Abstract
Host immune systems impose natural selection on pathogen populations, which respond by evolving different antigenic signatures. Like many evolutionary processes, pathogen evolution reflects an interaction between different levels of selection; pathogens can win in between-strain competition by taking over individual hosts (within-host level) or by infecting more hosts (population level). Vaccination, which intensifies and modifies selection by protecting hosts against one or more pathogen strains, can drive the emergence of new dominant pathogen strains—a phenomenon called vaccine-induced pathogen strain replacement. Here, we review reports of increased incidence of subdominant variants after vaccination campaigns and extend the current model for pathogen strain replacement, which assumes that pathogen strain replacement occurs only through the differential effectiveness of vaccines against different pathogen strains. Based on a recent theoretical study, we suggest a broader range of possible mechanisms, some of which allow pathogen strain replacement even when vaccines are perfect—that is, they protect all vaccinated individuals completely against all pathogen strains. We draw an analogy with ecological and evolutionary explanations for competitive dominance and coexistence that allow for tradeoffs between different competitive and life-history traits.
Keywords: multiple pathogen strains; vaccination; strain (serotype, serogroup) replacement; pathogen evolution
1. Introduction
Infectious diseases are a leading cause of death worldwide. As public health systems devise and implement strategies to control known diseases, newly emerging diseases (e.g. AIDS, hantavirus pulmonary syndrome, SARS) and re-emerging diseases (e.g. malaria, pertussis, tuberculosis) continue to challenge epidemiologists. Of the fourteen Grand Challenges in Global Health, six are related to vaccine-preventable diseases (Varmus et al. 2003). The re-emerging diseases are largely driven by social and economic changes, but can also reflect evolutionary changes in pathogens; understanding such evolution is critical to controlling re-emergence.
Host responses have always imposed strong selective pressures on pathogens. Advantages of micro-organisms in this coevolutionary race include high mutation rates, large population sizes and short generations; vertebrate hosts counter with high levels of genetic diversity, recombination and adaptive immunity. Recently, humans have added vaccination (which pre-arms adaptive immune responses to reject pathogens at first encounter) and chemotherapy (which supplements immunity by blocking key biochemical processes in pathogens) to their defensive arsenal. These new weapons can destabilize the existing host–pathogen evolutionary equilibria or accelerate pathogen evolution (Gandon et al. 2001; Lee & Suarez 2004; Mackinnon & Read 2004), leading to the phenomenon of treatment-induced pathogen strain replacement, in which vaccination or chemotherapy (box 1) drives emergence and dominance of a once-rare pathogen strain (we use pathogen type, strain and variant synonymously in the remainder of this article).
Box 1. Glossary of terms.
- Antigen
a foreign substance that elicits an immune response.
- Chemotherapy
the use of chemical agents to treat or control disease.
- Co-infection
simultaneous infection of a host by multiple strains.
- Conjugate vaccine
a vaccine created by attaching a poor antigen that cannot elicit a T-cell immune response to a carrier protein that can do so.
- Cross-immunity
partial immune protection against a strain generated by an antigenically similar strain.
- Differential effectiveness
difference in the reduction of disease transmission (level of protection) to vaccinated individuals. In the two extremes, vaccine can be completely effective and protect all vaccinated individuals or be completely ineffective when all vaccinated individuals are as susceptible as non-vaccinated individuals.
- Dominant strain
strain that either persists alone or has the highest prevalence in the population.
- Subdominant strain
a strain that persists in the population with lower prevalence than the dominant strain.
- Herd immunity
resistance of the entire population to the spread of an infectious disease due to the immunity of a high proportion of that population.
- Incidence
number of new disease cases per unit of time.
- Perfect vaccine
a vaccine uniformly and completely effective against all strains of a pathogen (article usage). The general usage signifies vaccines that are fully protective against the targeted strains.
- Prevalence
number (or proportion) of cases.
- Superinfection
the process of a strain taking over a host already infected by a different strain. In general, the second infecting micro-organism may arise from the existing infecting strain by mutation (endogenous superinfection) or may come as a second infection from an external source (exogenous superinfection). In this article superinfection means exogenous superinfection. We also limit superinfection to the case where the second strain displaces the first, not simply (as in some medical literature), non-simultaneous co-infection by two strains.
- Trade off mechanism
any process that allows a strain with a lower reproduction number to coexist with a strain with a higher reproduction number. In the absence of such a mechanism, the strain with the higher reproduction number must (eventually) exclude the strain with the lower reproduction number.
To reach high prevalence, pathogens must escape from the host immune system and/or resist chemotherapeutic agents; to persist, they must also compete successfully with other pathogen strains at both within-host and population levels of interaction. Strains that dominate competition within hosts can cause sickness in individual hosts; strains that successfully transmit to new hosts can increase in prevalence. When anthropogenic changes in selection alter the competitive balance among pathogens, novel pathogen strains can (re)emerge in the population (pathogen strain replacement). One example of great public health concern is the re-emergence of multi-drug-resistant tuberculosis (Espinal et al. 2001). Other potential examples include vaccine-resistant pertussis (Mooi et al. 2001), but are less firmly established. Given the huge population sizes and high mutation rates of many pathogenic agents, a concern with pathogen strain replacement should be a component of any long-term vaccination programme.
The standard explanation for treatment-induced pathogen strain replacement rests on the frequent observation that different strains respond differently to treatment; if a dominant strain is strongly affected by treatment while a rare strain is not (differential effectiveness), it is no surprise that the previously rare strain can then dominate both individual hosts and the host population (although, typically, at a lower prevalence than the original dominant strains, Bonhoeffer et al. 1997). Here, however, we suggest that treatment can lead to strain replacement through a variety of changes in the competitive environment, even when treatment affects all pathogen strains equally (e.g. vaccination protects all vaccinated individuals completely against all strains; here we consider only vaccines that prevent infection, as opposed to vaccines that change other within-host characteristics of infection such as infectious period, transmissibility or virulence; Gandon et al. 2001).
While chemotherapy and vaccination share many characteristics, they may differ in their action (e.g. chemotherapy is usually a response to symptoms, while vaccination is usually prophylactic; chemotherapy complements the immune response, while vaccination enhances it). We focus here on vaccination, although we revisit briefly strain replacement driven by chemotherapy in the discussion.
2. What is strain replacement?
It is useful to start with a more precise definition of strain replacement. Generally speaking, strain replacement is the phenomenon of substitution through time of one or more initially dominant strains of a pathogen by another strain or strains. It occurs through the interaction of dynamics at two levels.
Within-host (individual-level) strain replacement is the replacement of a strain that dominated the initial infection of a particular host by a new strain, without any intervening recovery period (Bogaert et al. 2004; Smith et al. 2005). While mechanisms of within-host competition and strain replacement are important (Mackinnon & Read 2004), we will focus here on population-level strain replacement.
Between-host (population-level) strain replacement, in its simplest form, occurs when a once-common strain in the population becomes rare, while a second (previously rare) strain increases to a prevalence greater than that of the first strain, due to a deterministic process (rather than, say, ecological drift in a rare host population).
By definition, strain replacement is a consequence of increased absolute fitness (at the population level) of the replacement strain, and/or decreased fitness of the initial strain. The deployment of a treatment such as vaccination changes conditions (e.g. the proportion of hosts susceptible to one or the other strain), which in turn changes the competitive balance between strains and hence their absolute fitnesses, ultimately shifting their relative and absolute abundances.
3. What causes strain replacement?
The currently accepted model of population level strain replacement relies on the selective nature of vaccine protection. Consider a pathogen with negligible co-infection: for simplicity, lump together vaccine strains (and all strains that suffer significant cross-immunity) as strain 1 and all other non-vaccine strains—strains that at least partially escape the vaccine—as strain 2. Since vaccines are typically targeted against dominant strains, strain 1 will be the dominant strain before vaccination, while strain 2 will have low prevalence. After a successful vaccination campaign, the prevalence of strain 1 should drop (indeed, this is how one measures ‘success’); however, the prevalence of strain 2 may simultaneously increase to become a new public health problem. Strain replacement has then occurred at the population level and it is vaccine induced—driven by a vaccine-induced reduction in the susceptible pool for strain 1. This replacement occurs because strain 2 can still infect vaccinated individuals and no longer has to compete with strain 1 for them. Those unvaccinated individuals who have contacts only with vaccinated individuals are protected by a population-level mechanism called herd immunity; herd immunity against strain 1 further reduces competition between strains for susceptible individuals. By reducing the prevalence of the dominant strain (strain 1), a differentially effective vaccine frees ‘resources’—available susceptible hosts—allowing strain 2 to proliferate (Lipsitch 1999) and driving population-level strain replacement (Porco & Blower 1998; Lipsitch 1999).
4. Is it a strain replacement?
Researchers use widely varying criteria to infer strain replacement. The only common element is an observed decline in the prevalence of vaccine strains accompanied by an increased prevalence of at least one non-vaccine strain. Many studies now report such increases (table 1). However, some researchers reject the observed increases in non-vaccine-type prevalence alone as evidence of vaccine-induced strain replacement. Increases in observed prevalence can be caused by ‘unmasking’—an increased ability to detect a non-vaccine strain within individual patients in the absence of the vaccine strain increases the apparent prevalence of the non-vaccine strain at the population level (Lipsitch 2001). Even without unmasking, one may need information on prior prevalence of non-vaccine strains, or evidence that increasing prevalence is not part of a general trend, to infer strain replacement (Urwin et al. 1996; Lipsitch 1999; Ribeiro et al. 2003).
Table 1.
Clinical trials and epidemiological surveys reporting increases in prevalence. (Reported increases in non-vaccine strains after vaccination.)
| disease | vaccine | increase in | region | references |
|---|---|---|---|---|
| H. influenzae | Hib | non-type b | Alaska | Centers for Disease Control (1996, 2002) and Perdue et al. (2000) |
| Hib | type f | multiple states, US | Urwin et al. (1996) | |
| conjugate Hib | type a | Brazil | Ribeiro et al. (2003) | |
| conjugate Hib | non-capsulated | UK | Slack et al. (1998) and Sarangi et al. (2000) | |
| S. pneumoniae | PCV-7a | NVTb | Finland | Eskola et al. (2001) |
| PCV-7 | NVT (carriage) | community-level, US | Sprat & Greenwood (2000) and Huang et al. (2005) | |
| PCV-7 | serogroups 15 and 33 | US PMPSG, US | Gonzalez et al. (2006) | |
| PCV-7 | NVT (AOMc) | hospitals, Pittsburgh | McEllistrem et al. (2003, 2005) | |
| PPV-23d | 12Fe, 7F, 22F, 7C | Alaska | Centers for Disease Control (2005) | |
| N. meningitidis | A-C vaccine | serogroup B | Austria | Biebl et al. (2005) |
| A-C vaccine | serogroup B | Europe | van Looveren et al. (2001), Pérez-Trallero et al. (2002) and Schrijver & Maes (2003) | |
| A-C vaccine | serogroup B | Cuba | Rodriguez et al. (1999) | |
| B. pertussis | pertussis | various | the Netherlands | Mooi et al. (1998, 2001) and van Loo et al. (1999) |
| pertussis | various | US | Hardwick et al. (2002) | |
| WCV/ACVf | various | Sweden | Hallander et al. (2005) |
7-Valent pneumococcal conjugate vaccine.
Non-vaccine types.
Acute otitis media.
23-Valent polysaccharide vaccine.
An outbreak of a strain included in the PPV-23.
Whole-cell/acellular vaccines used with a period of time between them.
Increased rates of carriage (i.e. presence of a micro-organism in an individual with or without clinical disease) of non-vaccine strains among vaccinated individuals in a clinical trial, relative to non-vaccinated controls, suggest vaccine-induced strain replacement in the treatment subjects (Lipsitch 2001). However, since strain replacement is at least partly a population-level phenomenon, such an observation is neither a necessary nor a sufficient condition to establish whether strain replacement will be a problem in the population at large. A vaccination programme restricted to a small experimental group will not generate the herd immunity expected if the programme extended to a large fraction of the population. Consequently, small-scale vaccination programmes have a lower probability of controlling the vaccine strain and of causing strain replacement. Conversely, in a case–control study of vaccinated and unvaccinated individuals during a mass vaccination effort, an increase in prevalence of non-vaccine strains in the vaccinated group could spill over to the control group, decreasing the differences between case and control groups and leading to a failure to infer strain replacement, even when it has occurred. For this reason, most surveillance studies monitor increases in non-vaccine strains in the general population without regard to the individuals' vaccine status, but such an approach leaves the causal link to vaccination uncertain. Furthermore, classical experimental designs based on randomization and replication are logistically unfeasible at the population level. While tools from economics and environmental science can facilitate analysis of large, unreplicated ‘experiments’ (Box & Tiao 1975; Stewart-Oaten et al. 1992; Underwood 1994), these tools have not yet been applied to this problem.
Many of the studies in table 1 report only modest increases in non-vaccine strains; even if they represent statistically significant strain replacement, such increases may be epidemiologically insignificant. Public health officials are most concerned with the overall level of disease mortality and morbidity. They will certainly care if strain replacement erases gains from vaccination and leads to constant (or even increased) overall prevalence under vaccination (Huang et al. 2005); or if non-vaccine strains become absolutely more prevalent (not just a higher proportion of infected hosts; Sarangi et al. 2000); or if the non-vaccine strains have higher average virulence, either by chance or owing to selection for virulence (Gandon et al. 2001; the general public may react badly to a vaccine-induced increase in a virulent strain, even if the overall disease burden is lower than before vaccination). Mathematical models can in principle illuminate all these forms of strain replacement. In the remainder of this article, we will consider a limiting case: strain 1 is dominant and strain 2 absent (competitively excluded) before vaccination, while after vaccination strain 1 disappears while strain 2 invades. Thus, strain replacement reflects a reversal of competitive dominance.
5. Do we really understand the mechanism(s) of strain replacement?
Differential effectiveness of vaccine is widely accepted as the causal mechanism of strain replacement. Many mathematical models have investigated this mechanism and its potential impact on disease control and eradication. These models assume that a vaccine has strain-specific effects because (i) parameters estimated from data can support the assumption (e.g. Blower et al. 2005) or (ii) differential effectiveness is simply assumed to be the causal mechanism of strain replacement (Lipsitch 1997, 1999; Porco & Blower 1998, 2000).
If we believe that differential effectiveness drives strain replacement, we should direct our efforts towards improving the breadth of vaccine effectiveness. One strategy is to include more and more strains in the vaccine, as for example in vaccines developed against Streptococcus pneumoniae. For many pathogens, however, this approach is not technically feasible. Another strategy is to design vaccines that target some vulnerable feature common to all strains (e.g. conserved surface proteins; Jedrzejas 2001; Miller 2003; ID Biomedical 2005).
A recent theoretical study, however, suggests that our knowledge of the causal mechanism of strain replacement may be incomplete. Iannelli et al. (2005) analyse a model of vaccination against a pathogen with two strains. The vaccine is assumed ‘perfect’ in the sense that it confers full immunity against both strains. If strain replacement can be driven only by differential effectiveness, then making the vaccine perfect should prevent strain replacement. Surprisingly, the authors find that strain replacement can occur if strains differ sufficiently in their within-host competitive abilities—for example through differential ability to super- or co-infect.
6. The causal mechanism of strain replacement
To understand such strain replacement, we have to understand what factors control strain dominance. The reproduction numbers and invasion reproduction numbers of the strains quantitatively integrate these factors to determine competitive outcomes. The reproduction number (also called the basic reproduction number) of a strain is defined as the expected number of secondary infections one infectious individual can produce during its lifetime as an infectious individual placed in an entirely susceptible population. Often, the strain with the largest reproduction number dominates in the population (Bremermann & Thieme 1989). This rule of dominance holds when exclusion is the only possible population level outcome of the competition. However, if population level coexistence is possible, the strain with the highest reproduction number may not even persist (Nowak & May 1994; Martcheva & Pilyugin 2006).
The reproduction number describes the initial dynamics of a pathogen introduced into a completely susceptible host population. However, dominance of a strain in the host population is determined not just by its ability to invade a completely susceptible host population, but also by its ability to invade an established population of another strain, described by its invasion reproduction number. The invasion reproduction number of strain i (i=1, 2), , measures the ability of strain i to invade a host population where the other strain is already present and at equilibrium: it is the expected number of secondary infections produced by one infectious individual containing strain i, introduced into a population where the other strain is at equilibrium. This definition makes invasion reproduction numbers as special cases of what is known as the ‘effective reproduction number’. Typically, strain 1 dominates when strain 2 cannot invade its equilibrium . This dominance is absolute, that is, it holds for all initial conditions, if strain 1 can also invade the equilibrium of strain 2 ; analogous conditions apply to strain 2. When neither strain can invade an equilibrium population of the other strain ( and ), which strain dominates depends on the initial conditions (‘founder control’ or ‘priority effect’). When each strain can invade the other's equilibrium ( and ), they are expected to coexist.
Reproduction numbers and invasion reproduction numbers depend on the vaccination rate ψ (appendix A). Vaccination always decreases reproduction numbers. However, a vaccine that protects equally against both strains, and even a vaccine that protects completely against all strains, can have very different effects on the invasion capabilities of different strains. Depending on the biological characteristics of the strains and their interdependence in the absence of vaccination, increasing vaccination rates may decrease the invasion reproduction number of one strain while increasing the invasion reproduction number of the other (figure 2; see appendix A). Suppose strain 1 dominates in the population in the absence of vaccination, while strain 2 cannot persist ( and ). Increasing vaccination rates may decrease and increase ; at a critical vaccination level ψ*, may drop below 1 while rises above 1. If this happens, strain 2 can now invade and outcompete strain 1, and strain replacement occurs (figure 1 shows an example of how prevalence of both strains changes with vaccination rate). Thus, strain replacement can occur if vaccination has reciprocal effects on the invasion capabilities of the two strains (decreasing the invasion number of dominant strains and increasing the invasion number of inferior strains; figure 2).
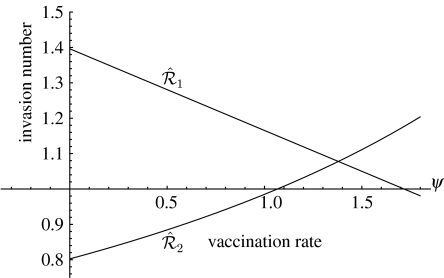
Figure 2.
Invasion numbers as a function of vaccination rate: the case of superinfection. The figure assumes that vaccinated individuals are completely protected against both strains. Note that is a decreasing function, while is an increasing function. For ψ<1.1, the invasion reproduction numbers satisfy and , so strain 1 will competitively exclude strain 2. For 1.1<ψ<1.75, the invasion reproduction numbers satisfy and and the two strains coexist. For ψ>1.75, the invasion reproduction numbers satisfy and , so strain 2 prevails. The reproduction numbers in the absence of vaccination are and .
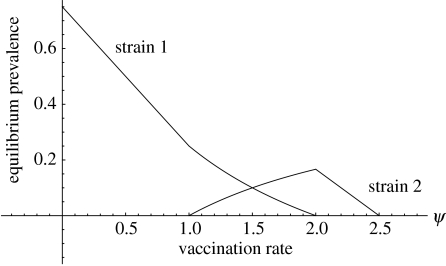
Figure 1.
Equilibrium prevalence as a function of vaccination rate for the system in appendix A. The figure assumes that vaccinated individuals are completely protected against both strains. Strain 1 dominates for vaccination rates between zero (no vaccination) and 1. For vaccination rates between 1 and 2 the two strains coexist. Strain 2 dominates for vaccination rates between 2 and 2.5. For higher vaccination rates, both strains are eliminated. The reproduction numbers of the strains in the absence of vaccination are respectively and .
If competitive exclusion is the only possible outcome of population-level competition between strains, then vaccination by an equally effective (or perfectly effective) vaccine will not affect the invasion reproduction numbers (consider the invasion reproduction numbers in appendix A with δ=0, i.e. no superinfection), and vaccine-induced strain replacement (in the narrow sense we consider here) is impossible. Therefore, the possibility of vaccine-induced strain replacement depends on the details of competitive outcomes at the within-host level. In particular, it depends on the existence of some asymmetry in the mode of transmission or within-host competitive effectiveness of the strain, an asymmetry whose impact on population-level competition changes as the vaccination level increases. This is a clear indication that models will be necessary to understand how population-level consequences emerge from individual-level pathogen interactions.
7. Why does differential effectiveness of the vaccine lead to strain replacement?
Suppose the vaccine is differentially effective, preventing infection by vaccine strain(s) but only partially protecting against non-vaccine strains. Even in the absence of other differences between the strains, vaccination can allow coexistence of multiple strains, in contrast with equally effective vaccines (even those that are not fully protective). This conclusion follows from mathematical models (Mclean 1995), but empirical studies have also found an increased genetic diversity of pathogens following a vaccination campaign (Schouls et al. 2005).
As vaccine levels increase, the invasion capability of the vaccine strain decreases, eventually dropping to zero (i.e. ). In contrast, the non-vaccine strain can infect vaccinated individuals, so its invasion capability increases with vaccination rate (from the combination of increasing numbers of vaccinated individuals and increased herd immunity against the vaccine strain). Thus, the differential effectiveness of the vaccine leads to reciprocal and opposite effects on the invasion capabilities of the two strains. If the vaccine strain dominates in the absence of vaccination, the vaccine reduces its invasion capabilities and eliminates it. If the non-vaccine strain is excluded in the absence of vaccination, the vaccine allows it to invade.
8. Other trade off mechanisms
A trade off mechanism is a difference between organisms (species or strains) that can allow coexistence; for example, some plant species' dispersal ability may require traits that lower their ability to compete for local resources, leading to a competition–colonization trade off (Holmes & Wilson 1998). In the epidemiological context, many different trade off mechanisms can lead to reciprocal effects of vaccination on invasion numbers and drive strain replacement, even if the vaccine is equally and perfectly effective against both strains. In this section, we will consider such a best-case scenario of perfect vaccines.
Many pathogen strains are known to have differential capabilities for superinfection (Nowak & May 1994), where one strain can rapidly take over hosts already infected with another strain (Smith et al. 2005). Strain replacement in Iannelli et al.'s study (2005) arises from the assumption that strain 1 can superinfect individuals carrying strain 2, but strain 2 can rarely or never superinfect individuals harbouring strain 1. (In fact, the model equations can be reinterpreted to allow some superinfection by strain 2, with the strain 1 superinfection rate really representing the difference between the superinfection rates of strain 2 by 1 and vice versa.) The vaccine targets both strains, and vaccinated individuals are completely removed from the population (from the pathogens' perspective). Increasing vaccination levels decrease the equilibrium prevalence of each pathogen when alone. Since strain 1 can infect hosts infected with strain 2, when strain 2 is at equilibrium, vaccination reduces the resources for strain 1 and decreases its invasion capabilities. Conversely, when strain 1 is at equilibrium, decreasing its equilibrium prevalence reduces the rate of superinfection, thereby increasing strain 2's invasion capabilities. This asymmetry in superinfection causes opposite effects of the vaccine on the potential for invasion by the two strains.
Co-infection (May & Nowak 1995), where hosts can harbour both strains for significant time periods, provides a milder form of tradeoff. Since co-infection acts symmetrically—it removes jointly infected individuals from both strains' susceptible pools—it is natural to ask whether co-infection can allow strain replacement via perfect, equal vaccination. The answer is yes, if co-infection ability is asymmetric; for example, if strain 1 can co-infect individuals infected with strain 2 but not vice versa. To further weaken strain 2, assume that jointly infected individuals cannot transmit strain 2. Vaccination decreases the equilibrium prevalence of strain 1, leading to fewer jointly infected individuals who can spread strain 1, and thereby decreasing the invasion capabilities of strain 1. Vaccination also decreases the equilibrium prevalence of strain 2, but since strain 2 suffers more severely from competition, the net effect of vaccination (through the positive effect of decreased co-infection) can be an increase in its invasion capabilities. Figure 3 shows how the invasion reproduction numbers of each species depend on vaccination rate for this scenario.
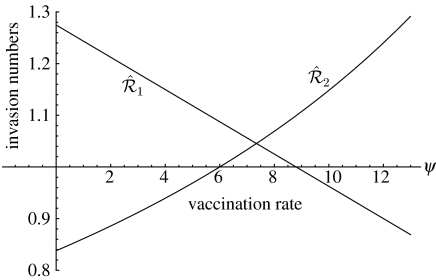
Figure 3.
Invasion numbers as a function of vaccination rate: the case of co-infection. The figure assumes that vaccinated individuals are completely protected against both strains. As before, is a decreasing function, while is an increasing function. For ψ<6, the invasion reproduction numbers satisfy and , so strain 1 will competitively exclude strain 2. For 6<ψ<9, the invasion reproduction numbers satisfy and and the two strains coexist. For ψ>9, the invasion reproduction numbers satisfy and , so strain 2 prevails. The reproduction numbers in the absence of vaccination are and .
The strong assumptions in the scenario above (perfectly asymmetric co-infectiveness, absence of strain 2 transmission from co-infected individuals) simplify analysis, but are not necessary to allow strain replacement. Figure 4 shows a case of strain replacement in a co-infection model with asymmetric, but not perfectly asymmetric, co-infection rates (Martcheva 2007).
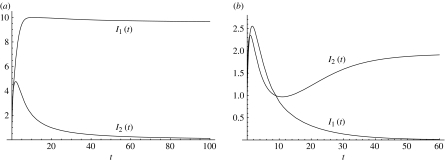
Figure 4.
Strain replacement in a co-infection model without perfect asymmetry (Martcheva 2007). (a) Strain 1 dominates and strain 2 is eliminated in the absence of vaccination (ψ=0). (b) Strain 2 dominates while strain 1 is eliminated at vaccination rate ψ=12. With this vaccination rate, the fraction vaccinated in a disease-free population will be f=0.96. The reproduction numbers of the two strains in the absence of vaccination are and .
9. Discussion and conclusions
Pathogen strain replacement weakens disease control. Understanding how it works is of paramount importance for future success in the control and eradication of infectious diseases. Developing this understanding requires bringing together concepts from immunology, epidemiology, ecology and evolution.
Modelling can lead the way to the integration of all these perspectives. Dynamical, immunological or epidemiological models have long had an ecological perspective, and more recently they have been linked to trait evolution (mostly through adaptive dynamics, e.g. Dieckmann et al. 2002). The mutual infusion of immunological and epidemiological modelling has only begun recently, but it is currently developing rapidly.
However, models can only determine logical possibilities. In order to quantify the relative importance of different within-host mechanisms, we will need data on the asymmetries of superinfection and co-infection of different strains. At present, such data are typically available either from small numbers of closely observed patients (Laskus et al. 2001; Ramos et al. 2002) or from in vitro or animal-model studies (Turner et al. 1999; Hirano 2001; Novella et al. 2004; Zhao et al. 2006). Statistical analyses of strain co-occurrence may be able to quantify some forms of within-host competition (Turner et al. 1999), but we are unlikely ever to have the level of information available from ecological studies where infected individuals can be released and recaptured to determine within-host strain turnover rates (Sousa 1993). Nevertheless, epidemiologists should try to determine how much information on superinfection and co-infection rates can be determined from population-level data.
The general idea of tradeoffs in local competition determining population-level coexistence and replacement has previously been explored in the ecological literature, in the context of habitat destruction (analogous to vaccination; Nee & May 1992; Tilman et al. 1994). Theoretical biologists have long appreciated the analogy between metapopulation ecology, in which suitable habitat patches can be empty or occupied by one or more plant or animal species, and epidemiology, in which hosts can be susceptible or infected by one or more pathogen strains (May & Nowak 1994). Klausmeier & Tilman (2002, see their fig. 3.2) in an ecological context describe the main model discussed here (with neither co-infection nor superinfection) as ‘local founder control’; the superinfection model as ‘hierarchical competition’; and the co-infection case (approximately) as ‘1 better than 2’. Ecologists are well aware that the detailed assumptions about biological mechanisms and asymmetries in models of competition, especially the details of within-patch interactions, can determine population-level outcomes (Pacala & Rees 1998). The match between disciplines is not perfect; ecologists have focused on species coexistence, while epidemiologists tend to focus on strain replacement. Habitat destruction and vaccination differ in their effects on the dynamics of single patches/hosts, e.g. are patches destroyed permanently or temporarily? Do patch turnover rate or the mortality of individuals within the patch change? The immunological details needed to understand within-host competition in detail will differ greatly from the ecological details needed to understand within-patch competition. Nonetheless, models of habitat modification and pathogen treatment (by vaccination or chemotherapy) give broadly similar conclusions and focus attention on the importance of scaling within-host/patch outcomes to the population level.
In this article, we have critically examined the proposition that the differential effectiveness of vaccine is the causal mechanism for strain replacement. This perception is contradicted by theoretical studies that suggest strain replacement may occur even if the vaccine is equally and fully effective against all strains. Replacement occurs when vaccination has opposing effects on the fitness of the two strains—it decreases the fitness of the stronger strain and increases the fitness of the weaker strain (quantified by the invasion reproduction number). Differential effectiveness can lead to replacement, but other trade off mechanisms can also be responsible for such replacement, even for broad-spectrum vaccines. Superinfection and co-infection are two such mechanisms covered here. Future studies should examine other trade off mechanisms involving cross-immunity, vertical transmission and virulence tradeoffs (Gandon et al. 2001). Understanding strain replacement requires a close analysis of the interplay of within-host and population-level processes.
Mathematical models can crystallize the basic mechanisms driving strain replacement and highlight unappreciated extensions to these mechanisms. Here we have focused on strain replacement in the strong sense—when one of the strains persists and the other goes extinct—so that they exchange their roles as a result of vaccination. This focus on persistence and extinction is typical of theoretical explorations in ecology, because persistence and extinction are much simpler to characterize theoretically than predicting the prevalence of disease at an endemic multi-strain equilibrium (but see Dushoff 1999). Future modelling should focus on the relative prevalence of coexisting pathogen variants and on vaccine-induced changes in relative prevalence. This will permit comparison with a wide range of observed disease systems where different strains change in prevalence but manage to coexist both before and after vaccination.
Vaccines that are equally and highly effective against all variants of a given pathogen, such as the one currently being developed by ID Biomedical for S. pneumoniae (ID Biomedical 2005), will surely decrease the probability of strain replacement. However, even if we make our vaccines completely effective against all pathogen strains, we cannot necessarily prevent strain replacement (Iannelli et al. 2005), particularly if the population is vaccinated at a rate below the persistence thresholds of individual strains.
In practice, an integrative approach is required. Making our vaccines capable of protecting against all known strains of a pathogen should be coupled with a sufficiently high vaccination rate if we hope to eradicate particular infectious diseases. Many diseases now are represented by multiple strains, often distinct from the ones used in the vaccines. For instance, the rising incidence of Bordetella pertussis may reflect the spread of more strains (Pereira et al. 2005). Even in the absence of vaccine strategies that can minimize the probability of pathogen strain replacement, we should update vaccines more regularly. This updating is now done only for influenza, based on thorough surveillance and intricate predictive techniques which allow for the preparation of the annual vaccine before the start of the flu season. Using mathematical theory to strengthen strategies of vaccine delivery, however, is still in its infancy despite many studies on both the population dynamics (Nokes & Swinton 1995; Moneim & Greenhalgh 2005) and evolutionary dynamics (Gandon et al. 2001; Lipsitch 2002) of pathogens under the influence of vaccine programmes. The perspective presented here emphasizes the need to consider within-host processes such as superinfection and co-infection when attempting to unravel the causes driving strain replacement due to vaccination.
Acknowledgments
The authors thank three anonymous referees in the Department of Zoology, UF for their valuable comments on an early version of this manuscript. M.M. thanks Mike Barfield for his editing and Manojit Roy for a helpful discussion. M.M. was supported by NSF grant DMS-0408230 when this paper was being written. R.D.H was partially supported by NIH grant 7 R01 GM060792-05 and the University of Florida Foundation.
Appendix A. The baseline model
The model considered in Iannelli et al. (2005) is structured by the time since vaccination. We include here its ordinary differential equation version that is based on a classical SIS epidemic model with superinfection (Nowak & May 1994). The number of susceptible individuals at time t is given by S(t), the number of individuals infected with strain 1 is given by I(t), and the number of individuals infected with strain 2 is given by J(t). The model takes the form
where V(t) is the number of vaccinated individuals at time t and N=S+I+J+V is the total population. The parameters are as follows: Λ is the birth/recruitment rate, μ is the natural death rate and βi is the contact rate of strain i. The parameter δ describes the coefficient of reduction or enhancement of infection at superinfection; e.g. δ could be less than 1 because cross-immunity from the immune system's encounter with strain 1 affects strain 2. Finally, γi is the recovery rate from strain i, and ψ is the vaccination rate (Hadeler & Castillo-Chavez 1995). As stated in the text, we assume that vaccine prevents infection rather than lowering the virulence or transmission rate of vaccinated individuals. Rather than modelling vaccination at birth (where vaccination would be represented as a proportional reduction in the birth rate Λ between 0 and 1), we assume a constant rate of vaccination of susceptible individuals ψ. In this case, the fraction of individuals vaccinated in a disease-free population is given by f=ψ/(ψ+μ). This model assumes that individuals infected with strain 2 can get superinfected with strain 1, but not vice versa. Vaccinated individuals are completely protected from both strains.
The reproduction numbers of the two strains are
both decreasing with increasing vaccination rate ψ (Iannelli et al. 2005). Reproduction numbers are independent of the parameters governing superinfection, as superinfection does not lead to infection of susceptible individuals.
The corresponding invasion reproduction numbers, however, are not independent of the superinfection process, since they measure the number of secondary infections one strain i-infected individual will produce in a population where strain j is at equilibrium. The invasion reproduction number of strain 1 is given by
As the vaccination rate ψ increases, decreases. Thus, vaccination decreases the invasion capabilities of strain 1. In contrast, the invasion reproduction number of strain 2 is
A close look at this invasion reproduction number reveals that it is an increasing function of the vaccination rate. Hence, vaccination increases the invasion capabilities of strain 2. The reason for this effect is that when the two strains coexist, increasing the vaccination rate decreases the number of individuals infected with strain 1. This in turn reduces the rate of superinfections, benefiting strain 2. This produces an overall increase in infections with strain 2 (figure 1). Comparable phenomena can emerge in other models with superinfection (e.g. Hochberg & Holt 1990, which uses density-dependent transmission).
References
- Biebl A, et al. Vaccine strategies of meningococcal disease: results of a 10-year population-based study. Eur. J. Pediatr. 2005;164:735–740. doi: 10.1007/s00431-005-1719-7. [DOI] [PubMed] [Google Scholar]
- Blower S, Bodine E.N, Grovit-Ferbas K. Predicting the potential public health impact of disease-modifying HIV vaccines in South Africa: the problem of subtypes. Curr. Drug Targets Infect. Disord. 2005;5:179–192. doi: 10.2174/1568005054201616. [DOI] [PubMed] [Google Scholar]
- Bogaert D, Veenhoven R, Sluijter M, Sanders E, de Groot R, Hermans P.W.M. Colony blot assay: a useful method to detect multiple pneumococcal serotypes within clinical specimens. FEMS Immunol. Med. Microbiol. 2004;41:259–264. doi: 10.1016/j.femsim.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Bonhoeffer S, Coffin J.M, Nowak M.A. Human immunodeficiency virus drug therapy and virus load. J. Virol. 1997;71:3275–3278. doi: 10.1128/jvi.71.4.3275-3278.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box G.E.P, Tiao G.C. Intervention analysis with applications to economic and environmental problems. J. Am. Stat. Assoc. 1975;70:70–79. doi: 10.2307/2285379. [DOI] [Google Scholar]
- Bremermann H.J, Thieme H.R. Competitive exclusion principle for pathogen virulence. J. Math. Biol. 1989;27:179–190. doi: 10.1007/BF00276102. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control. Progress toward elimination of Haemophilus influenzae type b disease among infants and children—United States, 1987–1995. MMWR Morb. Mortal. Wkly. Rep. 1996;45:901–906. [PubMed] [Google Scholar]
- Centers for Disease Control. Progress toward elimination of Haemophilus influenzae type b disease among infants and children—United States, 1998–2000. MMWR Morb. Mortal. Wkly. Rep. 2002;51:234–237. [PubMed] [Google Scholar]
- Centers for Disease Control. Outbreak of invasive pneumococcal disease—Alaska, 2003–2004. MMWR Morb. Mortal. Wkly. Rep. 2005;54:72–75. [PubMed] [Google Scholar]
- Dieckmann U, Metz J.A.J, Sabelis M.W, Sigmund K. Cambridge University Press; Cambridge, UK: 2002. Adaptive dynamics of infectious diseases: in pursuit of virulence management. [Google Scholar]
- Dushoff J. Host heterogeneity and disease endemicity: a moment-based approach. Theor. Popul. Biol. 1999;56:325–335. doi: 10.1006/tpbi.1999.1428. [DOI] [PubMed] [Google Scholar]
- Eskola J, et al. Efficacy of a pneumococcal conjugate vaccine against acute otitis media. N. Engl. J. Med. 2001;344:403–409. doi: 10.1056/NEJM200102083440602. [DOI] [PubMed] [Google Scholar]
- Espinal M.A, et al. Global trends in resistance to antituberculosis drugs. N. Engl. J. Med. 2001;344:1294–1303. doi: 10.1056/NEJM200104263441706. [DOI] [PubMed] [Google Scholar]
- Gandon S, Mackinnon M, Nee S, Read A.F. Imperfect vaccines and the evolution of pathogen virulence. Nature. 2001;414:751–756. doi: 10.1038/414751a. [DOI] [PubMed] [Google Scholar]
- Gonzalez B.E, Hulten K.G, Lamberth L, Kaplan S.L, Mason E.O, the U.S. Pediatric Multicenter Pneumococcal Surveillance Group Streptococcus pneumoniae serogroups 15 and 33: an increasing cause of pneumococcal infections in children in the United States after the introduction of the pneumococcal 7-valent conjugate vaccine. Pediatr. Infect. Dis. J. 2006;25:301–305. doi: 10.1097/01.inf.0000207484.52850.38. [DOI] [PubMed] [Google Scholar]
- Hadeler K, Castillo-Chavez C. A core group model for disease transmission. Math. Biosci. 1995;128:41–55. doi: 10.1016/0025-5564(94)00066-9. [DOI] [PubMed] [Google Scholar]
- Hallander H.O, Advani A, Donnelly D, Guftafsson L, Carlsson R.-M. Shifts of Bordetella pertussis variants in Sweden from 1970 to 2003 during the three periods marked by different vaccination programs. J. Clin. Microbiol. 2005;43:2856–2865. doi: 10.1128/JCM.43.6.2856-2865.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick T.H, Cassiday P, Weyant R.S, Bisgard K.M, Sanden G.N. Changes in predominance and diversity of genomic subtypes of Bordetella pertussis isolated in the United States, 1939–1999. Emerg. Infect. Dis. 2002;8:44–49. doi: 10.3201/eid0801.010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano A. Subacute sclerosing panencephalitis virus dominantly interferes with replication of wild-type measles virus in a mixed infection: implication for viral persistence. J. Virol. 2001;66:1891–1898. doi: 10.1128/jvi.66.4.1891-1898.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg M.E, Holt R.D. The coexistence of competing parasites: I. The role of cross-species infection. Am. Nat. 1990;136:517–541. doi: 10.1086/285111. [DOI] [Google Scholar]
- Holmes E, Wilson H. Running from trouble: long-distance dispersal and the competitive coexistence of inferior species. Am. Nat. 1998;151:578–586. doi: 10.1086/286143. [DOI] [PubMed] [Google Scholar]
- Huang S.S, Platt R, Rifas-Shiman S.L, Pelton S.I, Goldman D, Finkelstein J. Post-PCV7 changes in colonizing pneumococcal serotypes in 16 Massachusetts communities, 2001 and 2004. Pediatrics. 2005;116:e408–e413. doi: 10.1542/peds.2004-2338. [DOI] [PubMed] [Google Scholar]
- Iannelli M, Martcheva M, Li X.-Z. Strain replacement in an epidemic model with super-infection and perfect vaccination. Math. Biosci. 2005;195:23–46. doi: 10.1016/j.mbs.2005.01.004. [DOI] [PubMed] [Google Scholar]
- ID Biomedical 2005 ID Biomedical receives positive data from clinical testing of its Group-Common Pneumococcal Vaccine, 22 March 2005, Life Sciences BC, http://www.lifesciencesbc.ca/News/Member_Press_Releases/pr03220501.asp.
- Jedrzejas M.J. Pneumococcal virulence factors: structure and function. Microbiol. Mol. Biol. Rev. 2001;65:187–207. doi: 10.1128/MMBR.65.2.187-207.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausmeier, C. A. & Tilman, D. 2002 Spatial models of competition. In Competition and coexistence, vol. 161 (eds U. Sommer & B. Worm). Ecological studies, ch. 3, pp. 43–78. Berlin, Germany: Springer.
- Laskus T, Wang L.-F, Radkowski M, Vargas H, Nowicki M, Wilkinson J, Rakela J. Exposure of hepatitis C virus (HCV) RNA-positive recipients to HCV RNA-positive blood donors results in rapid predominance of a single donor strain and exclusion and/or suppression of the recipient strain. J. Virol. 2001;75:2059–2066. doi: 10.1128/JVI.75.5.2059-2066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Suarez D.A.S.D. Effect of vaccine use in the evolution of Mexican lineage H5N2 avian influenza virus. J. Virol. 2004;78:8372–8381. doi: 10.1128/JVI.78.15.8372-8381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M. Vaccination against colonizing bacteria with multiple serotypes. Proc. Natl Acad. Sci. USA. 1997;94:6571–6576. doi: 10.1073/pnas.94.12.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M. Bacterial vaccines and serotype replacement: lessons from Haemophilus influenzae and prospects for Streptococcus pneumoniae. Emerg. Infect. Dis. 1999;5:336–345. doi: 10.3201/eid0503.990304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M. Interpreting results from trials of pneumococcal conjugate vaccines: a statistical test for detecting vaccine-induced increase in carriage of nonvaccine serotypes. Am. J. Epidemiol. 2001;154:85–92. doi: 10.1093/aje/154.1.85. [DOI] [PubMed] [Google Scholar]
- Lipsitch M. Vaccination and serotype replacement. In: Dieckmann U, Metz J.A.J, Sabelis M.W, Sigmund K, editors. Adaptive dynamics of infectious diseases: in pursuit of virulence management. Cambridge University Press; Cambridge, UK: 2002. pp. 362–374. [Google Scholar]
- Mackinnon M.J, Read A.F. Immunity promotes virulence evolution in a malaria model. PLoS Biol. 2004;2:1286–1292. doi: 10.1371/journal.pbio.0020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martcheva M. On the mechanism of strain replacement in epidemic models with vaccination. In: Mahdavi K, Culshaw R, Boucher J, editors. Current developments in mathematical biology. Proc. Conf. on Mathematical Biology and Dynamical Systems. World Scientific Press; Hackensack, NJ: 2007. pp. 149–172. [Google Scholar]
- Martcheva M, Pilyugin S. The role of coinfection in multi-disease dynamics. SIAM J. Appl. Math. 2006;66:843–872. doi: 10.1137/040619272. [DOI] [Google Scholar]
- May R, Nowak M. Superinfection, metapopulation dynamics, and the evolution of diversity. J. Theor. Biol. 1994;170:95–114. doi: 10.1006/jtbi.1994.1171. [DOI] [PubMed] [Google Scholar]
- May R, Nowak M. Coinfection and the evolution of parasite virulence. Proc. R. Soc. B. 1995;261:209–215. doi: 10.1098/rspb.1995.0138. [DOI] [PubMed] [Google Scholar]
- McEllistrem M.C, Adams J.M, Mason E, Wald E. Epidemiology of acute otitis media caused by Streptococcus pneumoniae before and after licensure of the 7-valent pneumococcal protein conjugate vaccine. J. Infect. Dis. 2003;188:1679–1684. doi: 10.1086/379665. [DOI] [PubMed] [Google Scholar]
- McEllistrem M.C, et al. Acute otitis media due to penicillin-nonsusceptible Streptococcus pneumoniae before and after the introduction of the pneumococcal conjugate vaccine. Clin. Infect. Dis. 2005;40:1738–1744. doi: 10.1086/429908. [DOI] [PubMed] [Google Scholar]
- Mclean A. Vaccination, evolution and the changes in efficacy of vaccines: a theoretical framework. Proc. R. Soc. B. 1995;261:389–393. doi: 10.1098/rspb.1995.0164. [DOI] [PubMed] [Google Scholar]
- Miller J.D. The replacement effect. Scientist. 2003;4:20030523–20030605. [Google Scholar]
- Moneim I, Greenhalgh D. Use of a periodic vaccination strategy to control the spread of epidemics with seasonally varying contact rate. Math. Biosci. Eng. 2005;2:591–611. doi: 10.3934/mbe.2005.2.591. [DOI] [PubMed] [Google Scholar]
- Mooi F.R, van Oirschot H, Heuvelman K, van der Heide H.G.J, Gaastra W, Willems R.J.L. Polymorphism in the Bordetella pertussis virulence factors P.69/pertactin and pertussis toxin in The Netherlands: temporal trends and evidence for vaccine-driven evolution. Infect. Immunol. 1998;66:670–675. doi: 10.1128/iai.66.2.670-675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooi F.R, van Loo I.H.M, King A.J. Adaptation of Bordetella pertussis to vaccination: a cause for its reemergence? Emerg. Infect. Dis. 2001;7:526–528. doi: 10.3201/eid0707.017708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nee S, May M. Dynamics of metapopulations: habitat destruction and competitive coexistence. J. Anim. Ecol. 1992;61:37–40. doi: 10.2307/5506. [DOI] [Google Scholar]
- Nokes D.J, Swinton J. The control of childhood viral infections by pulse vaccination. IMA J. Math. Appl. Med. Biol. 1995;12:29–53. doi: 10.1093/imammb/12.1.29. [DOI] [PubMed] [Google Scholar]
- Novella I.S, Reissig D.D, Wilke C.O. Density-dependent selection in vesicular stomatitis virus. J. Virol. 2004;78:5779–5804. doi: 10.1128/JVI.78.11.5799-5804.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M, May A. Superinfection and the evolution of parasite virulence. Proc. R. Soc. B. 1994;255:81–89. doi: 10.1098/rspb.1994.0012. [DOI] [PubMed] [Google Scholar]
- Pacala S.W, Rees M. Field experiments that test alternative hypotheses explaining successional diversity. Am. Nat. 1998;152:729–737. doi: 10.1086/286203. [DOI] [PubMed] [Google Scholar]
- Perdue D, Bulkow L, Gellin B, Davidson M, Petersen K.M, Singleton R, Parkinson A. Invasive Haemophilus influenzae disease in Alaskan residents aged 10 years and older before and after infant vaccination program. J. Am. Med. Assoc. 2000;283:3089–3094. doi: 10.1001/jama.283.23.3089. [DOI] [PubMed] [Google Scholar]
- Pereira A, Pereira A.S.P, Moreira-Filho C.A, Bando S.Y, Tambourgi D.V. Comperative analysis of a Bordetella pertussis patient isolated strain and classical strains used in pertussis vaccine. Vaccine. 2005;23:4353–4358. doi: 10.1016/j.vaccine.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Pérez-Trallero E, Vicente D, Montes M, Cistema R. Positive effect of meningococcal C vaccination on serogroup replacement in Neisseria meningitidis. Lancet. 2002;360:953. doi: 10.1016/S0140-6736(02)11061-0. [DOI] [PubMed] [Google Scholar]
- Porco T.C, Blower S. Designing HIV vaccination policies: subtypes and cross-immunity. Interfaces. 1998;28:167–190. [Google Scholar]
- Porco T.C, Blower S. HIV vaccines: the effect of the mode of action on the coexistence of HIV subtypes. Math. Popul. Stud. 2000;8:205–229. [Google Scholar]
- Ramos A, et al. Intersubtype human immunodeficiency virus type 1 superinfection following seroconversion to primary infection in two injection drug users. J. Virol. 2002;76:7444–7452. doi: 10.1128/JVI.76.15.7444-7452.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro G, et al. Prevention of Haemophilus influenzae type b meningitis and emergence of serotype replacement with type a strains after introduction of Hib immunization in Brazil. J. Infect. Dis. 2003;187:109–116. doi: 10.1086/345863. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Dickson F, Baly A, Martinez R. The epidemiological impact of anti-meningococcal B vaccination in Cuba. Mem. Inst. Oswaldo Cruz, Rio de Janeiro. 1999;94:433. doi: 10.1590/s0074-02761999000400002. [DOI] [PubMed] [Google Scholar]
- Sarangi J, Cartwright K, Stuart J, Brookers S, Morris R, Slack M. Invasive Haemophilus influenzae disease in adults. Epidemiol. Infect. 2000;124:441–447. doi: 10.1017/S0950268899003611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouls L.M, van der Ende A, van de Pol I, Schot C, Spanjaard L, Vauterin P, Wilderbeek D, Witteveen S. Increase in genetic diversity of Haemophilus influenzae serotype b (Hib) strains after introduction of Hib vaccination in The Netherlands. J. Clin. Microbiol. 2005;43:2741–2749. doi: 10.1128/JCM.43.6.2741-2749.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrijver K.D, Maes I. An outbreak of serogroup C meningococcal disease in the province Antwerp (Belgium) in 2001–2002. Eur. J. Epidemiol. 2003;18:1073–1077. doi: 10.1023/A:1026100321871. [DOI] [PubMed] [Google Scholar]
- Slack H.J.A.M, Hagreaves R.M, Ramsey M.E. Enhanced surveillance of invasive Haemophilus influenzae disease in England, 1990 to 1996: impact of conjugate vaccines. Pediatr. Infect. Dis. J. 1998;17:S204–S207. doi: 10.1097/00006454-199809001-00026. [DOI] [PubMed] [Google Scholar]
- Smith D.M, Wong J.K, Hightower G.K, Ignacio C.C, Koelsch K.K, Petropoulos C.J, Richman D.D, Little S.J. HIV drug resistance acquired through superinfection. AIDS. 2005;19:1251–1256. doi: 10.1097/01.aids.0000180095.12276.ac. [DOI] [PubMed] [Google Scholar]
- Sousa W.P. Interspecific antagonism and species coexistence in a diverse guild of larval trematode parasites. Ecol. Monogr. 1993;63:103–128. doi: 10.2307/2937176. [DOI] [Google Scholar]
- Sprat B.G, Greenwood B. Prevention of pneumococcal disease by vaccination: does serotype replacement matter? Lancet. 2000;356:1210–1211. doi: 10.1016/S0140-6736(00)02779-3. [DOI] [PubMed] [Google Scholar]
- Stewart-Oaten A, Bence J.R, Osenberg C.W. Assessing effects of unreplicated perturbations: no simple solutions. Ecology. 1992;73:1396–1404. doi: 10.2307/1940685. [DOI] [Google Scholar]
- Tilman D, May R.M, Lehman C.L, Nowak M.A. Habitat destruction and the extinction debt. Nature. 1994;371:65–66. doi: 10.1038/371065a0. [DOI] [Google Scholar]
- Turner P.E, Burch C.L, Hanley K.A, Chao L. Hybrid frequencies confirm limit to coinfection in the RNA bacteriophage ϕ6. J. Virol. 1999;73:2420–2424. doi: 10.1128/jvi.73.3.2420-2424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood A.J. On beyond BACI: sampling designs that might reliably detect environmental disturbances. Ecol. Appl. 1994;4:3–15. doi: 10.2307/1942110. [DOI] [Google Scholar]
- Urwin G, Krohn J, Deaver-Robinson K, Wenger J, Farley M. Invasive disease due to Haemophilus influenzae serotype f: clinical and epidemiological characteristics in the H. influenzae serotype b vaccine era. Clin. Infect. Dis. 1996;22:1069–1076. doi: 10.1093/clinids/22.6.1069. [DOI] [PubMed] [Google Scholar]
- van Loo I.H, van der Heide H.G.J, Nagelkerke N.J.D, Verhoef J, Mooi F.R. Temporal trends in the population structure of Bordetella pertussis during 1949–1996 in a highly vaccinated population. J. Infect. Dis. 1999;179:915–923. doi: 10.1086/314690. [DOI] [PubMed] [Google Scholar]
- van Looveren M, Caugant D, Chapelle S, Carion F, Goossens H. Interpreting the rising incidence of meningococcal disease in Belgium: the contribution of molecular typing. J. Med. Microbiol. 2001;50:986–990. doi: 10.1099/0022-1317-50-11-986. [DOI] [PubMed] [Google Scholar]
- Varmus H, Klausner R, Zerhouni E, Acharya T, Daar A.D, Singer P.A. Grand challenges in global health. Science. 2003;302:398–399. doi: 10.1126/science.1091769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Enmei L, Chen F.-P, Sullender W.M. In vitro and in vivo fitness of respiratory syncytial virus monoclonal antibody escape mutants. J. Virol. 2006;80:11 651–11 657. doi: 10.1128/JVI.01387-06. [DOI] [PMC free article] [PubMed] [Google Scholar]