Abstract
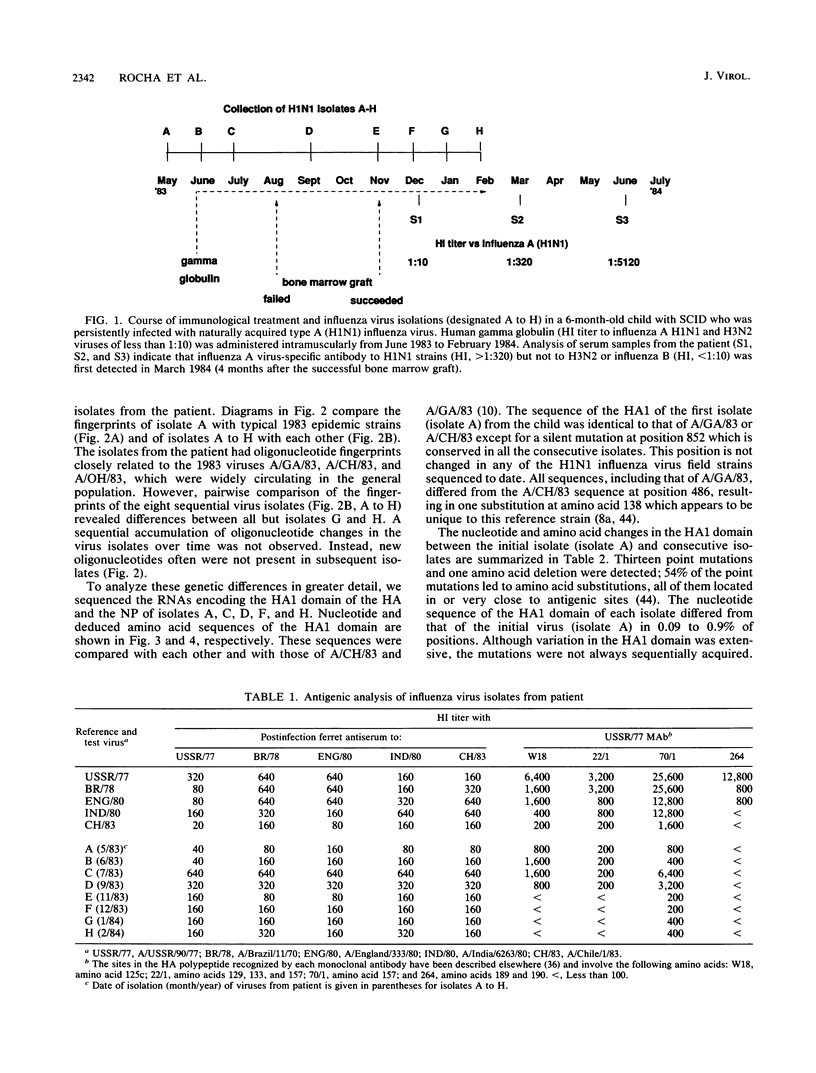
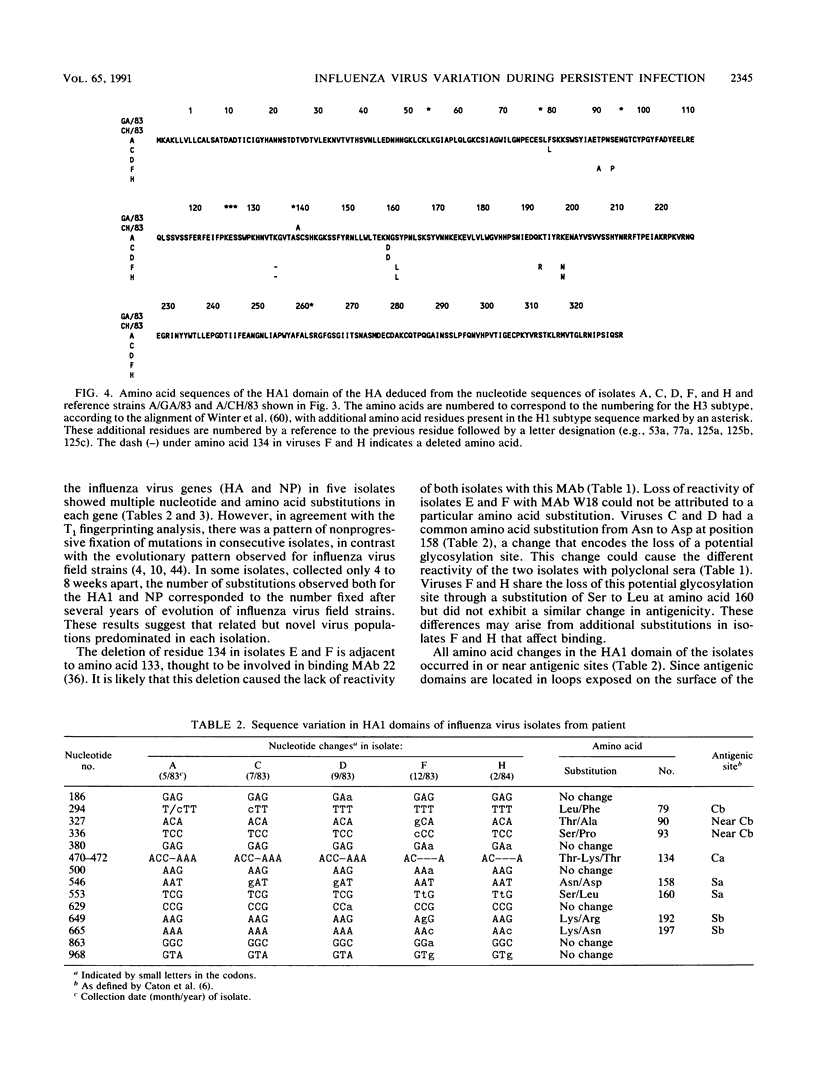
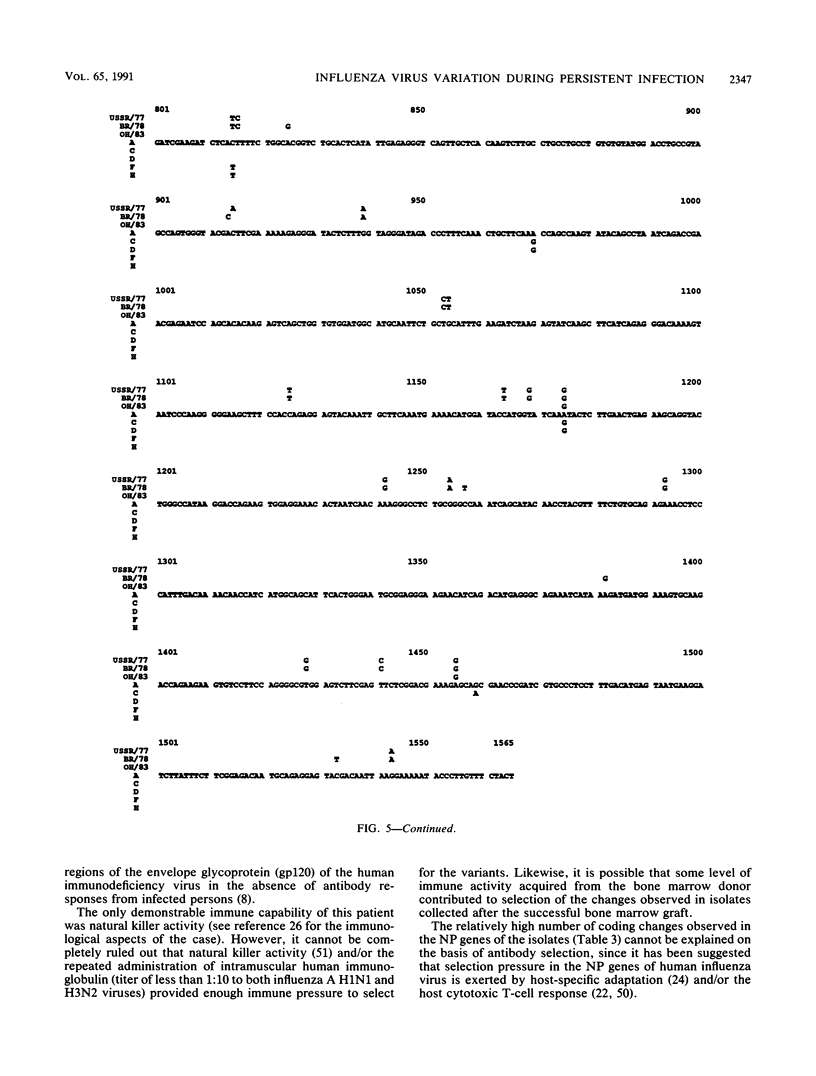
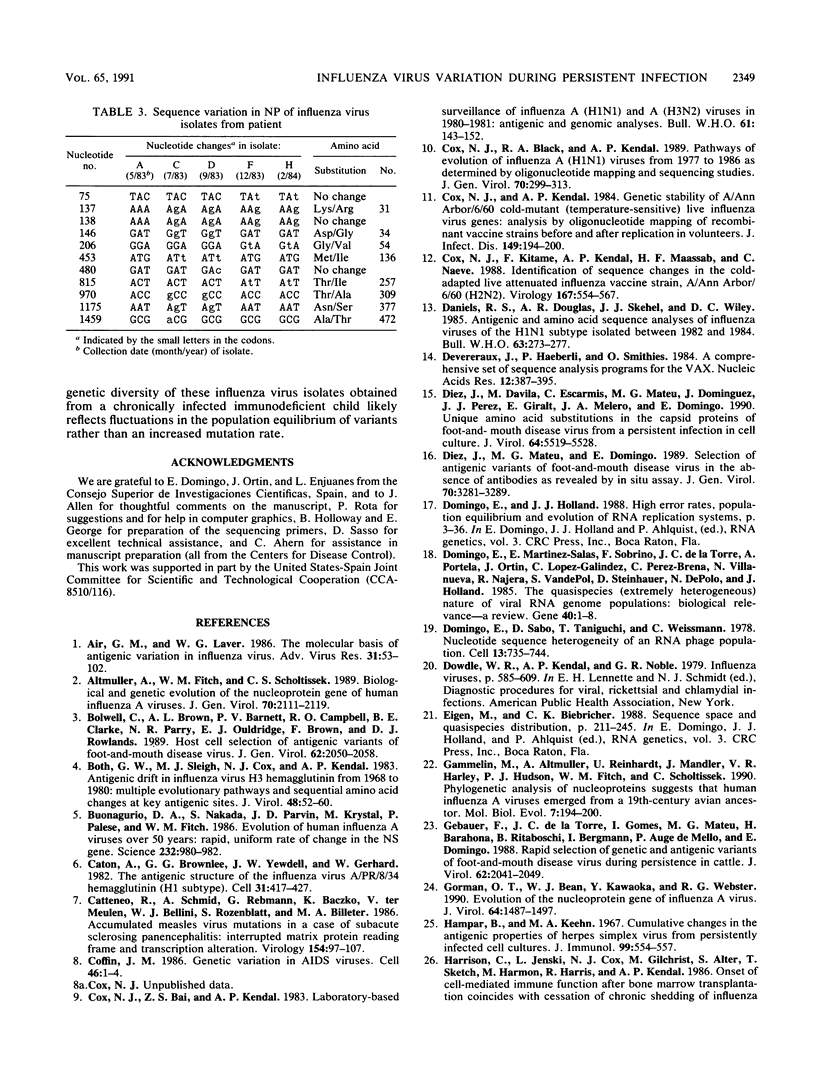
Antigenic and genetic variations have been analyzed in eight consecutive isolates recovered from a child with severe combined immunodeficiency syndrome persistently infected with naturally acquired type A (H1N1) influenza virus over a 10-month period. Hemagglutination inhibition reactions and T1 oligonucleotide fingerprinting demonstrated that these viruses were related to strains causing outbreaks in the United States at that time (1983 to 1984) but that antigenic and genetic differences between consecutive isolates could be detected. This variation between isolates was examined further by sequencing the RNAs encoding the HA1 region of the hemagglutinin (HA) and the nucleoprotein (NP) in five of the consecutive isolates. Multiple point mutations were detected in both genes, and a deletion of one amino acid was detected in the HA. Depending on the isolates compared, 5.8 x 10(-3) to 17 x 10(-3) substitutions per nucleotide site per year were detected in the RNAs encoding the HA1, and 3.5 x 10(-3) to 24 x 10(-3) substitutions per nucleotide site per year were detected in the NP gene. Fifty-four percent of the base changes in the HA1 and 73% in the NP led to amino acid substitutions. A progressive accumulation of mutations over time was not observed, suggesting that the genetic diversity of these viruses may best be interpreted as the result of shifts in the population equilibrium (quasi-species) of replicating variant genomes.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Air G. M., Laver W. G. The molecular basis of antigenic variation in influenza virus. Adv Virus Res. 1986;31:53–102. doi: 10.1016/s0065-3527(08)60262-6. [DOI] [PubMed] [Google Scholar]
- Altmüller A., Fitch W. M., Scholtissek C. Biological and genetic evolution of the nucleoprotein gene of human influenza A viruses. J Gen Virol. 1989 Aug;70(Pt 8):2111–2119. doi: 10.1099/0022-1317-70-8-2111. [DOI] [PubMed] [Google Scholar]
- Both G. W., Sleigh M. J., Cox N. J., Kendal A. P. Antigenic drift in influenza virus H3 hemagglutinin from 1968 to 1980: multiple evolutionary pathways and sequential amino acid changes at key antigenic sites. J Virol. 1983 Oct;48(1):52–60. doi: 10.1128/jvi.48.1.52-60.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonagurio D. A., Nakada S., Parvin J. D., Krystal M., Palese P., Fitch W. M. Evolution of human influenza A viruses over 50 years: rapid, uniform rate of change in NS gene. Science. 1986 May 23;232(4753):980–982. doi: 10.1126/science.2939560. [DOI] [PubMed] [Google Scholar]
- Caton A. J., Brownlee G. G., Yewdell J. W., Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell. 1982 Dec;31(2 Pt 1):417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- Cattaneo R., Schmid A., Rebmann G., Baczko K., Ter Meulen V., Bellini W. J., Rozenblatt S., Billeter M. A. Accumulated measles virus mutations in a case of subacute sclerosing panencephalitis: interrupted matrix protein reading frame and transcription alteration. Virology. 1986 Oct 15;154(1):97–107. doi: 10.1016/0042-6822(86)90433-2. [DOI] [PubMed] [Google Scholar]
- Coffin J. M. Genetic variation in AIDS viruses. Cell. 1986 Jul 4;46(1):1–4. doi: 10.1016/0092-8674(86)90851-2. [DOI] [PubMed] [Google Scholar]
- Cox N. J., Bai Z. S., Kendal A. P. Laboratory-based surveillance of influenza A(H1N1) and A(H3N2) viruses in 1980-81: antigenic and genomic analyses. Bull World Health Organ. 1983;61(1):143–152. [PMC free article] [PubMed] [Google Scholar]
- Cox N. J., Black R. A., Kendal A. P. Pathways of evolution of influenza A (H1N1) viruses from 1977 to 1986 as determined by oligonucleotide mapping and sequencing studies. J Gen Virol. 1989 Feb;70(Pt 2):299–313. doi: 10.1099/0022-1317-70-2-299. [DOI] [PubMed] [Google Scholar]
- Cox N. J., Kendal A. P. Genetic stability of A/Ann Arbor/6/60 cold-mutant (temperature-sensitive) live influenza virus genes: analysis by oligonucleotide mapping of recombinant vaccine strains before and after replication in volunteers. J Infect Dis. 1984 Feb;149(2):194–200. doi: 10.1093/infdis/149.2.194. [DOI] [PubMed] [Google Scholar]
- Cox N. J., Kitame F., Kendal A. P., Maassab H. F., Naeve C. Identification of sequence changes in the cold-adapted, live attenuated influenza vaccine strain, A/Ann Arbor/6/60 (H2N2). Virology. 1988 Dec;167(2):554–567. [PubMed] [Google Scholar]
- Daniels R. S., Douglas A. R., Skehel J. J., Wiley D. C. Antigenic and amino acid sequence analyses of influenza viruses of the H1N1 subtype isolated between 1982 and 1984. Bull World Health Organ. 1985;63(2):273–277. [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez J., Mateu M. G., Domingo E. Selection of antigenic variants of foot-and-mouth disease virus in the absence of antibodies, as revealed by an in situ assay. J Gen Virol. 1989 Dec;70(Pt 12):3281–3289. doi: 10.1099/0022-1317-70-12-3281. [DOI] [PubMed] [Google Scholar]
- Domingo E., Martínez-Salas E., Sobrino F., de la Torre J. C., Portela A., Ortín J., López-Galindez C., Pérez-Breña P., Villanueva N., Nájera R. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance--a review. Gene. 1985;40(1):1–8. doi: 10.1016/0378-1119(85)90017-4. [DOI] [PubMed] [Google Scholar]
- Domingo E., Sabo D., Taniguchi T., Weissmann C. Nucleotide sequence heterogeneity of an RNA phage population. Cell. 1978 Apr;13(4):735–744. doi: 10.1016/0092-8674(78)90223-4. [DOI] [PubMed] [Google Scholar]
- Díez J., Dávila M., Escarmís C., Mateu M. G., Dominguez J., Pérez J. J., Giralt E., Melero J. A., Domingo E. Unique amino acid substitutions in the capsid proteins of foot-and-mouth disease virus from a persistent infection in cell culture. J Virol. 1990 Nov;64(11):5519–5528. doi: 10.1128/jvi.64.11.5519-5528.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammelin M., Altmüller A., Reinhardt U., Mandler J., Harley V. R., Hudson P. J., Fitch W. M., Scholtissek C. Phylogenetic analysis of nucleoproteins suggests that human influenza A viruses emerged from a 19th-century avian ancestor. Mol Biol Evol. 1990 Mar;7(2):194–200. doi: 10.1093/oxfordjournals.molbev.a040594. [DOI] [PubMed] [Google Scholar]
- Gebauer F., de la Torre J. C., Gomes I., Mateu M. G., Barahona H., Tiraboschi B., Bergmann I., de Mello P. A., Domingo E. Rapid selection of genetic and antigenic variants of foot-and-mouth disease virus during persistence in cattle. J Virol. 1988 Jun;62(6):2041–2049. doi: 10.1128/jvi.62.6.2041-2049.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman O. T., Bean W. J., Kawaoka Y., Webster R. G. Evolution of the nucleoprotein gene of influenza A virus. J Virol. 1990 Apr;64(4):1487–1497. doi: 10.1128/jvi.64.4.1487-1497.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampar B., Keehn M. A. Cumulative changes in the antigenic properties of herpes simplex virus from persistently infected cell cultures. J Immunol. 1967 Sep;99(3):554–557. [PubMed] [Google Scholar]
- Hayashida H., Toh H., Kikuno R., Miyata T. Evolution of influenza virus genes. Mol Biol Evol. 1985 Jul;2(4):289–303. doi: 10.1093/oxfordjournals.molbev.a040352. [DOI] [PubMed] [Google Scholar]
- Holland J. J., Grabau E. A., Jones C. L., Semler B. L. Evolution of multiple genome mutations during long-term persistent infection by vesicular stomatitis virus. Cell. 1979 Mar;16(3):495–504. doi: 10.1016/0092-8674(79)90024-2. [DOI] [PubMed] [Google Scholar]
- Holland J., Spindler K., Horodyski F., Grabau E., Nichol S., VandePol S. Rapid evolution of RNA genomes. Science. 1982 Mar 26;215(4540):1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- Hundley F., McIntyre M., Clark B., Beards G., Wood D., Chrystie I., Desselberger U. Heterogeneity of genome rearrangements in rotaviruses isolated from a chronically infected immunodeficient child. J Virol. 1987 Nov;61(11):3365–3372. doi: 10.1128/jvi.61.11.3365-3372.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz J. M., Webster R. G. Antigenic and structural characterization of multiple subpopulations of H3N2 influenza virus from an individual. Virology. 1988 Aug;165(2):446–456. doi: 10.1016/0042-6822(88)90588-0. [DOI] [PubMed] [Google Scholar]
- Kendal A. P., Joseph J. M., Kobayashi G., Nelson D., Reyes C. R., Ross M. R., Sarandria J. L., White R., Woodall D. F., Noble G. R. Laboratory-based surveillance of influenza virus in the United States during the winter of 1977-1978. I. Periods of prevalence of H1N1 and H3N2 influenza A strains, their relative rates of isolation in different age groups, and detection of antigenic variants. Am J Epidemiol. 1979 Oct;110(4):449–461. doi: 10.1093/oxfordjournals.aje.a112826. [DOI] [PubMed] [Google Scholar]
- Kendal A. P., Noble G. R., Dowdle W. R. Swine influenza viruses isolated in 1976 from man and pig contain two coexisting subpopulations with antigenically distinguishable hemagglutinins. Virology. 1977 Oct 1;82(1):111–121. doi: 10.1016/0042-6822(77)90037-x. [DOI] [PubMed] [Google Scholar]
- Krystal M., Buonagurio D., Young J. F., Palese P. Sequential mutations in the NS genes of influenza virus field strains. J Virol. 1983 Feb;45(2):547–554. doi: 10.1128/jvi.45.2.547-554.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima S., Cox N. J., Kendal A. P. Antigenic and genomic analyses of influenza A(H1N1) viruses from different regions of the world, February 1978 to March 1980. Infect Immun. 1981 Apr;32(1):287–294. doi: 10.1128/iai.32.1.287-294.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima S., Nakajima K., Kendal A. P. Identification of the binding sites to monoclonal antibodies on A/USSR/90/77 (H1N1) hemagglutinin and their involvement in antigenic drift in H1N1 influenza viruses. Virology. 1983 Nov;131(1):116–127. doi: 10.1016/0042-6822(83)90538-x. [DOI] [PubMed] [Google Scholar]
- O'Neil K. M., Pallansch M. A., Winkelstein J. A., Lock T. M., Modlin J. F. Chronic group A coxsackievirus infection in agammaglobulinemia: demonstration of genomic variation of serotypically identical isolates persistently excreted by the same patient. J Infect Dis. 1988 Jan;157(1):183–186. doi: 10.1093/infdis/157.1.183. [DOI] [PubMed] [Google Scholar]
- Ortín J., Nájera R., López C., Dávila M., Domingo E. Genetic variability of Hong Kong (H3N2) influenza viruses: spontaneous mutations and their location in the viral genome. Gene. 1980 Nov;11(3-4):319–331. doi: 10.1016/0378-1119(80)90072-4. [DOI] [PubMed] [Google Scholar]
- Oxford J. S., Abbo H., Corcoran T., Webster R. G., Smith A. J., Grilli E. A., Schild G. C. Antigenic and biochemical analysis of field isolates of influenza B virus: evidence for intra- and inter-epidemic variation. J Gen Virol. 1983 Nov;64(Pt 11):2367–2377. doi: 10.1099/0022-1317-64-11-2367. [DOI] [PubMed] [Google Scholar]
- Oxford J. S., Salum S., Corcoran T., Smith A. J., Grilli E. A., Schild G. C. An antigenic analysis using monoclonal antibodies of influenza A (H3N2) viruses isolated from an epidemic in a semi-closed community. J Gen Virol. 1986 Feb;67(Pt 2):265–274. doi: 10.1099/0022-1317-67-2-265. [DOI] [PubMed] [Google Scholar]
- Palese P., Young J. F. Variation of influenza A, B, and C viruses. Science. 1982 Mar 19;215(4539):1468–1474. doi: 10.1126/science.7038875. [DOI] [PubMed] [Google Scholar]
- Parvin J. D., Moscona A., Pan W. T., Leider J. M., Palese P. Measurement of the mutation rates of animal viruses: influenza A virus and poliovirus type 1. J Virol. 1986 Aug;59(2):377–383. doi: 10.1128/jvi.59.2.377-383.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond F. L., Caton A. J., Cox N. J., Kendal A. P., Brownlee G. G. The antigenicity and evolution of influenza H1 haemagglutinin, from 1950-1957 and 1977-1983: two pathways from one gene. Virology. 1986 Jan 30;148(2):275–287. doi: 10.1016/0042-6822(86)90325-9. [DOI] [PubMed] [Google Scholar]
- Robertson J. S., Bootman J. S., Newman R., Oxford J. S., Daniels R. S., Webster R. G., Schild G. C. Structural changes in the haemagglutinin which accompany egg adaptation of an influenza A(H1N1) virus. Virology. 1987 Sep;160(1):31–37. doi: 10.1016/0042-6822(87)90040-7. [DOI] [PubMed] [Google Scholar]
- Salinovich O., Payne S. L., Montelaro R. C., Hussain K. A., Issel C. J., Schnorr K. L. Rapid emergence of novel antigenic and genetic variants of equine infectious anemia virus during persistent infection. J Virol. 1986 Jan;57(1):71–80. doi: 10.1128/jvi.57.1.71-80.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. V., Stowring L., Haase A. T., Narayan O., Vigne R. Antigenic variation in visna virus. Cell. 1979 Oct;18(2):321–327. doi: 10.1016/0092-8674(79)90051-5. [DOI] [PubMed] [Google Scholar]
- Steinhauer D. A., Holland J. J. Rapid evolution of RNA viruses. Annu Rev Microbiol. 1987;41:409–433. doi: 10.1146/annurev.mi.41.100187.002205. [DOI] [PubMed] [Google Scholar]
- Tobita K., Sugiura A., Enomote C., Furuyama M. Plaque assay and primary isolation of influenza A viruses in an established line of canine kidney cells (MDCK) in the presence of trypsin. Med Microbiol Immunol. 1975 Dec 30;162(1):9–14. doi: 10.1007/BF02123572. [DOI] [PubMed] [Google Scholar]
- Vandepol S. B., Holland J. J. Evolution of vesicular stomatitis virus in athymic nude mice: mutations associated with natural killer cell selection. J Gen Virol. 1986 Mar;67(Pt 3):441–451. doi: 10.1099/0022-1317-67-3-441. [DOI] [PubMed] [Google Scholar]
- Webster R. G., Kendal A. P., Gerhard W. Analysis of antigenic drift in recently isolated influenza A (H1N1) viruses using monoclonal antibody preparations. Virology. 1979 Jul 15;96(1):258–264. doi: 10.1016/0042-6822(79)90189-2. [DOI] [PubMed] [Google Scholar]
- Webster R. G., Laver W. G., Air G. M., Schild G. C. Molecular mechanisms of variation in influenza viruses. Nature. 1982 Mar 11;296(5853):115–121. doi: 10.1038/296115a0. [DOI] [PubMed] [Google Scholar]
- Weis W., Brown J. H., Cusack S., Paulson J. C., Skehel J. J., Wiley D. C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988 Jun 2;333(6172):426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- Wiley D. C., Skehel J. J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- Wiley D. C., Wilson I. A., Skehel J. J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981 Jan 29;289(5796):373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- Wilson I. A., Cox N. J. Structural basis of immune recognition of influenza virus hemagglutinin. Annu Rev Immunol. 1990;8:737–771. doi: 10.1146/annurev.iy.08.040190.003513. [DOI] [PubMed] [Google Scholar]
- Wilson I. A., Skehel J. J., Wiley D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981 Jan 29;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Winter G., Fields S., Brownlee G. G. Nucleotide sequence of the haemagglutinin gene of a human influenza virus H1 subtype. Nature. 1981 Jul 2;292(5818):72–75. doi: 10.1038/292072a0. [DOI] [PubMed] [Google Scholar]
- Young J. F., Palese P. Evolution of human influenza A viruses in nature: recombination contributes to genetic variation of H1N1 strains. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6547–6551. doi: 10.1073/pnas.76.12.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]



