 | 5.74 | 1.28 | 2.54 |
| Vibcho | 2.96 | 1.05 | 2.4 | 5.17 | 3.9 | 4.93 | 5.02 | 6.2 | 1.32 | 2.63 |
| Psying | 3.16 | 1.11 | 2.13 | 5.98 | 4.92 | 6.25 | 5.47 | 4.54 | 1.16 | 2.45 |
*Esccol = Escherichia coli MG1655, Shewone = Shewanella oneidensis MR-1, Vibcho = Vibrio cholerae, Psying = Psychromonas ingrahamii 37
However, like the genome, the codons for these residues are GC rich, a factor that must be taken into account. Codon usage for P. ingrahamii is shown in Table 2 for all CDSs (coding DNA sequences) and for the highly expressed genes tuf, tsf, fusA+RP genes (P. Sharp, personal communication). Thus when amino acid content is examined as a function of GC3s, correcting for GC content at synonymously variable third positions, the overall amino acid composition of P. ingrahamii was found not to be remarkable. At the value of 34.2 determined for P. ingrahamii, amino acid contents fall on the curves generated for GC3s dependency in 80 other organisms (P. Sharp, personal communication) (data not shown) [9].
Table 2.
Number of codons in highly expressed and in all genes of Psychromonas ingrahamii
| High | All | High | All | High | All | High | All | ||||||||
| Phe | UUU | 107 | 41275 | Ser | UCU | 139 | 14451 | Tyr | UAU | 47 | 25785 | Cys | UGU | 35 | 8165 |
| Phe | UUC | 103 | 10226 | Ser | UCC | 3 | 7455 | Tyr | UAC | 86 | 10612 | Cys | UGC | 5 | 4681 |
| Leu | UUA | 128 | 50155 | Ser | UCA | 73 | 16739 | Ter | UAA | 32 | 2366 | Ter | UGA | 1 | 571 |
| Leu | UUG | 44 | 17564 | Ser | UCG | 16 | 7210 | Ter | UAG | 7 | 608 | Trp | UGG | 31 | 13405 |
| Leu | CUU | 150 | 20860 | Pro | CCU | 98 | 13581 | His | CAU | 56 | 17366 | Arg | CGU | 408 | 17987 |
| Leu | CUC | 9 | 7631 | Pro | CCC | 5 | 9132 | His | CAC | 67 | 7202 | Arg | CGC | 74 | 11043 |
| Leu | CUA | 87 | 9170 | Pro | CCA | 110 | 10292 | Gln | CAA | 167 | 29958 | Arg | CGA | 12 | 5132 |
| Leu | CUG | 52 | 21318 | Pro | CCG | 32 | 9734 | Gln | CAG | 50 | 22366 | Arg | CGG | 3939 | |
| Ile | AUU | 180 | 48789 | Thr | ACU | 189 | 17262 | Asn | AAU | 91 | 39859 | Ser | AGU | 48 | 19260 |
| Ile | AUC | 208 | 20430 | Thr | ACC | 31 | 19451 | Asn | AAC | 128 | 16887 | Ser | AGC | 60 | 14213 |
| Ile | AUA | 22 | 17588 | Thr | ACA | 113 | 16391 | Lys | AAA | 452 | 57148 | Arg | AGA | 14 | 6404 |
| Met | AUG | 188 | 28244 | Thr | ACG | 41 | 10226 | Lys | AAG | 134 | 14939 | Arg | AGG | 0 | 2583 |
| Val | GUU | 347 | 29210 | Ala | GCU | 328 | 26162 | Asp | GAU | 228 | 49174 | Gly | GGU | 412 | 33290 |
| Val | GUC | 32 | 13155 | Ala | GCC | 41 | 21828 | Asp | GAC | 83 | 13878 | Gly | GGC | 119 | 20816 |
| Val | GUA | 198 | 15326 | Ala | GCA | 273 | 30437 | Glu | GAA | 354 | 47661 | Gly | GGA | 33 | 12381 |
| Val | GUG | 52 | 18068 | Ala | GCG | 62 | 19459 | Glu | GAG | 91 | 21337 | Gly | GGG | 6 | 10497 |
Correspondence analysis (CA)
Lobry and Chessel [10] examined datasets of amino acid content and codon usage in thermophilic and mesophic bacteria and found evidence that the amino acid composition of thermophilic proteins was under the control of a pressure at the nucleic acid level, not a selection at the protein level. An extended study [11] produced similar results of no connection. However, the authors identified the most discriminating codon as being AGG for arginine, present in many thermophiles, not in either mesophiles or a psychrophile. P. ingrahamii fits this observation in that it does not use the AGG codon (Table 2).
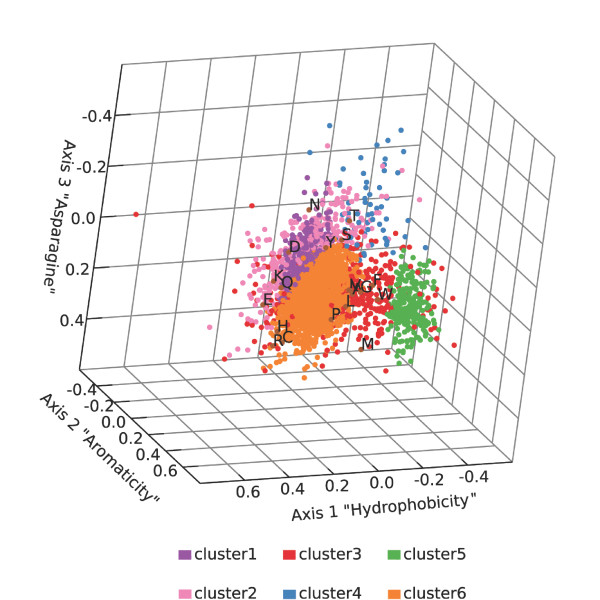
We analyzed the amino acid compositions of all individual proteins of P. ingrahamii by CA [12] after trimming the first 10 and last 5 residues because there is a strong nucleotide bias at these positions. Data for each protein is presented in Additional file 1: "Ping Correspondence Analysis.xls". Clustering of groups of proteins with similar composition was performed using a bayesian approach as proposed by Bailly-Bechet et al. [13] Figure 2 presents a plot of the first three most informative axes: hydrophobicity, aromaticity and asparagine content.
Figure 2.
Correspondence analysis of amino acid content of P. ingrahamii proteins. Proteins over 100 residues were subjected to correspondence anlysis by amino acid content, clustered and plotted on first three most informative axes. Amino acid frequencies are superimposed. (See Methods).
The proteins fell into six classes whereas most bacteria contain at most five well separated clusters with bayesian clustering and at most four with dynamic clouds clustering [14]. Proteins of two bacterial proteomes examined to date fall into four clusters, Aeropyrum pernix and Thermoplasma acidophilum [14]. Subsequently it was seen that psychrophilic P. haloplanktis has 5 clusters [7], exceeded now by P. ingrahamii, a more extreme psychrophile, which resolves into six. In P. ingrahamii, the bulk of the proteins fall into three clusters (clusters 1, 2, 6). The integral inner membrane proteins (IIMPs) fall largely into clusters 3 and 5 (Figure 2). In other bacteria the IIMPs are distinctly separate from the bulk of other proteins [14]. In P. ingrahamii, however, separation is poor and there is some continuity between the IIMPs and the bulk proteins. Evidently the property of hydrophobicity is not distributed in psychrophile proteins as in mesophiles.
Not seen in any other bacteria viewed to date by CA is the sixth group, cluster 4. This group is of 57 proteins characterized by an excess of threonine that forms a bleb protruding from one of the core protein clusters. Examining the annotations of these proteins reveals that almost half are hypothetical proteins with no homologs in other bacteria examined at threshold Pam 150. It is tempting to suggest this group of proteins could be involved in facilitating low temperature growth.
As to amino acid content, one sees that asparagine occurs more frequently than expected for a random distribution, and on the other hand that cysteine, methionine, arginine and histidine are relatively rare (Figure 2). This same asparagine-driven bias has been seen in other psychrophiles, notably P. haloplanktis [7]. Deamidation via cyclization into aspartate threatens integrity of asparagine, a process which is sensitive to higher temperature, providing a rationale for an asparagine excess in psychrophiles. The corresponding lower amounts of cysteine, methionine, arginine and histidine can be understood as decreasing proportions of these oxygen-sensitive residues. Oxygen concentrations are higher in the liquid medium at low temperatures. Although similar composition gradients were seen in another psychrophile, P. haloplanktis, they are stronger in P. ingrahamii, perhaps correlated with its lower growth temperatures.
Thus, the main features that emerged from the CA of P. ingrahamii protein compositons (Figure 2) are that (1) there are more classes of proteins than have been seen in other bacteria, (2) one of these classes, cluster 4, has a high proportion of "orphan" hypothetical proteins, (3) IIMPs merge into bulk proteins rather than occupying a separate space, possibly due to IIMPs having lower hydrophobic character, and (4) one notes the strong opposition between asparagine, sensitive to heat, and the amino acids methionine, arginine, cysteine and histidine, sensitive to oxygen.
Annotation
Of the 3545 genes for proteins of length 83 or more, 41 are fusions of two genes which in other organisms are separate and independent. They are distributed as 21 fused enzymes, 9 fused regulator components, 4 fused ABC transporter components, 4 fused phosphotransferase system (PTS) components and 3 mixed enzyme-regulator combinations. Fused genes can cause problems with annotation based on results of sequence similarity algorithms. To avoid such problems, we split fused genes for purposes of sequence comparisons in order to be able to identify orthologs of both parts independently.
The first round of annotation was carried out at Oak Ridge National Laboratory, posted for public access August 2006 [15]. We have here supplemented this data with manual analysis using the dynamic search programs of the Darwin system [16]. In this system after first approximations of sequence similarities, amino acid substitution tables are recalculated appropriate to degree of similarity and to the codon usage, and then pairwise alignments are produced. Degree of similarity between two sequences is expressed as percent identity and as Pam values [The Pam score (point accepted mutations) is an inverse measure of sequence differences] [17].
We first processed the P. ingrahamii protein sequences in relation to the then-completed 111 bacterial genomes. We extracted from this data the match with best (lowest) Pam score for each P. ingrahamii protein. The descriptions of the orthologs were retrieved from RefSeq and/or Genbank at the National Center for Biotechnology Information (NCBI) Web site [18]. The results allowed us to add some predicted protein products to the initial JGI annotation results.
Since the set of 111 bacterial genomes we first used was not balanced in respect to types of bacteria, we also identified orthologs in a reference set of 53 genomic sequences of organisms which were chosen to span the breadth of bacterial species. To include other sequenced psychrophiles, the set includes two additional marine species, C. psychrerythraea 34 H and I. loihiensis. C. psychrerythraea 34 H grows over the range -1C to 10C [5]; I. loihiensis has a broad temperature range from 4C to 46C [6].
Annotations and function information are attached in Additional file 2: "Ping Annotations 2.xls". The rank order of similar sequences and number of "best hits" are shown in Table 3.
Table 3.
Organisms with similarity to greatest number of P. ingrahamii proteins*
| Organism | Number of "Best Hits" |
| Vibrio cholerae | 697 |
| Shewanella oneidensis | 539 |
| Colwellia psychrerythraea | 499 |
| Escherichia coli | 285 |
| Idiomarina loihiensis | 143 |
| Pseudomonas aeruginosa | 137 |
* sizes greater than 83 residues
Surprisingly, the organism with greatest similarity is V. cholerae. It is surprising because neither is V. cholerae one of the psychrophiles nor is it any member of the Order Alteromonadales. P. ingrahamii is a member of the Family Alteromonadacae in the Order Alteromondales. Psychromonas is related to other members of this family such as Shewanella, Colwellia and Idiomarina [20]. Like some other members of these families, P. ingrahamii is a marine organism and has gas vesicles. We found there are extensive similarities with proteins of other Alteromonads as expected, but there are even more similarities with V. cholerae, even though the the Order Vibrionales and Family Vibrionaceae are separate from the Alteromonadales [19]. This observation may be explained by the selected conservation of genes from a common ancestor between these two genera and their loss by their closest relatives or it may indicate confusion due to unexpected horizontal gene transfer of 16S rDNA within these lineages.
In manually curating, we used practices aimed both at discovery and at caution. Whenever a "best match" was annotated as a hypothetical protein, yielding no information, we looked to the next best match. Sometimes the next best match was a very good one which provided useful information based on the annotated gene product of that ortholog.
It is widely appreciated in the genomics community that annotation by transfer of stipulated annotations from other organisms becomes ever more problematic at lower levels of similarity and as the number of sequential steps of sequence matches increases between the query and an experimentally demonstrated gene product. We were careful to be conservative in attributing a gene product if the match was not very close. When Pam scores were low (excellent match), we transferred directly the annotation of the match; but when in a middle range, 75–125, we sometimes generalized, removing specificity (i.e. we stipulated "an aminotransferase" instead of "aspartate aminotransferase"); and when Pam scores were high, over 125, we often used the word "predicted" to indicate that the assignment was based on a less rigorous threshold than for other assignments.
When formulating words of description of the gene products, we adopted another practice. We made an effort to name similar products similarly. We have tried to standardize product descriptions to some extent in order to make information on like proteins easier to find, thus making a list of alphabetized product names useful.
Assignments
From the above two sets of ortholog matches combined with the work of the JGI scientists, we constructed a table of P. ingrahamii gene numbers and corresponding best-guess annotated gene products available in Additional file 2: "Ping Annotations 2". Included in the table are the gene number, the type of gene product (enzyme, regulator, etc) and the name of the protein. (Gene number throughout this report is the number of the locus_tag which, as submitted December 2006 to Genbank, CP000510.1, has the prefix "Ping"). Altogether 217 proteins originally characterized as unknown gained an annotation by the manual process, some suggestive, others substantive.
Kinds of proteins
Table 4 gives classification of all proteins of P. ingrahamii by imputed function. The distribution of sizes of classes is similar to that for Escherichia coli K-12 [20] with enzymes being the largest class followed by transporters, then regulators, the rest divided into smaller categories.
Table 4.
Distribution of functional types among all P. ingrahamii proteins*
| Type of function | Number |
| Enzymes | 1317 |
| Unknown | 761 |
| Transporters | 443 |
| Regulators | 252 |
| Domains known | 220 |
| Factors | 203 |
| Structural | 122 |
| Horizontal | 81 |
| Carriers | 47 |
| Membrane | 38 |
| Cell process | 38 |
| Lipoproteins | 21 |
| Total | 3545 |
* sizes greater than 83 residues
Horizontal transfer
Eighty-one genes were identifiable as currently known horizontally transferrable loci such as transposases of insertion sequences and integrases of phages. Thus clearly horizontal exchange of mobile elements into the chromosome has taken place. Among the bacteria tested, most such loci of P. ingrahamii matched elements of Shewanella oneidensis MR1. As is the case with so many genome sequences, we cannot know with currently available information how much of the genome of P. ingrahamii was formed by horizontal transfer and how much vertically inherited.
RNA
There are 10 ribosomal RNA clusters containing 5S, 16S, 23S RNAs (see Additional file 2). The relatively high number could reflect the need for high capacity of translation at cold temperatures and an ability to adapt quickly to changing conditions of nutrient availability [21]. 86 tRNA genes support translation.
Gene clusters
There are 100 contiguous clusters of two or more related genes such as subunits of an enzyme or pathway-related proteins. The largest cluster, for septum formation and peptidogylcan synthesis enzymes, comprises 14 genes. These are genes 1140 through 1153. Altogether 850, or 25% of CDSs, reside in clusters related by function.
Paralogous groups
Enumeration of sets of paralogs within a genome requires definition of degree of relatedness. In the initial analysis done at Oak Ridge National Laboratory, 1799 proteins were identified as belonging to 510 paralogous clusters. Using Darwin analysis [22] and a more conservative threshold of relatedness (Pam =< 175), 965 proteins were identified as grouped into 273 paralogous families of sizes ranging from two to 66. This threshold is comparable to that used to enumerate E. coli paralogs [23] and permits comparison. Functions of paralogous groups of size 7 or above are shown in Table 5. Just as had been found previously for E. coli, the largest paralogous groups are transporters and regulators. Even though enzymes are present in the genome in the largest numbers, they fall into smaller, more differentiated families. For transporters and also regulators, evidently a limited number of mechanisms have evolved for these functions, creating large groups of similar proteins of either transporters or regulators that differ in specificity but not in mechanism of action. Enzymes are more diverse, use a larger variety of mechanisms, thus they fall into a larger number of smaller groups of similar proteins.
Table 5.
Distribution of functional types among largest P. ingrahamii paralogous groups
| Group size | Protein function |
| 66 | ATP-binding subunits of ABC transporters |
| 51 | Cyclic-diGMP regulation, diguanylate cyclases |
| 24 | Transcriptional regulators, LysR type |
| 23 | Substrate-binding subunits of ABC transporters |
| 15 | Two-component response regulators |
| 15 | Transcriptional regulators, LacI type |
| 12 | Peptide-binding subunits of ABC transporters |
| 11 | ATP-dependent RNA helicases |
| 11 | Short-chain alcohol dehydrogenase family |
| 10 | Tripartite C4-dicarboxylate transporter, DctM-type subunits |
| 9 | Unknown |
| 9 | Fused ATP-binding/substrate-binding subunits of ABC transporters |
| 9 | IS4 transposases |
| 8 | Two-component sensor histidine kinases |
| 7 | IS30 transposases |
| 7 | Aldehyde dehydrogenases |
| 7 | Oxidoreductases |
| 7 | Tripartite C4-dicarboxylate transporter, DctQ-type subunits |
| 7 | Crotonase-like epimerase/dehydratases |
| 7 | Extracellular amino acid-binding subunits of ABC transporters |
| 7 | Aminotransferases |
Intermediary metabolism
Almost half of all enzymes annotated in P. ingrahamii (634/1317) were identified as enzymes of small molecule metabolism. Not all enzymes of every pathway were found, not unexpected since some orthologs may not reach the threshold of similarity employed, nevertheless there is strong evidence for standard pathways of intermediary metabolism being present.
P. ingrahamii is a facultative anaerobe capable of both respiratory and fermentative metabolism [24]. In agreement we find in its enzyme sequences that pathways are present for fermentation, glycolysis, pentose phosphate pathway, the TCA cycle and gluconeogenesis. For respiration, electron transfer agents are present such as iron-sulfur centers, flavodoxin and flavoproteins, ubiquinone, and cytochromes; for anaerobic respiration, sequences of reductases for fumarate, nitrate, nitrite and sulfite are present (Table 6). Nitrate reduction has been observed experimentally [24]. In addition there are 10 members of the short-chain oxidoreductase family and 8 oxidoreductases identified by domain. Any one of these could be either a primary dehydrogenase or a terminal reductase not yet characterized (Table 7). The varieties of oxidants that appear to be used by P. ingrahamii suggest that its capabilities in this regard are comparable to that of Shewanella species.
Table 6.
Genes and enzymes of glucose and energy metabolism
| Pathway | Gene | Enzyme |
| Glycolysis | ||
| 591 | 6-phosphofructokinase | |
| 1316 | 6-phosphofructokinase | |
| 669 | enolase | |
| 3617 | fructose-1,6-bisphosphatase, class II | |
| 2682 | fructose-bisphosphate aldolase | |
| 372 | fructose-bisphosphate aldolase, class II | |
| 2359 | glucokinase | |
| 2382 | glucokinase | |
| 2932 | glucokinase, thermoresistant | |
| 324 | glucose-6-phosphate isomerase | |
| 2004 | glyceraldehyde-3-phosphate dehydrogenase, | |
| 2367 | glyceraldehyde-3-phosphate dehydrogenase, | |
| 3636 | glyceraldehyde-3-phosphate dehydrogenase, | |
| 879 | phosphoglucomutase | |
| 769 | phosphoglucomutase | |
| 371 | phosphoglycerate kinase | |
| 249 | phosphoglycerate mutase | |
| 2320 | phosphoglycerate mutase | |
| 3211 | phosphoglycerate mutase, cofactor-independent | |
| 2361 | pyruvate kinase | |
| 2879 | pyruvate kinase | |
| 2199 | triose-phosphate isomerase | |
| Metabolic connections | ||
| 3617 | fructose-1,6-bisphosphatase, class II | |
| 93 | phosphoenolpyruvate carboxykinase | |
| 537 | malic enzyme | |
| 304 | isocitrate lyase and phosphorylmutase | |
| 303 | malate synthase | |
| Pentose pathway | ||
| 2752 | 6-phosphogluconate dehydrogenase, decarboxylating | |
| 2937 | 6-phosphogluconate dehydrogenase, decarboxylating | |
| 2753 | 6-phosphogluconolactonase | |
| 167 | ribulose-phosphate 3-epimerase | |
| 2754 | glucose-6-phosphate 1-dehydrogenase | |
| 3554 | ribose 5-phosphate isomerase | |
| 601 | ribose 5-phosphate isomerase | |
| 2054 | transaldolase | |
| 86 | transaldolase B | |
| 339 | transketolase | |
| 3086 | glucose dehydrogenase | |
| 2936 | gluconate kinase | |
| Pyruvate dehydrogenase | ||
| 2782 | pyruvate dehydrogenase complex, E1 beta subunit | |
| 3602 | pyruvate dehydrogenase complex, E1 beta subunit | |
| 3601 | pyruvate dehydrogenase complex, E1 acetate transfer subunit | |
| 3603 | pyruvate dehydrogenase complex, E2 subunit | |
| 2779 | dihydrolipoamide dehydrogenase E3 subunit | |
| 2780 | dihydrolipoamide dehydrogenase E3 subunit | |
| 2925 | dihydrolipoamide dehydrogenase E3 subunit | |
| Tricarboxylic acid cycle | ||
| 2927 | 2-oxo-acid dehydrogenase E1 subunit | |
| 2252 | 2-oxoglutarate dehydrogenase, E1 subunit | |
| 2926 | 2-oxoglutarate dehydrogenase E2 subunit | |
| 2251 | 2-oxoglutarate dehydrogenase, E2 subunit | |
| 2899 | aconitate hydratase | |
| 2120 | aconitate hydratase 1 | |
| 800 | adenylyl-sulfate kinase | |
| 2257 | citrate synthase I | |
| 2617 | citrate synthase I | |
| 1738 | fumarate hydratase | |
| 1977 | fumarate hydratase | |
| 983 | isocitrate dehydrogenase, NADP-dependent | |
| 297 | malate dehydrogenase, NAD-dependent | |
| 3376 | oxaloacetate decarboxylase alpha subunit | |
| 3375 | oxaloacetate decarboxylase, beta subunit | |
| 2253 | succinate dehydrogenase catalytic subunit SdhB | |
| 2254 | succinate dehydrogenase, flavoprotein subunit SdhA | |
| 2256 | succinate dehydrogenase, cytochrome b-binding subunit sdhC | |
| 2255 | succinate dehydrogenase, cytochrome b-binding subunit sdhD | |
| 2249 | succinyl-CoA synthetase, alpha subunit | |
| 2250 | succinyl-CoA synthetase, beta subunit | |
| Anaerobic respiration | ||
| 3279 | fumarate reductase iron-sulfur subunit | |
| 3281 | fumarate reductase, D subunit | |
| 3278 | fumarate reductase, flavoprotein subunit | |
| 3280 | fumarate reductase, subunit C | |
| 2175 | nitrate reductase accessory periplasmic protein NapD | |
| 2172 | nitrate reductase periplasmic cytochrome c-type protein NapC | |
| 2173 | nitrate reductase periplasmic cytochrome c-type subunit NapB | |
| 2174 | nitrate reductase, periplasmic large subunit | |
| 1024 | nitrite reductase [NAD(P)H], large subunit | |
| 1023 | nitrite reductase [NAD(P)H], small subunit | |
| 3435 | sulfite reductase (NADPH) hemoprotein, beta-component | |
| 3434 | sulfite reductase [NADPH] flavoprotein, alpha chain | |
| 3436 | phosphoadenosine phosphosulfate reductase (PAPS reductase) | |
| Oxidoreductases of unknown substrate | ||
| 45 | short-chain dehydrogenase/reductase SDR | |
| 223 | short-chain dehydrogenase/reductase SDR | |
| 951 | short-chain dehydrogenase/reductase SDR | |
| 989 | short-chain dehydrogenase/reductase SDR | |
| 1000 | short-chain dehydrogenase/reductase SDR | |
| 1973 | short-chain dehydrogenase/reductase SDR | |
| 2106 | short-chain dehydrogenase/reductase SDR | |
| 2109 | short-chain dehydrogenase/reductase SDR | |
| 2778 | short-chain dehydrogenase/reductase SDR | |
| 3154 | short-chain dehydrogenase/reductase SDR | |
| 272 | oxidoreductase FAD/NAD(P)-binding domain protein | |
| 3187 | oxidoreductase FAD/NAD(P)-binding domain protein | |
| 2122 | oxidoreductase alpha (molybdopterin) subunit | |
| 1244 | oxidoreductase domain protein | |
| 1809 | oxidoreductase domain protein | |
| 2666 | oxidoreductase domain protein | |
| 3535 | oxidoreductase domain protein | |
| 576 | oxidoreductase, molybdopterin binding | |
| Fermentation indications | ||
| 91 | fermentative D-lactate dehydrogenase | |
| 2123 | formate dehydrogenase, subunit FdhD | |
| 1217 | hydrogenase, NADP-reducing subunit C | |
| many alcohol dehydrogenases |
Table 7.
Metabolism of fatty acids
| Pathway | Gene | Enzyme |
| Degradation | 3600 | acetyl-CoA synthetase |
| 2603 | acyl-CoA dehydrogenase domain protein | |
| 1208 | enoyl-CoA hydratase/isomerase | |
| 2604 | fused 3-hydroxyacyl-CoA dehydrogenase, NAD-binding and enoyl-CoA hydratase/isomerase | |
| 2401 | acyl-CoA thiolase (acetyl-CoA transferase) | |
| Synthesis | 1090 | acyl carrier protein |
| 1088 | malonyl CoA-acyl carrier protein transacylase | |
| 1995 | 3-oxoacyl-(acyl-carrier-protein) synthase I | |
| 1087 | 3-oxoacyl-(acyl-carrier-protein) synthase III | |
| 1997 | 3-oxoacyl-[acyl-carrier-protein) synthase III | |
| 1091 | beta-ketoacyl synthase | |
| 1982 | beta-hydroxyacyl-(acyl-carrier-protein) dehydratase | |
| 1089 | 3-oxoacyl-(acyl-carrier-protein) reductase | |
| 188 | lauroyl (or palmitoleoyl)-ACP acyltransferase | |
| 2965 | (3R)-hydroxymyristoyl-ACP dehydratase | |
| Unsaturation | 1684 | polyunsaturated fatty acid synthase |
| 1685 | polyunsaturated fatty acid synthase | |
| 1686 | polyunsaturated fatty acid synthase | |
| 1687 | polyunsaturated fatty acid synthase |
Enzymes are present for pathways of utilization of both carbohydrates and amino acids as carbon and energy sources. Glycerol was the carbon source provided in the medium for the below-freezing temperature growth experiments [8]. In agreement, genes for glycerol uptake (genes 3166, 3169), glycerol kinase (3168) and dehydrogenase (3207) are present.
Carbon sources besides glucose that were found experimentally to be utilized by P. ingrahamii [24] were compared with the list of enzyme orthologs. One finds in P. ingrahamii genes for enzymes for utilization of fructose (971, 3552), galactose (2016, 2017), mannitol (89), N-acetylglucosamine (488–490), ribose (344) and sucrose (974), in agreement with experimental findings. We also found genetic evidence for the capability to utilize lactose (2019) and glucuronate (130,131, 132 and 136).
However comparing to E. coli enzymes, many of the sequences of enzymes for utilization of other carbohydrates are not present in P. ingrahamii. These include enzymes for utilization of arabinose, xylose, sorbitol or galactitol. Again the absence is in agreement with experimental observations [24]. Also missing are orthologs of E. coli enzymes for utilization of tagatose, fucose, rhamnose, glucarate, galactarate, altronate or idonate. These carbon sources have not yet been tested in culture. This picture is similar to the one found in S. oneidensis, a relatively poor capacity to utilize a variety of monomeric carbohydrates [25].
Although capability for utilization of carbohydrates is restricted, by contrast P. ingrahamii does have sequences for enzymes for utilization of most amino acids, and many of these have been verified experimentally. Transaminases convert to the corresponding keto acids, decarboxylases to amines. The amino group can be removed with dehydratases or lyases.
Fatty acids, breakdown
The four major enzymes of degradation of fatty acids are present, capable of generating acetyl-CoA for general metabolism (Table 7).
Fatty acids, biosynthesis
It has long been known that proteobacteria at low temperatures adjust fatty acid composition to the more flexible unsaturated and/or branched types. P. ingrahamii has genes for enzymes of fatty acid biosynthesis (Table 7). Close similarity is found for all E. coli enzymes starting with malonyl-CoA, proceeding via malonyl-ACP through the fatty acid biosynthesis cycle, each cycle adding 2-carbon moieties. At the 10-carbon level, the pathway of unsaturated fatty acids commences. Orthologs for all E. coli enzymes of the unsaturation pathway are present except for the last enzyme, FabI. Final steps for synthesis of unsaturated fatty acids by P. ingrahamii must differ from those in E. coli. The principal fatty acids that were detected in the organism experimentally are the saturated 16:0 (18.7%) and the singly unsaturated 16:1ω7c (67.0%) [24].
P. ingrahamii has sequences of the four subunits of a polyunsaturase closely similar to the polyunsaturase in psychrophile C. psychrerythraea 34H (Table 8). It is reasonable to suppose that at the very low temperatures of the environment P. ingrahamii might require fatty acids with more than one unsaturated bond. Also branched chain fatty acids are often found at cold temperatures. Based on enzyme sequences, precursors for synthesis of the branched-chain fatty acids could be generated as intermediates in breakdown of the amino acids leucine, isoleucine and valine. However, neither polyunsaturated nor branched chain fatty acids were detected in P. ingrahamii cultures grown at about 6 to 8°C (i.e. refrigerator) conditions [25]. Perhaps the polyunsaturated fatty acids are produced at lower temperatures or were not detected by the fatty acid analysis procedure used.
Table 8.
Enzymes of macromolecule hydrolysis
| Macromolecule | Gene | Enzyme |
| Peptides, protein | 3027 | D-alanyl-D-alanine carboxypeptidase, serine-type |
| 3293 | D-alanyl-D-alanine carboxypeptidase/D-alanyl-D-alanine-endopeptidase | |
| 314 | O-sialoglycoprotein endopeptidase | |
| 3325 | prepilin peptidase type 4 | |
| 894 | aminoacyl-histidine dipeptidase | |
| 1331 | aminopeptidase | |
| 2545 | aminopeptidase M24 | |
| 2344 | aminopeptidase N | |
| 1777 | aspartyl aminopeptidase | |
| 1034 | carboxypeptidase thermostable | |
| 2765 | deacylase/carboxypeptidase family member, Zn-dependent | |
| 409 | dipeptidase | |
| 395 | leucyl aminopeptidase | |
| 2026 | metallopeptidase M24 family | |
| 3003 | methionine aminopeptidase, type I | |
| 212 | oligopeptidase A | |
| 633 | oligopeptidase B | |
| 2690 | peptidase | |
| 1673 | peptidase C1A, papain | |
| 2671 | peptidase C26 | |
| 268 | peptidase M14, carboxypeptidase A | |
| 2457 | peptidase M14, carboxypeptidase A | |
| 2444 | peptidase M15B | |
| 3480 | peptidase M16 domain protein | |
| 2127 | peptidase M19, renal dipeptidase | |
| 2301 | peptidase M22, glycoprotease | |
| 1544 | peptidase M23B | |
| 3212 | peptidase M23B | |
| 3225 | peptidase M23B | |
| 676 | peptidase M23B, peptidoglycan-binding | |
| 2784 | peptidase M24 | |
| 1800 | peptidase M48, Ste24p | |
| 926 | peptidase M48, Ste24p, Zn-dependent, TPR repeats | |
| 686 | peptidase M50 | |
| 3180 | peptidase M50 | |
| 1350 | peptidase M56, BlaR1 | |
| 2144 | peptidase M6, immune inhibitor A | |
| 2723 | peptidase S1 and S6, chymotrypsin/Hap | |
| 994 | peptidase S16, LON domain protein | |
| 925 | peptidase S49 | |
| 1972 | peptidase S49, N-terminal domain protein | |
| 1301 | peptidase U32 | |
| 2189 | peptidase U32 family | |
| 2460 | peptidase dimerization domain protein | |
| 410 | peptidase domain protein | |
| 478 | prepilin peptidase dependent protein D | |
| 606 | proline aminopeptidase P II | |
| 253 | proline iminopeptidase | |
| Polysaccharides | 1954 | alpha amylase, catalytic region |
| 3067 | predicted glucoamylase I (alpha-1,4-glucan glucosidase) | |
| 2381 | alpha-D-1,4-glucosidase | |
| 2383 | dextran glucosidase | |
| 554 | glucan endo-1,3-beta-D-glucosidase | |
| 893 | glycogen debranching enzyme | |
| 2363 | glycogen debranching enzyme | |
| 3070 | glycogen debranching enzyme | |
| 558 | glycoside hydrolase family | |
| 2014 | glycoside hydrolase family | |
| 2529 | glycoside hydrolase family | |
| 2841 | glycoside hydrolase family | |
| Murein | 1782 | gamma-D-glutamate-meso-diaminopimelate muropeptidase |
| 1075 | lytic murein transglycosylase | |
| 499 | lytic murein transglycosylase, catalytic | |
| 293 | lytic transglycosylase, catalytic protein | |
| 3319 | lytic transglycosylase, catalytic protein | |
| 293 | lytic transglycosylase, catalytic protein | |
| 367 | lytic transglycosylase, catalytic protein | |
| 3319 | lytic transglycosylase, catalytic protein | |
| Lipids | 1779 | esterase/lipase/thioesterase family protein |
| 1892 | lipase, class 3 | |
| 2631 | lipase-like | |
| 2470 | lipase/acylhydrolase family protein, GDSL-lilke | |
| 258 | phospholipase | |
| 3493 | phospholipase A(1) | |
| 2455 | phospholipase D/transphosphatidylase | |
| 1334 | phospholipase family, patatin-like protein | |
| 1844 | phospholipase family, patatin-like protein | |
| 3290 | predicted lysophospholipase | |
| Nucleic acids | 2776 | 5'-3' exonuclease |
| 201 | ATP-dependent endonuclease of the OLD family | |
| 501 | DNA mismatch repair endonuclease mutH | |
| 2451 | HNH endonuclease | |
| 317 | TatD-related deoxyribonuclease | |
| 807 | TatD-related deoxyribonuclease | |
| 716 | crossover junction endodeoxyribonuclease | |
| 1096 | deoxyribonuclease of TatD family | |
| 732 | endonuclease III | |
| 1020 | endonuclease/exonuclease/phosphatase | |
| 1518 | endonuclease/exonuclease/phosphatase | |
| 2456 | endonuclease/exonuclease/phosphatase | |
| 2694 | endonuclease/exonuclease/phosphatase | |
| 3327 | endonuclease/exonuclease/phosphatase | |
| 416 | endoribonuclease L-PSP | |
| 2129 | endoribonuclease L-PSP | |
| 2646 | endoribonuclease L-PSP | |
| 369 | excinuclease ABC, A subunit | |
| 2085 | excinuclease ABC, A subunit | |
| 1082 | excinuclease ABC, B subunit | |
| 1193 | excinuclease ABC, C subunit | |
| 2436 | exodeoxyribonuclease I | |
| 1319 | exodeoxyribonuclease III | |
| 1460 | exodeoxyribonuclease V, alpha subunit | |
| 1459 | exodeoxyribonuclease V, beta subunit | |
| 1458 | exodeoxyribonuclease V, gamma subunit | |
| 2951 | exodeoxyribonuclease VII, large subunit | |
| 2238 | exodeoxyribonuclease VII, small subunit | |
| 1303 | exonuclease SMC domain protein | |
| 1302 | exonuclease SbcCD, D subunit | |
| 2586 | exonuclease of the beta-lactamase domain protein | |
| 2270 | exoribonuclease II | |
| 2580 | extracellular deoxyribonuclease | |
| 53 | formamidopyrimidine-DNA glycosylase | |
| 2029 | predicted endoribonuclease L-PSP | |
| 2233 | predicted exonuclease | |
| 3302 | single-stranded-DNA-specific exonuclease | |
| 1283 | uracil-DNA glycosylase | |
| 1819 | predicted ribonuclease BN | |
| 3482 | predicted ribonuclease BN | |
| 1668 | ribonuclease D | |
| 496 | ribonuclease H | |
| 2962 | ribonuclease HII | |
| 640 | ribonuclease III | |
| 3609 | ribonuclease P protein component | |
| 3479 | ribonuclease PH | |
| 3417 | ribonuclease R | |
| 2450 | ribonuclease T | |
| 1126 | ribonuclease, Rne/Rng family | |
| 2208 | ribonuclease, Rne/Rng family | |
| 2214 | tRNA-guanine transglycosylase | |
| 2567 | nuclease (SNase domain protein) | |
| 2838 | nuclease (SNase domain protein) | |
| 265 | exonuclease, RNase T and DNA polymerase III | |
| 968 | exonuclease, RNase T and DNA polymerase III | |
| 3335 | exonuclease, RNase T and DNA polymerase III |
Interestingly, two polyunsaturated fatty acids, 20:5 (eicosapentaenoic acid) and 22:6 (docosahexaenoic acid), have been reported from two other species of Psychromonas, P. kaikoae and P. marina[25].
Glycogen storage
There are genes for 6 glucose-1-phosphate adenyltransferases (299, 1296, 2063, 3033,3034 and 3464) any one of which could serve for the first step in synthesis of glycogen, and there are 2 glycogen/starch synthases (2348 and 3035). One or more of the 15 glycosyl transferases could be involved in synthetic reactions.
Digestion of macromolecules
Like other marine organisms, P. ingrahamii appears to have the capability of utilizing macromolecules in the environment for nutrition and energy. P. ingrahamii has genes for a relatively large number of 48 peptidases and proteases (Table 8). Some of these are no doubt required for internal turnover, but some seem likely to be exported out of the cell in order to hydrolyze environmental proteins, thus providing small molecular weight nutrients for uptake. P. ingrahamii has a complete general secretion system capable of excreting such degradative enzymes to the environment. To take up the digestion products of proteolysis, there are ABC-type transporters for peptides, many for amino acids. Not consistent with this prediction is the experimental observation that gelatin is not hydrolysed [24].
Storage glycogen as well as external polysaccharides including starch could be hydrozysed for production of sugars to supply energy. For utilization of some polysaccharides there are amylases, glucosidases, debranching enzymes, and glycosyl hydrolases (Table 8), some of which may be intracellular and others extracellular. There are 7 lytic transglycosylases which, in cleaving peptidoglycan links could be involved in modellingof the cell or could break up environmental cell wall fragments. Capability to hydrolyse fats also exists as there are 3 lipases, 5 phospholipases and a lyso-lipase. There are many enzymes hydrolyzing nucleic acids with different specificities and functions, many with vital internal metabolic roles. In addition, some may be used to hydrolyze external nucleic acid debris (Table 8).
Other hydrolases are encoded in the genome whose physiological roles are not currently known, for example there are 11 HAD-superfamily hydrolases, 7 alpha/beta hydrolase fold proteins, 5 metal-dependent phosphohydrolases.
Chaperones and stress proteins
Multiple chaperone proteins are encoded in P. ingrahamii, suggesting that folding of proteins is an important process (Table 9). There are 4 proteins like DnaK, 4 like DnaJ, 3 GroEL monomers and 2 GroES monomers There are 12 peptidylprolyl isomerases (trigger factors that act at nascent polypeptide chains), and a ClpB protein disaggregating complex. One can speculate that the the role of the chaperones is to guide nascent polypeptides into functional three-dimensional configurations permitting activity at low temperatures. Future characterization of some of these chaperones could reveal what kinds of folding are required to retain protein function at sub-zero temperatures. Ferrer et al. [26] found that GroEL of Oleispira antartica RB8 functioned as a single ring of 7 units at cold temperatures, but as a double ring of 7 over 7 at warm temperatures. They pinpointed two residues as critical to the transition from double to single ring. However on inspection these residues do not occur at the comparable positions in P. ingrahamii GroEL proteins. Actions of P. ingrahamii chaperones at cold temperatures remain to be explored.
Table 9.
Chaperones and stress proteins
| Category | Gene numbers | Protein |
| Chaperones | 917, 1232, 1233, 1328 | DnaK-like |
| 918, 1039, 2621, 2499 | DnaJ-like | |
| 843, 2494 | GroES | |
| 844, 2493, 2791 | GroEL | |
| 919, 1049, 1080, 1469, 1619, | Peptidyl prolyl isomerases | |
| 1856, 1917, 2185, 3116, 3199, | (trigger factors) | |
| 3257, 3269 | ||
| 1040, 3623 | ClpB disaggregator | |
| Stress proteins | 279, 755, 1097, 1881, 1953, | Cold Shock |
| 2158, 2543, 2698,2701, 3095, | ||
| 3098, 3704 | ||
| 3, 95, 202, 956, 1039, 1051, | Heat shock | |
| 1246, 1533, 1806, 1916, 2499, | ||
| 2692 | ||
| 125, 930, 954, 955, 959, 1234, | Universal stress proteins | |
| 2734 | ||
| 378, 1265, 1264, 1265, 1266, | Tellurite resistance | |
| 2005, 2574, 2575 | (anti_superoxide) |
P. ingrahamii has genes for a variety of known types of stress proteins: There are 12 cold shock proteins, 9 heat shock proteins, 7 UspA-type stress proteins as well as 9 "tellurite resistance" proteins now known to protect against superoxide formation [27] (Table 9). Experimental work will be needed to determine which if any of these have functions directed specifically at living in cold temperatures and if there are other types of stress proteins or any other cold-associated functions among the open reading frames of unknown function.
Transporters
Compared to E. coli, P. ingrahamii has few transporters specific for sugars and sugar alcohols, but does have many transporters for amino acids (see Additional file 2). In this respect, it seems that P. ingrahamii is like S. oneidensis and other Alteromondales in a capacity to utilize environmental amino acids as carbon (and nitrogen) sources, contrasted to less capacity for sugars.
As to types of transporters, the ABC type of ATP-driven multisubunit transporter is most common in the P. ingrahamii genome, as is the case for other bacteria. Also, as for many other bacteria, conventional secondary transporters follow in frequency. However, P. ingrahamii differs from many in the fact that in there are 11 sets of the dctM, dctP and dctQ genes for the tripartite ATP-independent periplasmic transporter systems (TRAP) [28,29] (Table 10), more than are found in the three mesophiles whose whole genomes were compared (E. coli, S. oneidensis and V. cholerae). TRAP systems specialize in transport of C4-dicarboxylic organic acids such as fumarate, perhaps for anaerobic respiration purposes. The number of 3-gene TRAP systems in bacteria is variable. E. coli has one, Pseudomonas aeruginosa has 6 whereas P. ingrahamii has 11 of these three-protein systems. Recently 15 TRAP systems have been identified in Sinorhizobium meliloti 1021 [30]. TRAP transporters use a proton motive force energized system, simpler and possibly more primitive than the ATP-utilizing ABC systems. A connection between the TRAP transporters and low temperature growth is not currently known.
Table 10.
Tripartite ATP-independent C4-dicarboxylate transporters
| DctM-like | DctQ-like | DctP-like |
| IIM* subunit | IIM* subunit | Periplasmic subunit |
| Gene | Gene | Gene |
| 133 | 134 | 135 |
| 538 | 539 | 540 |
| 572 | 571 | 570 |
| 646 | 647 | 648 |
| 710 | 709 | 708 |
| 2033 | 2034 | 2035 |
| 2595 | 2595 | 2594 |
| 2935 | 2934 | 2933 |
| 3148 | 3149 | 3150 |
| 3544 | 3545 | 3546 |
| 3673 | 3674 | 3675 |
*IIM = integral inner membrane
Regulators
Many types of regulation mechanisms have been identified in P. ingrahamii: transcriptional activators and repressors, cyclic-AMP regulation, chemotaxis systems, two-component sensor-response regulators of several types including the twin-arginine translocation (Tat) pathway signal sequence domain proteins, also synthesis/breakdown of cyclic-diGMP signalling second messengers associated with GGDEF and/or EAL domains (see Additional file 2). There are 61 regulators of the cyclic-diGMP signalling second messenger in the genome, compared to 29 in E. coli K-12 MG1655. Cyclic-diGMP concentrations are controlled by either a diguanylate cyclase or a specific phosphodiesterase or both, together governing synthesis or hydrolysis of the cyclic-GMP. The types of genes and corresponding physiology regulated by cyclic diguanylate systems so far identified are motility, adhesion factors, fimbriae and biofilm formation.
Given the finding of a large number of cyclic-diGMP signalling systems, we might guess that P. ingrahamii lives within the matrix of a biofilm. Altogether 16 glycosyl transferases are encoded (by genes 326, 327, 328, 329, 331, 336, 440, 449, 454, 456, 457, 779, 792, 1794, 3458, and 3647), suggesting that polysaccharide, perhaps exopolysaccharide, is a major synthetic product. For export, many efflux proteins are present. An ortholog is present (1200) of the quorum sensing HapR (LuxR) regulator that is involved in controlling biofilm formation in V. cholerae [31,32]. An extracellular matrix, a major physiological feature of the related S. oneidensis [33], could well be important to life in the cold, providing stability and resilience to the population. Since P. ingrahamii lives in sea ice, it is possible that extracellular polysaccharide (EPS) may be part of the sea ice microbial community (SIMCO) biofilm although it should be noted that P. ingrahamii was isolated from the ice column above the major biofilm of the SIMCO.
Alternatively, the production of EPS may serve a role in sequestering water from ambient saltwater at lower temperatures or actually lowering the freezing point. In this regard it is interesting to note that in the -12°C growth experiments, none of the tubes froze after the first week of growth following inoculation suggesting that a product, possibly EPS, produced by the bacterium may have lowered the freezing point of the growth medium.
Regulation of expression of certain classes of genes is moderated by the sigma factors of the RNA polymerase holoenzyme. P. ingrahamii is well endowed, appearing to have genes for sigma factors 24 (RpoE), 32 (RpoH), 38 (RpoS), 54 (RpoN) and 70(RpoD) [gene numbers are, respectively, 65, 626, 677, (424, 712, 2892 and 3175 for sigma-54) and (310, 946 and 995 for sigma-70)]. RpoE and RpoH are both stress-responding factors. RpoS operates in stationary phase and also under stress. RpoN is concerned with nitrogen metabolism and in V. cholerae regulates flagellin gene transcription. RpoD for sigma 70 is the primary factor in E. coli.
Ribosomes
There are 58 ribosomal proteins annotated, 5 of which are duplicated, therefore 53 unique. Of these, 38 were found as orthologs with Pam values less than 40 among the two sets of genomes we examined. Organisms with closest matches were, in descending order, V. cholerae, V. parahaemolyticus, S. oneidensis, H. influenzae = I. loihiensis, C. psychrerythraea. Again, the close relationship of P. ingrahamii with Vibrio species is evident.
Osmotic stability
P. ingrahamii grows well over the range 1 to 12% NaCl [25]. To manage the potential for osmotic imbalance, the genes for enzymes to synthesize the osmolyte glycine betaine from choline are present (genes 2071, 2072), as well as a transporter to take up choline (gene 969) or, bypassing synthesis, specific ABC transport systems for uptake of glycine betaine are present, (genes 614–616, 2073–2075). This capability may explain how the organism is able to survive and perhaps grow in the salt pockets that are formed within the sea ice.
Motility
There is a large cluster of flagellar genes in P. ingrahamii in the region between gene numbers 3562 to 3598 and four copies of sigma factor rpoN which is known in V. cholerae to regulate flagellin genes. Yet the bacteria in culture have been observed to be non-motile [8]. There may be a defect in one of the essential flagellar proteins or in the expression or assembly processes. Alternatively, the organism may not always express flagella formation and motility. Interestingly, the original description of the genus indicates that other members of the genus are motile [34].
Gas vesicles
Two kinds of gas vesicles have been observed in the cell under culture (24). Genes orthologous to known gas vesicle genes are present in two clusters, the ranges 1248 to 1262 and 1748 and 1750. Although the two types can be expressed simultaneously in cells, they may be differentially expressed under preferential conditions in the environment.
Different kinds of enzyme orthologs in different bacterial relatives
Of the 53 bacteria chosen to represent the breadth of the currently characterized eubacterial world, the three bacteria bearing the largest number of protein sequences scored as "best matches" with P. ingrahamii proteins are V. cholerae, S. oneidensis and C. psychrerythraea 34H (Table 3).
About 3/4 of the P. ingrahamii proteins annotated as enzymes, transporters or regulators have orthologs at the level of Pam <= 125 in either V. cholerae, S. oneidensis or C. psychroerythraea. By contrast, only 19% of P. ingrahamii CDSs of unknown function have orthologs in these bacteria. That the vast majority, 4/5, of P. ingrahamii CDSs of unknown function do not have good matches in the most closely related organisms suggests that many proteins in P. ingrahamii are qualitatively different. New, unique functions will not be revealed by annotations that depend on similarity to known proteins, but their existence suggests that further experimental study of this extremophile is worthwhile and could bring new biology to light.
Looking within the enzyme category, it is striking that most of the best hit homologs of enzymes of central metabolism are proteins of V. cholerae. This is particularly puzzling since V. cholerae is a mesophile and it is phylogenetically in a different Order and Family from P. ingrahamii. Could this be an example of massive horizontal transfer of genes from Vibrio to Psychromonas? It seems unlikely since the many genes for central metabolism are not in a few clusters similar to pathogenicity islands, rather they map throughout the chromosome, and the majority of the transposase-type genes have closest orthologs in S. oneidensis. Nevertheless, in spite of expectations, we have found that the sequences of enzymes of basic metabolism that function at low temperature are more similar to those of a mesophile than they are to those of another psychrophile, and even more surprising, a mesophile of a different taxonomic Order and Family. A feature that may suggest global genome reorganization and account for an apparent scrambling of otherwise conserved genes is that the Vibrios usually comprise two chromosomes, in contrast to P. ingrahamii which has only one.
Different from V. cholerae, one sees that enzyme homologs in S. oneidensis and C. psychrerythraea 34H are largely the enzymes of peripheral and macromolecule metabolism, not the enzymes of central metabolism.
We narrowed our question, looking for a commonality in enzymes among the psychrophiles. We identified the 566 proteins in C. psychrerythraea and I. hoiliensis that have highest similarity to P. ingrahamii proteins at Pam values =< 125. In this set, there are few enzymes important to central metabolism; many more are involved in macromolecule synthesis and maintenance. There are enzymes dealing with nucleic acids: exo and endo-nucleases, RNA helicases, DNA polymerase and helicases, transcriptional and translational factors, and recombinases. Psychrophily may require differences in the proteins of nucleic acid metabolism since even though the salt content of the sea ice and the low GC content of the DNA (40.1%) should act to lower the melting point of double stranded polynucleotides. However, handling nucleic acid structures at such cold temperatures may also require differences in the interacting proteins. Similarly, transcription and translation factors of a particular kind may be required to enable the processes at low temperatures.
Unique unknown proteins
We identified a few small groups of proteins that could perform novel functions related to psychrophily, warranting further investigation in the future. A contiguous set of nine genes that could comprise an operon, gene numbers 3053 through 3061 have no orthologs in current databases with Pam value <150. Four of the proteins are paralogous, with similar sequences among themselves. As we are looking for functions in psychrophiles not found in other bacteria, experimental characterization of this group of proteins might be worthwhile.
There are 3 groups of genes that could be contiguous operons whose members reside either in cluster 4 of the Correspondence Analysis (vide supra), or are unknown in other bacteria to date, or both. These are clusters 1672 through 1676, 1960 through 1964, and 2315 through 2319. Could any or all of these relate to the ability to live and grow at -12C or lower temperatures? We identify these seemingly unique sets of proteins with the thought that when results of proteomic expression experiments on this organism are available, it might be useful to characterize the proteins and to know under what circumstances they are synthesized.
Conclusion
The P. ingrahamii genome, although it is 25% smaller than the genome of E. coli K-12 MG1655 and has many more CDSs labeled hypothetical, is similar to E. coli in the distribution of functions among annotated proteins, with enzymes by far the largest category followed by transporters, then regulators. Other categories follow distantly. Similar to E. coli, the largest paralogous groups within the genome were transporters and regulators, the smaller ones enzymes. Although transporters and regulators are fewer in number than enzymes, they form larger paralogous groups. They belong to fewer families employing fewer mechanisms than is the case for enzymes which belong to smaller families of greater variety.
Unexpectedly, P. ingrahamii protein sequences were similar to more V. cholerae proteins than to proteins from S. oneidensis or C. psychrerythraea. This is in spite of the fact that Vibrio species belong to a different Family and Order than do Psychromonadaceae, Shewanellaceae and Collwelliaceae, and in spite of the fact that C. psychrerythraea is also a psychrophile.
In a comparison of gross properties of all the proteins of P. ingrahamii differences were not found from those of three mesophiles in several respects: distribution of lengths, total amino acid composition, codon usage when corrected for genomic GC content.
However, correspondence analysis (CA) of the amino acid content of the proteins showed that they cluster in ways that differ from most other bacteria, falling into more clusters that are not as well separated from one another as in other bacteria. This may be a consequence of an unusual distribution of hydrophobicity. One of the clusters is composed almost half of unidentified, unknown CDSs. P. ingrahamii contains proteins with relatively high asparagine content, and low content of amino acids potentially sensitive to the higher concentration of oxygn present in cold waters. These properties would seem to be appropriate for an extreme psychrophile.
As to metabolism, P. ingrahamii is a facultative anaerobe capable of both fermentation and respiration. In agreement, proteins similar to the corresponding metabolic pathways and proteins for synthesis and harvest of glycogen storage compound were identified. Most enzymes of central small molecule metabolism were most closely related to those of V. cholerae. Enzymes of macromolecular synthesis and maintenance are most closely related to those of S. oneidensis and C. psychrerythraea. There seems to be a preference of amino acids over sugars as sources of carbon and energy, perhaps harvested by extracellular degradation of environmental proteins by exported peptidases. To create a more flexible lipid layer adjusted to cold temperatures, sequences of a heteropolymeric polyunsaturase were found. However, the enzyme may not always be active as no polyunsaturated fatty acids were detected in culture. Perhaps, however, in nature they are expressed at temperatures lower than the lowest normally used for laboratory cultivation, 6–8°C.
In addition to transporters of types common to other proteobacteria were 11 three-component TRAP systems for C4-dicarboxylic acid transport. In addition to regulators of types common to other proteobacteria were 61 regulators of cyclic-diGMP second messengers, suggesting that biofilms and their regulation could be an important part of the life style of this psychrophile. Alternatively, P. ingrahamii may produce EPS to lower the freezing point in the surrounding environment, thereby making water available for low temperature growth.
Looking for particular proteins that might be unique to this psychrophile but are not yet in the genomic databases we searched, we have pointed out some sets of gene products that could be starting points for discovering new functions or new types of proteins. We noticed 9 contiguous genes that are all unknown hypotheticals, 4 of them sequence-related to one another. We also noted three apparent contiguous operons, members of which are unknown hypotheticals and/or members of the unique set of proteins in cluster 4. In future, we recommend that proteomic experiments should be used to explore the shift to cold temperature with a view of keeping an eye out for expression of any of these proteins, potentially part of the mechanisms of life at very cold temperatures.
Methods
Genome sequencing and finishing
The genome of Psychromonas ingrahamii was sequenced at the Joint Genome Institute Lawrence Laboratories, Walnut Creek, CA using a combination of (4 kb, 6.8 kb and 36 kb) DNA libraries. All general aspects of library construction and sequencing performed can be found on line [35]. Draft assemblies were based on 53473 total reads. All three libraries provided 11X coverage of the genome. For closing and finishing, the Phred/Phrap/Consed software package [36,37] was used for sequence assembly and quality assessment [38,39]. After the shotgun stage, 53473 reads were assembled with parallel phrap (High Performance Software, LLC). Possible mis-assemblies were corrected with Dupfinisher [41] or transposon bombing of bridging clones (Epicentre Biotechnologies, Madison, WI). Gaps between contigs were closed by editing in Consed, custom primer walks, or PCR amplification. A total of 952 primer walk reactions and 3 transposon bombs were necessary to close gaps, to resolve repetitive regions, and to raise the quality of the finished sequence. The completed genome sequences of Psychromonas ingrahamii contain 53,592 reads, achieving an average of 11-fold sequence coverage per base with an error rate less than 1 in 100,000. The sequence of P. ingrahamii can be accessed using the GenBank accession number CP000510.1.
Annotation
Annotation was initially carried out at Oak Ridge National Laboratory using methods detailed in [41,42]. Further annotation and analysis of protein sequence similarities used the Darwin system (Data Analysis and Retrieval With Indexed Nucleotide/peptide sequence package), version 2.0, developed at the ETHZ in Zurich, Switzerland [16,22]. Pairwise sequence alignments and scores were generated using the AllAllDb program of Darwin. Maximum likelihood alignments are generated with an initial global alignment by dynamic programming (Smith and Waterman algorithm) followed by dynamic local alignments (Needleman and Wunsch algorithm). A single scoring matrix is used for these steps. After the initial alignment, the scoring matrix is adjusted to fit the approximate distance between each protein pair to produce the minimum Pam value. Pam units are defined as the numbers of point mutations (base pair differences) per 100 residues [17]. The Darwin system's ability to apply scoring matrices according to the distance between each protein pair ensures a data set of highly accurate similarity calculations for distantly as well as closely related protein pairs. The identification of distantly related proteins is valuable in finding divergent but related protein functions.
To extract homolog matches from initial data, we required that the alignments with P. ingrahamii proteins be at least 83 residues long [43], and that the alignment must represent over 40% of both proteins. Unless otherwise stated we required Pam scores to be 150 or less. To assemble groups of paralogs, the Pam threshold for pairs was raised to 250, and then pairs were grouped by a transitive process as previously described [23].
Correspondence Analysis
The amino acid composition of the proteins of P. ingrahamii was analyzed using correspondence analysis (CA) [12-14] with the FactoMineR R package [44]. Each protein was truncated for the first 10 and last 5 amino acid residues and each is represented by its normalized aminoacid content. The representation is in a 19-dimension space where specific statistical distances are measured by the chi-square method. The 3 most informative dimensions are used to plot the position of each individual protein. The individual amino acid distributions are superimposed in this same space. The axes are numbered in order of the amount of information they carry. A bayseian method, as in Bailly-Bechet et al. [41], was used to cluster proteins of similar composition using the Mclust R package [45], and the best classification, which is associated with the largest BIC (Bayesian Information Criterion) value, was selected for further analysis.
Codon usage and selection bias were determined by methods of Sharp et al. [9].
Authors' contributions
MR used Darwin-generated data for further annotation and analysis and wrote the paper. JTS made biological determinations and made significant contributions to writing the paper. AD and TZW carried out and interpreted the corresponence analysis. AD made significant contributions to writing the paper. JST and TSB closed and finalized the sequence. MLL and LJH carried out the initial annotation and submitted to GenBank.
Supplementary Material
Ping Correspodence Analysis. Correspondence analysis of amino acid content of Psychromonas ingrahamii proteins. Gene number, Cluster number, Product
Ping Annotations 2. Further annotation of Psychromonas ingrahamii proteins. Gene number, Gi identifier, predicted protein product, type of product
Acknowledgments
Acknowledgements
Thanks to Dr. Paul M. Sharp for carrying out the codon usage analyses. Thanks to Dr. Margrethe Serres for suggestions on reading the manuscript and, with Daniella Wilmot, for database and library support. Dr. John L. Ingraham generated the list of 53 representative bacteria. MR acknowledges support from DE-FG02-04ER63940. JTS acknowledges the support from the University of Washington NASA NAI program and the NSF Astrobiology IGERT program. TZW acknowledges support from a grant from the Fondation Fourmentin-Guilbert and AD acknowledges support from the European Union BioSapiens Network of Excellence, Grant LSHG CT-2003-503265. Sequencing and first round of annotation was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University ofCalifornia, Lawrence Livermore National Laboratory under Contract No. W-7405-Eng-48, Lawrence Berkeley National Laboratory under Contract No. DE-AC02-05CH11231, Los Alamos National Laboratory under Contract No. W-7405-ENG-36, and Oak Ridge National Laboratory under Contract No. DE-AC05-00OR22725.
Contributor Information
Monica Riley, Email: mriley@mbl.edu.
James T Staley, Email: jtstaley@u.washington.edu.
Antoine Danchin, Email: adanchin@pasteur.fr.
Ting Zhang Wang, Email: wangtz@pasteur.fr.
Thomas S Brettin, Email: brettin@lanl.gov.
Loren J Hauser, Email: hauserlj@ornl.gov.
Miriam L Land, Email: landml@ornl.gov.
Linda S Thompson, Email: lthompsonnm@comcast.net.
References
- Margesin R, Neuner G, Storey KB. Cold-loving microbes, plants, and animals-fundamental and applied aspects. Naturwissenschaften. 2006;94:77–99. doi: 10.1007/s00114-006-0162-6. [DOI] [PubMed] [Google Scholar]
- Jakosky BM, Nealson KH, Bakermans C, Ley RE, Mellon MT. Subfreezing activity of microorganisms and the potential habitability of Mars' polar regions. Astrobiology. 2003;3:343–50. doi: 10.1089/153110703769016433. [DOI] [PubMed] [Google Scholar]
- Marx J-C, Collins T, D'Amico S, Feller G, Gerday C. Cold-adapted enzymes from marine antarctic microorganisms. Marine Biotech. 2006;9:293–304. doi: 10.1007/s10126-006-6103-8. [DOI] [PubMed] [Google Scholar]
- Siddiqui KS, Cavicchioli R. Cold-adapted enzymes. Annu Rev Biochem. 2006;75:403–33. doi: 10.1146/annurev.biochem.75.103004.142723. [DOI] [PubMed] [Google Scholar]
- Methe BA, Nelson KE, Deming JW, Momen B, Melamud E, Zhang X, Moult J, Madupu R, Nelson WC, Dodson RJ, Brinkac LM, Daugherty SC, Durkin AS, DeBoy RT, Kolonay JF, Sullivan SA, Zhou L, Davidsen TM, Wu M, Huston AL, Lewis M, Weaver B, Weidman JF, Khouri H, Utterback TR, Feldblyum TV, Fraser CM. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc Natl Acad Sci USA. 2005;102:10913–10918. doi: 10.1073/pnas.0504766102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S, Saw JH, Lee KS, Freitas TA, Belisle C, Kawarabayasi Y, Donachie SP, Pikina A, Galperin MY, Koonin EV, Makarova KS, Omelchenko MV, Sorokin A, Wolf YI, Li QX, Keum YS, Campbell S, Denery J, Aizawa S, Shibata S, Malahoff A, Alam M. Genome sequence of the deep-sea gamma-proteobacterium Idiomarina loihiensis reveals amino acid fermentation as a source of carbon and energy. Proc Natl Acad Sci USA. 2004;101:18036–18041. doi: 10.1073/pnas.0407638102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medigue C, Krin E, Pascal G, Barbe V, Bernsel A, Bertin PN, Cheung F, Cruveiller S, D'Amico S, Duilio A, Fang G, Feller G, Ho C, Mangenot S, Marino G, Nilsson J, Parrilli E, Rocha EP, Rouy Z, Sekowska A, Tutino ML, Vallenet D, von Heijne G, Danchin A. Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 2005;15:1325–1335. doi: 10.1101/gr.4126905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breezee J, Cady N, Staley JT. Subfreezing growth of the sea ice bacterium "Psychromonas ingrahamii". Microbial Ecology. 2004;47:300–304. doi: 10.1007/s00248-003-1040-9. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Bailes E, Grocock RJ, Peden JF, Sockett RE. Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 2005;33:1141–1153. doi: 10.1093/nar/gki242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry J, Chessel D. Internal correspondence analysis of codon and amino-acid usage in thermophilic bacteria. J Appl Genet. 2003;44:235–261. [PubMed] [Google Scholar]
- Lobry JR, Necsulea A. Synonymous codon usage and its potential link with optimal growth temperature in prokaryotes. Gene. 2006;385:128–136. doi: 10.1016/j.gene.2006.05.033. [DOI] [PubMed] [Google Scholar]
- Hill MO. Correspondence analysis: a neglected multivariate method. Appl Statist. 1974;23:340–354. [Google Scholar]
- Bailly-Bechet , Danchin A, Iqbal M, Marsili M, Vergassola M. Codon usage domains over bacterial chromosomes. PloS Computational Biology. 2006;2:e37. doi: 10.1371/journal.pcbi.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal G, Medigue C, Danchin A. Persistent biases in the amino acid composition of prokaryotic proteins. Bioessays. 2006;28:726–738. doi: 10.1002/bies.20431. renumber. [DOI] [PubMed] [Google Scholar]
- Computational Biology at Oak Ridge National Laboratory http://genome.ornl.gov/microbial/ping/
- Gonnet GH, Hallett MT, Korostensky C, Bernardin L. Darwin v. 2.0: an interpreted computer language for the biosciences. Bioinformatics. 2000;16:101–103. doi: 10.1093/bioinformatics/16.2.101. [DOI] [PubMed] [Google Scholar]
- Schwartz RM, Dayhoff MO. In: Atlas of Protein Sequence and Structure. Dayhoff MO, editor. Vol. 5. Washington: National Medical Research Foundation; 1978. pp. 353–358. [Google Scholar]
- National Center for Biotechnology Information http://ncbi.nlm.nih.gov/genomes/
- Ivanova EP, Flavier S, Christen R. Phylogenetic relationships among marine Alteromonas-like proteobacteria: emended description of the family Alteromonadaceae and proposal of Pseudoalteromonadaceae fam. nov., Colwelliaceae fam. nov., Shewanellaceae fam. nov., Moritellaceae fam. nov., Ferrimonadaceae fam. nov., Idiomarinaceae fam. nov. and Psychromonadaceae fam. nov. Int J Syst Evol Microbiol. 2004;54:1773–88. doi: 10.1099/ijs.0.02997-0. [DOI] [PubMed] [Google Scholar]
- Bowman JP, McMeekin TA. OrderX. Alteromonadales ord. nov. In: Garrity GM, editor. Bergey's Manual of Systematic Bacteriology The Proteobacteria. 2. Vol. 2. New York: Springer-Verlag; 2005. pp. 443–491. [Google Scholar]
- Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Computational Biochemistry Research Group http://www.cbrg.ethz.ch/darwin
- Serres MH, Riley M. Gene fusions and gene duplications: relevance to genomic annotation and functional analysis. BMC Genomics. 2005;6:33. doi: 10.1186/1471-2164-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auman AJ, Breezee JL, Gosink JJ, Kämpfer P, Staley JT. Psychromonas ingrahamii sp. nov., a novel gas vacuolate, psychrophilic bacterium isolated from Arctic polar sea ice. Int J Syst Evol Microbiol. 2006;56:1001–1007. doi: 10.1099/ijs.0.64068-0. [DOI] [PubMed] [Google Scholar]
- Serres MH, Riley M. Genomic analysis of carbon source metabolism of Shewanella oneidensis MR-1: Predictions versus experiments. J Bacteriol. 2006;188:4601–4609. doi: 10.1128/JB.01787-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer M, Lunsdorf H, Chernikova TN, Yakimov M, Timmis K, Golyshin PN. Functional consequences of single:double ring transitions in chaperonins: life in the cold. Mol Microbiol. 2004;53:167–182. doi: 10.1111/j.1365-2958.2004.04077.x. [DOI] [PubMed] [Google Scholar]
- Perez JM, Calderon IL, Arenas FA, Fuentes DE, Pradenas GA, Fuentes EL, Sandoval JM, Castro ME, Elias AO, Vasquez CC. Bacterial toxicity of potassium tellurite: unveiling an ancient enigma. PLoS ONE. 2007;2:e211. doi: 10.1371/journal.pone.0000211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DJ, Thomas GH. The tripartite ATP-independent periplasmic (TRAP) transporters of bacteria and archaea. FEMS Microbiol Rev. 2001;25:405–424. doi: 10.1111/j.1574-6976.2001.tb00584.x. [DOI] [PubMed] [Google Scholar]
- Janausch IG, Zientz E, Tran QH, Kroger A, Unden G. C4-dicarboxylate carriers and sensors in bacteria. Biochim Biophys Acta. 2002;1553:39–56. doi: 10.1016/s0005-2728(01)00233-x. [DOI] [PubMed] [Google Scholar]
- Mauchline TH, Fowler JE, East AK, Sartor AL, Zaheer R, Hosie AH, Poole PS, Finan TM. Mapping the Sinorhizobium meliloti 1021 solute-binding protein-dependent transportome. Proc Natl Acad Sci USA. 2006;47:17933–8. doi: 10.1073/pnas.0606673103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. Erratum in: Mol Microbiol. 2004 51:1521. [DOI] [PubMed] [Google Scholar]
- Mueller RS, McDougald D, Cusumano D, Sodhi N, Kjelleberg S, Azam F, Bartlett DH. Vibrio cholerae strains possess multiple strategies for abiotic and biotic surface colonization. J Bacteriol. 2007;189:5348–60. doi: 10.1128/JB.01867-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormann KM, Duttler S, Saville RM, Hyodo M, Shukla S, Hayakawa Y, Spormann AM. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J Bacteriol. 2006;188:2681–2691. doi: 10.1128/JB.188.7.2681-2691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountfort DO, Rainey FA, Burghardt J, Kaspar HF, Stackebrandt E. Psychromonas antarcticus gen. nov., sp. nov., A new aerotolerant anaerobic, halophilic psychrophile isolated from pond sediment of the McMurdo ice shelf, antarctica. Arch Microbiol. 1998;169:231–238. doi: 10.1007/s002030050566. [DOI] [PubMed] [Google Scholar]
- Joint Genomes Institute http://www.jgi.doe.gov/
- Codon Code Corporation http://www.phrap.com
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Research. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Research. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Research. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Han CF, Chain P. Finishing repeat regions automatically with Dupfinisher. In: Arabnia HR, Valafar H, Las Vegas NV, editor. Proceeding of the International Conference on Bioinformatics & Computational Biology. CSREA Press; 2006. pp. 141–146. 26-29 June 2006. [Google Scholar]
- Scott KM, Sievert SM, Abril FN, Ball LA, Barrett CJ, Blake RA, Boller AJ, Chain PS, Clark JA, Davis CR, Detter C, Do KF, Dobrinski KP, Faza BI, Fitzpatrick KA, Freyermuth SK, Harmer TL, Hauser LJ, Hugler M, Kerfeld CA, Klotz MG, Kong WW, Land M, Lapidus A, Larimer FW, Longo DL, Lucas S, Malfatti SA, Massey SE, Martin DD, McCuddin Z, Meyer F, Moore JL, Ocampo LH, Jr, Paul JH, Paulsen IT, Reep DK, Ren Q, Ross RL, Sato PY, Thomas P, Tinkham LE, Zeruth GT. The genome of deep-sea vent chemolithoautotroph Thiomicrospira crunogena XCL-2. PLoS Biol. 2006;12:e383. doi: 10.1371/journal.pbio.0040383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Integrated Microbial Genomes system http://img.jgi.doe.gov/
- Altschul SF. Amino acid substitution matrices from an information theoretic perspective. J Mol Biol. 1991;219:555–565. doi: 10.1016/0022-2836(91)90193-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FactoMineR R package http://factominer.free.fr/
- Mclust R package http://www.stat.washington.edu/mclust/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ping Correspodence Analysis. Correspondence analysis of amino acid content of Psychromonas ingrahamii proteins. Gene number, Cluster number, Product
Ping Annotations 2. Further annotation of Psychromonas ingrahamii proteins. Gene number, Gi identifier, predicted protein product, type of product