Abstract
The gap between the amount of genome information released by genome sequencing projects and our knowledge about the proteins' functions is rapidly increasing. To fill this gap, various ‘genomic-context’ methods have been proposed that exploit sequenced genomes to predict the functions of the encoded proteins. One class of methods, phylogenetic profiling, predicts protein function by correlating the phylogenetic distribution of genes with that of other genes or phenotypic characteristics. The functions of a number of proteins, including ones of medical relevance, have thus been predicted and subsequently confirmed experimentally. Additionally, various approaches to measure the similarity of phylogenetic profiles and to account for the phylogenetic bias in the data have been proposed. We review the successful applications of phylogenetic profiling and analyse the performance of various profile similarity measures with a set of one microsporidial and 25 fungal genomes. In the fungi, phylogenetic profiling yields high-confidence predictions for the highest and only the highest scoring gene pairs illustrating both the power and the limitations of the approach. Both practical examples and theoretical considerations suggest that in order to get a reliable and specific picture of a protein's function, results from phylogenetic profiling have to be combined with other sources of evidence.
Keywords: gene co-occurrence, phylogenetic profiles, genome evolution, genomic context, protein–protein interactions, pathway evolution
1. Introduction
The approach of modern biology to understand the complexity of organisms is to analyse the cooperation of their individual components. Nevertheless, extensive experimental studies are necessary just to identify the function of a single protein in its cellular context. Owing to this, even for well-studied model organisms, the functions of many proteins are yet unknown. For example, even for the economically and scientifically important budding yeast, Saccharomyces cerevisiae, approximately 20% of the open reading frames are completely uncharacterized (Saccharomyces Genome Database, January 2007; Cherry et al. 1998) and for many other proteins, we have only knowledge of certain aspects of their function, like their subcellular location, or the phenotype after knockout of its gene.
Owing to the wealth of data accumulating in databases and by means of powerful algorithms, e.g. for aligning sequences and inferring evolutionary relations between them, it becomes increasingly interesting to predict protein functions in a comparative approach. This comparative approach is essentially based on the observation that homologous proteins retain aspects of their function over long evolutionary times. This allows for a homology-based function prediction, i.e. the transfer of knowledge about the function between homologous proteins. However, although homology frequently implies a conservation of mechanistic aspects of function, it provides less information about the functional context, i.e. about the processes a protein is involved in. For example, homologous enzymes may catalyse similar reactions, but the substrates and products involved in this reaction may be part of different pathways. The function of a protein can only be fully understood if both its mechanistic and contextual aspects are considered. Hence, homology-based function prediction can be complemented by context-based function prediction that focuses on how a protein is linked to other proteins and that uses the ‘guilt by association’ principle (Aravind 2000) to transfer knowledge between non-homologous proteins, i.e. the function of a protein can be predicted by linking it to proteins of known function. As the examples discussed in this review show, the links predicted by context-based methods like phylogenetic profiling are not very specific and generally do not provide information on the exact role of the protein in a process but frequently they are decisive to guide further analysis.
The full predictive power of the guilt by association principle unfolds with high-throughput data on co-expression, or genetic and physical interaction. For instance, for about 90% of the genes of S. cerevisiae, associations to other genes have been found (BioGRID, v. 2.0.20; Stark et al. 2006). Nevertheless, comparison with information from the literature and comparison of different protein–protein interaction datasets indicate that despite this good coverage in terms of proteins, the coverage in terms of the interactions between proteins is low (Han et al. 2005; Reguly et al. 2006). Furthermore, high-throughput data have been suggested to contain considerable levels of noise (Bader & Hogue 2002; von Mering et al. 2002). Again, the comparative approach proved to enhance the data quality, for example by considering the evolutionary conservation of co-expression (Stuart et al. 2003; van Noort et al. 2003; Bergmann et al. 2004; Snel et al. 2004).
The so-called genomic-context methods are stricter in their use of the comparative approach because they exclusively rely on comparing sequenced and annotated genomes to predict functional associations. A functional link between genes is suggested by gene fusion and fission events (Rosetta stone method; Enright et al. 1999; Marcotte et al. 1999); conservation of gene neighbourhood (Dandekar et al. 1998; Overbeek et al. 1999; Korbel et al. 2004) correlating evolutionary rates (mirror tree method; Pazos & Valencia 2001) or correlating gene occurrence (phylogenetic profiling; Gaasterland & Ragan 1998; Huynen & Bork 1998; Pellegrini et al. 1999; Date & Marcotte 2003). Gene fusion and fission are relatively rare genomic events that indicate functional links with high reliability and usually affect genes that are functionally tightly coupled (Yanai et al. 2001; von Mering et al. 2003a). Conservation of gene neighbourhood uses the genomic proximity of genes that, in particular in prokaryotes, suggests co-regulation (Yanai et al. 2002; von Mering et al. 2003a).
Since its invention, phylogenetic profiling diversified into a large number of related approaches and no systematic attempt has yet been made to sort this diversity. The original form of phylogenetic profiling uses binary vectors—the phylogenetic profiles or phyletic patterns (Tatusov et al. 1997)—that indicate in which species a homologue is present or absent (Gaasterland & Ragan 1998; Huynen & Bork 1998; Pellegrini et al. 1999). The idea is that genes that are functionally related are gained and lost together from genomes during evolution, which results in a correlation of their occurrence vectors. Nevertheless, despite the obvious simplicity and straightforwardness of the concept, it is, as so often in bioinformatics, not immediately obvious how to optimally translate the concept into an algorithm that gives the best performance. For instance, in a variant of the method, a phylogenetic profile is defined as a vector of similarities of a query gene from a query genome to its highest scoring hit in a number of subject genomes (Date & Marcotte 2003), which, in turn, is related to the mirror tree method that correlates matrices of pair-wise similarities (Pazos & Valencia 2001). Here, we first give an overview of successful applications that illustrate the principle and role of phylogenetic profiling in a simple and intuitive way and classify phylogenetic profiling as a trait correlation method. Subsequently, we describe the different variants of phylogenetic profiling and how to account for the non-independence of profile values due to the evolutionary relation between species. This discussion is concluded by pointing out links of phylogenetic profiling to similarity-based methods such as the mirror tree method (Pazos & Valencia 2001). Finally, we discuss the assumptions and limitations of phylogenetic profiling and relate them to the structure and evolution of protein function and their interactions. Although some limitations have been overcome by recent technical advances others, such as a heterogeneous distribution of evolutionary modularity over different cellular processes (Snel & Huynen 2004; Campillos et al. 2006), may inherently limit the coverage of the method. Furthermore, these limitations underpin the importance of integrating results from phylogenetic profiling, as a versatile and easily applicable method, with other types of evidence for functional associations to get a comprehensive picture of cellular systems.
2. Successful applications of phylogenetic profiling
The predictions made by phylogenetic profiling usually hint at the functional role of a protein without giving precise information about the molecular mechanisms or the nature of the functional association (Huynen et al. 2000). Frequently, complementary evidence from other context-based, homology-based or experimental methods is consulted to get more specific predictions. A number of predictions concerning various cellular processes have been made by phylogenetic profiling and were experimentally confirmed (table 1). They underscore that the role of phylogenetic profiling is frequently a pinpointing of genes that could be involved in a process about which we have only incomplete knowledge. For example, phylogenetic profiling identified enzymes of the MEP/DOXP pathway, which in plant chloroplasts, apicomplexa, cyanobacteria and a number of other bacteria produces the building blocks of isoprenoids. Cunningham et al. (2000) determined the occurrence profiles of the first five known enzymes in the MEP/DOXP pathway and found two other proteins that co-occurred with the pathway, LytB and GcpE. Although the involvement of LytB in the pathway was supported by experimental evidence, eventually further genetic experiments were necessary to determine the exact positions of LytB and GcpE within the pathway (Altincicek et al. 2001a,b). An alternative approach to phylogenetic profiling stems from the observation that a gene may be substituted during evolution by another gene of the same or similar function. Such a non-orthologous gene displacement (Koonin et al. 1996) leads to an anti-correlation of the displaced and the substitute gene's occurrence profiles. This is illustrated by the folate-dependent thymidylate synthase ThyA and its flavin-dependent counterpart ThyX (Thy1). Dynes & Firtel (1989) showed that thyX complements thymidine prototrophy in Dictyostelium discoideum but a thymidylate synthase function could not be formally proven because the exact nature of the mutation in the D. discoideum strain they used was unknown. An additional hint on the function of thyX came from the observation that the occurrence profile of thyX is complementary to that of the known folate-dependent thymidylate synthase gene thyA (Galperin & Koonin 2000). Subsequently, further complementation experiments and a biochemical (Myllykallio et al. 2002) and structural (Kuhn et al. 2002) characterization showed that ThyX is indeed a new class of flavin-dependent thymidylate synthases.
Table 1.
Phylogenetic profiling-based function predictions that have been verified in the original or by subsequent publications. Complement: anti-correlating occurrence profiles; ibid.: verification in same publication as prediction.
| protein/gene | context | relation | function | prediction | verification |
|---|---|---|---|---|---|
| SelR | fusion/fission, gene order, co-occurrence | enzymatic activity | methionine sulphoxide reductase | Galperin & Koonin (2000); Huynen et al. (2000) | Kryukov et al. (2002) |
| Yfh1 | co-occurrence, biochemical data | process | iron–sulphur protein maturation | Huynen et al. (2001) | Muhlenhoff et al. (2002) |
| YchB | co-occurrence | metabolic pathway | terpenoid synthesis | Luttgen et al. (2000) | ibid. |
| SmpB | co-occurrence | process | trans-translation | Pellegrini et al. (1999) | Karzai et al. (1999) |
| ThyX | complement, knockout data | enzymatic activity | thymidilate synthase | Galperin & Koonin (2000) | Kuhn et al. (2002); Myllykallio et al. (2002) |
| ThiN | complement, fusion/fission | enzymatic activity | thiamine phosphate synthase | Rodionov et al. (2002) | Morett et al. (2003) |
| NrdR | co-occurrence, neighbourhood | physical interaction | regulator of nucleotide reductases | Rodionov & Gelfand (2005) | Borovok et al. (2004) |
| NM_029821 | co-occurrence, neighbourhood | metabolic pathway | 5-hydroxyisourate (HIU) hydrolase | Ramazzina et al. (2006) | Ramazzina et al. (2006); Lee et al. (2006) |
| NM_001039678 | fusion/fission, neighbourhood, co-occurrence | metabolic pathway | 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline (OHCU) decarboxylase | Ramazzina et al. (2006) | ibid. |
| B17.2L | co-occurrence, homology | physical interaction | Complex I assembly factor | Gabaldon et al. (2005) | Ogilvie et al. (2005) |
| LytB | co-occurrence | metabolic pathway | DOXP/MEP pathway | Cunningham et al. (2000) | Altincicek et al. (2001a) |
| GcpE | co-occurrence | metabolic pathway | DOXP/MEP pathway | Cunningham et al. (2000) | Altincicek et al. (2001b) |
| PRP43 | integrated (including: fusion/fission, co-occurrence) | physical interaction | RNA helicase also involved in ribosome biogenesis and rRNA processing | Lee et al. (2004) | Combs et al. (2006) |
| RLI | co-occurrence, conserved co-expression | — | RNase L inhibitor involvement in ribosome assembly | Gabaldon & Huynen (2004) | Yarunin et al. (2005); Kispal et al. (2005) |
| COG1980 | co-occurrence, complement | metabolic pathway | fructose-1,6-bisphosphatase | Makarova et al. (2003) | Sato et al. (2004) |
| Msa | genotype/phenotype correlation | — | mannose-specific adhesin | Pretzer et al. (2005) | ibid. |
| BBS5 | genotype/phenotype co-occurrence | — | flagella and basal body | Li et al. (2004) | ibid. |
| SUR2 (YDR297W) | genotype/metabolite co-occurrence | metabolic pathway | fungal sphingolipid C9-methyltransferase | Ternes et al. (2006) | ibid. |
| COG1206 (=trmFO, gid) | genotype/phenotype co-occurrence | metabolic pathway | flavin-dependent tRNA:m5U-54 MTase | Urbonavicius et al. (2005) | ibid. |
In many cases, we are not even aware of specific gaps in our knowledge about a cellular system. For example, a physical complex can have a different composition depending on the cellular state and an experimental exploration of all possible conditions remains impractical. Here, phylogenetic profiling can suggest new functional links. This was done, for instance, for NADH: ubiquinone oxidoreductase (Complex I), an energy-transducing multi-protein complex located in the mitochondrial inner membrane. Several of its members have paralogues that are members of Complex I itself or of other mitochondrial complexes. Gabaldon et al. (2005) noted that Complex I member N7BM (B17.2) and its paralogue B17.2L have similar occurrence profiles and were lost together from multiple independent lineages. Unfortunately, the function of N7BM within the complex was unknown and thus the co-occurrence- and homology-based link to this protein and to Complex I did not provide information on the specific function of B17.2L. It was an experimental study of Complex I assembly that showed that B17.2L is an assembly factor involved in this process and that its mutation can lead to a Complex I deficiency associated with a progressive encephalopathy in human patients (Ogilvie et al. 2005).
Generally, the examples illustrate that phylogenetic profiling derives most of its power from a large-scale approach that allows using it as an explorative method while being relatively easy to implement. After narrowing down the search space to a reasonable number of candidates, additional lines of evidence are needed. These may include results from other genomic-context methods, like gene order conservation, from published high-throughput experiments or from small-scale studies. In this process, towards the goal of discovering good candidates for further experiments, all these different sources should be considered and phylogenetic profiling is one of them. Notably, a number of web-based databases such as Predictome (Mellor et al. 2002), PLEX (Date & Marcotte 2005) or String (von Mering et al. 2007) integrate results from genomic context as well as small-scale and high-throughput experiments into unified, easily accessible interfaces and thus ease the process of gathering information for a broad public.
3. The link to the phenotype
In contrast to what may be suggested by the previous examples, co-occurrence profiling can be applied to any genotypic or phenotypic trait that can be coded as binary indicator vector. Examples of alternative genotypic traits are protein domains (Rodionov et al. 2002; Pagel et al. 2004; Liu et al. 2006), signal sequences (Haft et al. 2006), regulatory sites (Rodionov & Gelfand 2005) and restriction sites (Gelfand & Koonin 1997). Genotype/phenotype correlation has thus, for example, been applied to identify genes associated with pathogenicity (Huynen et al. 1997), hyperthermophily (Makarova et al. 2003; Jim et al. 2004), respiratory tract tropism, pili assembly (Jim et al. 2004), Gram-negativity, oxidative respiration, endospore formation, intracellular pathogenicity (Slonim et al. 2006) as well as eukaryotic and prokaryotic flagella (Jim et al. 2004; Li et al. 2004; Slonim et al. 2006). Similar to linking genes to each other via their co-occurrence in genomes, linking genes to phenotypes implicates them in a biological process. However, it does not require prior knowledge about the involvement of other genes in that process.
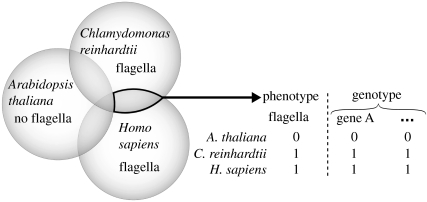
The link between genotype and phenotype was well exemplified by a study of the eukaryotic flagellum and basal body, which eventually identified BBS5 as a new disease gene. Li et al. (2004) selected three species—human, the aflagellate plant Arabidopsis thaliana and the flagellate alga Chlamydomonas reinhardtii. Blast searches identified genes present in human and the alga but lacking from the plant. This corresponds to a simple set union and intersection procedure that selects genes that have the same occurrence profile as the trait ‘having flagella and basal bodies’ (figure 1). Indeed, the resulting gene set included 88% of the known flagella and basal body genes of C. reinhardtii. Nevertheless, 92% of the resulting gene set was not known to be related to the flagellum and thus the reduction was not sufficient to draw conclusions about individual genes. A further intersection with a set of about 230 genes contained in a genomic locus associated with Bardet–Biedl syndrome (BBS), a deficiency of the basal body, resulted in a dramatic reduction to only two genes. Indeed, other evidence supports that one of these genes, BBS5, is related to the disease. This includes mutations that lead to premature termination of transcription of the BBS5 gene in four patients and cell-biological assays which show that that BBS5 localizes to basal bodies in flagellate cells of Caenorhabditis elegans.
Figure 1.
Genotype/phenotype profiling as exemplified by the study of the eukaryotic flagellum by Li et al. (2004).
Phenotype/genotype profiling illustrates the principle of phylogenetic profiling from a new perspective by linking the environment of an organism to its molecular evolution. The environment and its change are important factors that drive the evolution because they induce more or less specific adaptations in the organism. Examples of specific adaptation that involve changes of only few proteins are the resistance to pentachlorophenol by recruitment of proteins into a new degradative pathway in Sphingomonas chlorophenolica (Copley 2000), the agglutination of lactobacilli to budding yeast, which is based on a single mannose-dependent adhesin protein that was discovered by comparing the lactobacilli's phenotypes to their genomes (Pretzer et al. 2005), or the presence of specific metabolites, such as C9-methylated glucosylceramides in fungi (Ternes et al. 2006). In contrast, other environmental factors, e.g. stress induced by extreme temperature, require adaptation of various unrelated cellular processes (Felsenstein 1985). This suggests that the diversity, specificity and correlation structure of environmental factors active on a branch of the phylogeny will determine the value of coordinated gene gains and gene losses on this branch for phylogenetic profiling and that by increasing the resolution of the phylogenetic tree also the resolution in terms of selective factors is increased. This may be one reason for the observed improvement of results from phylogenetic profiling both by increasing the number of species and by a balanced choice of species (Sun et al. 2005). The value of genotype/phenotype profiling will increase with the availability of detailed phenotypic information for many species from, for instance, literature mining (Korbel et al. 2005; Liu et al. 2006). Furthermore, the combination of phenotypic information with genotypic data from metagenomic studies (e.g. Tyson et al. 2004; Venter et al. 2004; Tringe et al. 2005) or microarrays (e.g. Pretzer et al. 2005) will probably improve the prediction of functional links not only for already sequenced model organisms but also for organisms that cannot be cultivated under laboratory conditions.
4. Co-occurrence methods
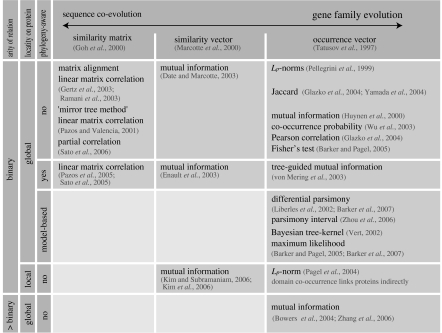
Most of the above presented success stories only considered identical profiles or used a simple distance measure, such as the number of occurrence values differing between two profiles. These ‘naive’ distance measures, however, ignore that the occurrence of a homologue in one species is not independent from its occurrence in another, probably closely related species. A number of ‘model-based’ approaches have been developed to account for this non-independence of profile values. Model-based phylogenetic profiling uses explicit models of evolution to infer gene gain and gene loss events and correlates the evolutionary processes rather than static absence/presence patterns. A complete overview and classification of all variants of phylogenetic profiling and of related methods discussed in this review is given in figure 2. For a discussion of similarity-based methods and a number of further extensions, such as methods that predict triples of functionally related genes instead of gene pairs or that focus on local regions on proteins, we refer to §§5 and 6, respectively.
Figure 2.
Overview of published phylogenetic profiling methods and related matrix-similarity methods. The ‘arity’ is the number of genes between which a functional relation is predicted. ‘Localized’ methods require only a coevolution of local region on two proteins to predict a functional link.
4.1 Naive co-occurrence profiling methods
In the first application of co-occurrence profiling, Pellegrini et al. (1999) used Hamming distance between two profile vectors as similarity measure. The Hamming distance is the number of species that do not have the same absence/presence value. It is a member of a family of distance measures that also includes the Euclidean distance (§8.4.6). Notably, although these measures differ in magnitude, they produce the same order of profile pairs: if a profile pair is the Nth-closest with Hamming distance, it will also be the Nth-closest pair with Euclidean distance or any other Lp-norm. Alternatively, profiles can be compared by statistical correlation measures, such as the Pearson correlation coefficient (Glazko & Mushegian 2004), Fisher's exact test (Barker & Pagel 2005) or mutual information (Huynen et al. 2000; Wu et al. 2003). The Pearson correlation coefficient quantifies the degree of linearity of two factors and is only zero if there is no linear correlation. In contrast, mutual information is a general correlation measure that detects any kind of correlations (Steuer et al. 2002) and measures the average amount of information that one profile conveys about the other and vice versa (MacKay 2005). Fisher's exact test and mutual information have been specifically designed for categorical data such as occurrence profiles. An alternative approach is taken by Wu et al. (2003, 2006) who have derived a formula for the co-occurrence probability of two genes. Note that anti-correlating profiles are treated differently by the different similarity measures: for instance, Lp-norms assign particularly high distances to complementary profiles while Pearson correlation gives them a negative correlation coefficient. In contrast, mutual information in principle does not distinguish anti-correlating from correlating profile pairs and both just appear as pairs with high mutual information. In practice, however, anti-correlating profiles are easy to identify and can be treated separately, and have actually been used successfully to predict protein function, e.g. in the case of ThyX (see §2). Finally, phylogenetic profiles have repeatedly been compared by the Jaccard coefficient (Jaccard 1912; Glazko & Mushegian 2004; Yamada et al. 2004, 2006). The Jaccard coefficient usually accounts only for the similarity generated by co-presence by ignoring the number of genomes that do not contain any of the compared orthologues in its calculation (see §8.4.5).
4.2 Phylogeny-aware profiling
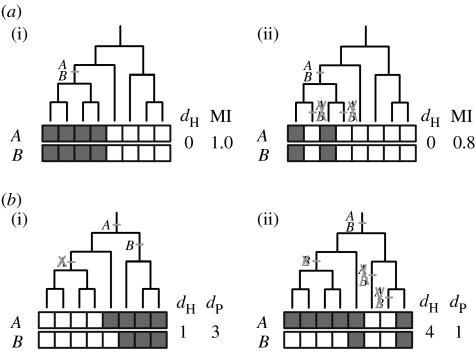
All above-mentioned statistical approaches to compare phylogenetic profiles assume statistical independence of the ‘sampled’ occurrence values in different species. However, because species are evolutionarily related the independence assumption is clearly violated, which may have negative effects on the quality of the predictions. Consistent with this, it was found that a substantial portion of modularity derived from the species distribution of orthologous genes is the results of such a phylogenetic signal (Snel & Huynen 2004). The effects of the non-independence of profile values may differ for the different naive profiling methods. For example, the similarity measures differ in how they score lineage-specific genes (table 2). Hamming distance always indicates high similarity for identical profiles independent of the lineage specificity, even for gene-families that occur only in a single genome. In contrast, mutual information is limited by the minimal information content of the profiles (see §8.4.8). Thus, for identical profiles with increasing lineage specificity, mutual information yields a lower correlation value. This results in a counter-intuitive behaviour. Consider two lineage-specific genes that are co-lost from some species within a clade (figure 3a). If a lineage encompasses less than half of the species, the additional co-losses will lead to lower mutual information, despite the additional evidence for functional linkage.
Table 2.
Naive co-occurrence measures differ in how they score ineage-specific genes. With pairs of identical profiles over 30 species both Hamming distance and Pearson correlation constantly yield scores that indicate high similarity. In contrast, Fisher's exact test (two-tailed) and mutual information yield less significance with a narrower and broader than 50% species distributions.
| presences | Hamming | Pearson | Fisher | mutual information |
|---|---|---|---|---|
| 0/30 | 0 | — | 1 | 0 |
| 3/30 | 0 | 1 | 0.02 | 0.7 |
| 15/30 | 0 | 1 | 0.008 | 1 |
| 27/30 | 0 | 1 | 0.02 | 0.7 |
| 30/30 | 0 | — | 1 | 0 |
Figure 3.
Negative influence of non-independence on some naive profiling methods. (a) (i) Two orthologous groups A and B occur in half of the species and have identical patterns of gains. Both Hamming distance (dH) and mutual information (MI) indicate high similarity. Although two additional co-losses in (ii) represent stronger evidence for a functional relation, the mutual information score is lower than in the previous situation (i). (b) (i) A and B are gained and lost independently but Hamming distance suggests high similarity (false positive). (ii) A single independent loss of B early in the phylogeny leads to high Hamming distance (false negative), despite two co-losses. In contrast, with differential Dollo parsimony (dP; see §8.4.3) the example of dependent evolution (ii) would correctly result in a better score than the example of independent evolution (i). dH, Hamming distance; MI, mutual information; dP, differential Dollo parsimony.
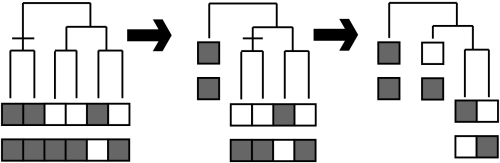
One way to reduce the effect of the phylogenetic bias is a reasonable, tree-guided selection of species that improves the prediction quality in comparison with a naive inclusion of all species (Sun et al. 2005). A related approach was taken for the STRING database, in which subtrees of the phylogenetic tree are collapsed and substituted by their ancestral state (von Mering et al. 2003a; figure 4). Nevertheless, both approaches still rely on naive correlation measures and thus only reduce the bias, but the remaining occurrence values are still statistically dependent.
Figure 4.
Tree-guided approach implemented in the String database (von Mering et al. 2003a). A subtree is collapsed only if all its leaves have the same presence/absence pattern, i.e. if the ancestral state at the subtree's root is known with high certainty.
4.3 Model-based co-occurrence methods
A completely different approach to handle the non-independence of the profile values is to use the phylogenetic tree to correlate the evolutionary processes rather than their observed outcomes. Model-based occurrence profiling methods use a model of gene content evolution to reconstruct ancestral genomes in a phylogenetic tree based on the observed occurrence patterns. Three methodological frameworks have been applied to phylogenetic profiling: the parsimony principle, maximum likelihood and a kernel-based method that models the evolutionary process by a Bayesian tree.
According to the parsimony principle, of many alternative evolutionary histories the one with the least costs, i.e. gene gains and losses, is the most credible. Liberles et al. (2002) used Fitch's parsimony algorithm to determine the presence or absence of each gene in the ancestral species of the used phylogeny (Fitch 1971). Fitch's parsimony model allows arbitrary and equally penalized changes between character states, i.e. gain and loss of homologous groups are considered equally probable. Ambiguities arising from equally parsimonious reconstructions were resolved by branch length weighting (D. Liberles 2006, personal communication). Barker et al. (2007) used Dollo parsimony for ancestral state reconstruction. Dollo parsimony allows a gene to be gained only once throughout a phylogenetic tree, which may require an arbitrary number of subsequent gene losses (Farris 1977). Based on the parsimonious reconstruction, gain/loss profiles for branches rather than occurrence profiles for genomes are constructed. The gains and losses on different branches can be assumed as reasonably independent, which justifies an application of similarity measures like Hamming distance, Pearson correlation coefficient or mutual information. Indeed, a simple similarity measure applied to Dollo-based gain/loss profiles yielded considerably better results on a eukaryotic dataset than a naive application of Hamming distance on occurrence profiles (Barker et al. 2007).
The parsimony approach usually treats the inferred ancestral states as if they are known without error. Nevertheless, there be can considerable uncertainty in the reconstructed ancestral states even if the parsimony solution is unambiguous. A requirement for parsimony methods to accurately reconstruct ancestral states is that the rates of change are low (Omland 1999). In eukaryotes, this requirement may be met because horizontal gene transfer (HGT) from non-endosymbiotic origin is rare and mainly confined to phagotrophic protists (reviewed in Andersson 2005). In contrast, in prokaryotes, HGT provides a mechanism of repeated gene gain and is estimated to affect a fraction of 40–60% (Kunin & Ouzounis 2003; Beiko et al. 2005) or even 90% of gene families (Mirkin et al. 2003).
One way to account for the uncertainty in the estimate of the ancestral state is to use ‘parsimony intervals’ that contain a number of suboptimal solutions (Schluter et al. 1997). Recently, Zhou et al. (2006) proposed a dynamic programming algorithm to calculate such parsimony intervals for phylogenetic profile comparison. The algorithm determines the best 100 suboptimal ancestral state reconstructions for each phylogenetic profile and compares them by a similarity measure that quantifies the number of correlated events while accounting for the degree of suboptimality of the reconstructions. However, a parsimony interval of 100 reconstructions is small in comparison with the number of possible reconstructions, which is exponential in the number of ancestral species. Two alternative model-based methods account for the uncertainty in the ancestral state estimation by considering all possible reconstructions: the maximum likelihood approach of Barker et al. (2005, 2007) and the tree-kernel method of Vert (2002).
The maximum likelihood method uses continuous-time Markov models to describe the evolutionary gain and loss of two genes (Barker & Pagel 2005). In order to quantify the probability that two genes have been gained and lost together, the likelihood (i.e. the goodness of fit) of a model of contingent evolution, in which the gain and loss of one gene is influenced by the presence and absence of the other, is compared to the likelihood of a model of independent evolution. In contrast to the parsimony approaches, maximum likelihood accounts for the branch lengths in the tree. Furthermore, the likelihood values are independent from a specific ancestral state reconstruction because they are calculated over all possible combinations of ancestral states. Although this can be done in linear time (Pagel 1994), the fitting of the model remains a computationally demanding task, which has to be solved by heuristic optimization. Recently, the method was considerably improved by assuming low gain rates instead of estimating these rates for each pair of occurrence profiles (Barker et al. 2007). The global gain-rate parameter depends on the branch lengths in the phylogenetic tree and has to be estimated from the dataset itself. Furthermore, the maximum likelihood method can also account for uncertainty in the tree topology (Barker et al. 2007) that is known to negatively affect predictions made by model-based phylogenetic profiling methods (Zhou et al. 2006).
The tree-kernel method circumvents the costly estimation of the rate parameters by considering both gain and loss probabilities as priors of a Bayesian tree (Vert 2002). Each branch of the tree has two associated prior probabilities—a gain probability p(1|0) and a loss probability p(0|1). Additionally, a probability for a gene to be present at the root has to be provided. A simplifying assumption of the original publication was that the gain and loss probabilities were the same for all branches of the tree, independent of the length of the branch. However, the Bayesian tree representation allows for more complex models with branch-specific change probabilities and differing gain and loss probabilities. The virtue of the method also comes from the combination of this Bayesian tree with a kernel-based approach that allows efficient calculation of a profile distance that accounts for all possible ancestral state reconstructions.
Although in recent applications model-based methods assumed that the phylogenetic tree and the patterns are known without error, this may not always be the case. The annotation of genomes sometimes misses genes and phylogenetic profiling has successfully been used to identify these (e.g. Natale et al. 2000; Mikkelsen et al. 2005). The uncertainty in the occurrence values can be expected to be even more pronounced in metagenomic data and in data from microarray genotyping (Molenaar et al. 2005; Pretzer et al. 2005). Both the maximum likelihood method and the tree-kernel method could be modified to account for such uncertainty.
5. Similarity methods
The original phylogenetic profiling approach, which uses the co-occurrence of genes, is related to another genomic-context method that identifies functionally associated gene pairs based on correlating rates of sequence evolution. For the purpose of this review, we will refer to these two alternative approaches as ‘co-occurrence profiling’ and ‘similarity profiling’, respectively. There are a number of reasons to include a discussion of similarity-based methods in a review about phylogenetic profiling. Both co-occurrence and similarity profiling methods use the idea of coevolution of functionally related genes to predict functional associations. Furthermore, there is a significant, positive correlation between the sequence evolutionary rate (estimated as average similarity to homologues in an outgroup species) and the rate of gene loss, although both mutually explain only about 10–15% of their variation (Krylov et al. 2003). This suggests that co-occurrence profiling and similarity profiling rely on distinct but not entirely independent signals. Finally, the phylogenetic profiling method of Date & Marcotte (2003) seamlessly integrates co-occurrence profiling and similarity profiling.
The phylogenetic profiling method of Date & Marcotte (2003) correlates vectors of transformed Blastp E-values that code the similarity of a protein in a query species to their best hits in a number of subject genomes (Marcotte 2000). To make the similarity values independent from the length and sequence composition of the query protein, they can be normalized by the score of the protein's self-alignment (Enault et al. 2003). The resulting similarity vectors are compared by mutual information, which requires a binning of values. The fact that the number and boundaries of the bins can be arbitrarily chosen illustrates that the similarity vectors also contain information about the occurrence of genes. If the subject genome does not contain a significant hit, the corresponding component of the vector is set to zero but is not excluded from the profile and thus contributes to the correlation value. In the extreme situation with only two bins representing worse than cut-off (absence) and better than cut-off (presence), the Date & Marcotte method and co-occurrence profiling are equivalent (Snitkin et al. 2006). According to our definition, the method of Date & Marcotte is thus both a similarity and co-occurrence profiling method.
The data structure of pair-wise similarities of a query gene to a number of best hits in subject genomes can be extended into a matrix of pair-wise similarities between homologous genes. Goh et al. (2000) demonstrated that the correlation coefficient between the resulting similarity matrices quantifies the degree of coevolution. Comparing similarity matrices has been used by two types of methods that have different fields of application. The matrix alignment methods of Gertz et al. (2003) and Ramani & Marcotte (2003) predict pairs of interacting partners between members of two groups of paralogues that reside in a single species. For each gene family, a matrix of pair-wise similarities is constructed. Two matrices are ‘aligned’ by shuffling the columns and rows in order to make them as similar as possible, i.e. to match members of both families that are most similar in their evolutionary rates. The matrix alignment is computationally costly, which restricts the method to small gene families, preferably ones of which some members are already known to interact. In contrast, the mirror tree method of Pazos & Valencia (2001) predicts associations between two groups of orthologous genes across different species. Each matrix contains the similarities between unique representatives of a gene family from each species. Different matrices are compared by matrix correlation coefficients in order to identify pairs of gene families that have similar evolutionary rates. In contrast to the similarity-vector method of Date & Marcotte, the mirror tree method can be considered as pure similarity profiling because it only accounts for the coevolution signals of those proteins that are present and ignores the loss or gain events; organisms that do not contain a homologue are treated as missing values by skipping the respective matrix rows and columns for the calculations of matrix similarity.
To our knowledge, no model-based similarity profiling methods have yet been proposed. Nevertheless, both the similarity vector method and the mirror tree method have been modified to correct for the dependence of similarity values on the distance of the compared species. In the similarity vector method of Date & Marcotte (2003), the similarity of the query protein to its best hits in a subject genome can be expected to decrease with increasing evolutionary distance of the query and subject species. This distance effect can be corrected for by dividing each vector component by the average similarity of all proteins' hits to the subject genome (Enault et al. 2003). Similarly, the mirror tree method can be corrected for the background similarity of the genomes either by using an algebraic projection operator (a linear map; Sato et al. 2005) or by subtracting a distance matrix based on a species tree from the homologous group distance matrices (Pazos et al. 2005).
6. Limitations and extensions
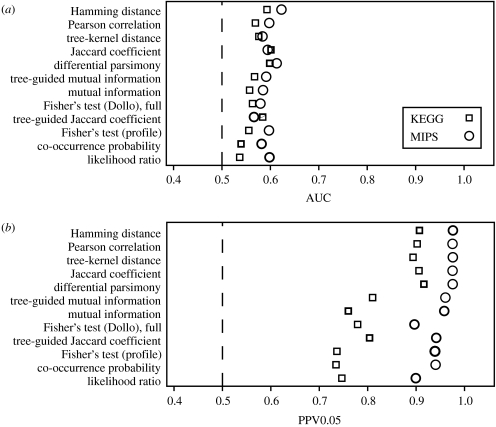
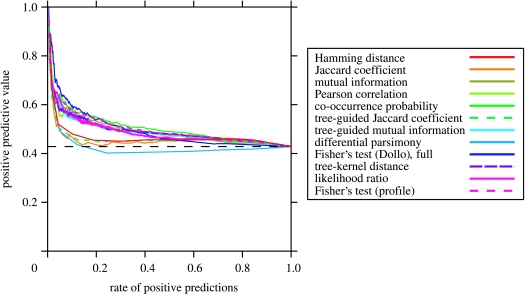
Although the successful small-scale studies using phylogenetic profiling provide an intuitive introduction to the method, phylogenetic profiling remains abstract, as long as one did not go through the process of using it on real data. We analysed a set of 25 fungi and the microsporidium Encephalitozoon cuniculi with 12 naive and model-based co-occurrence profiling methods. Although we observe a low overall performance with this dataset (figure 5a), highly reliable predictions can be made if a stringent score cut-off is used and only the highest scoring orthologous group pairs are considered as positive predictions (figure 5b), which indicates the presence of a signal that reflects functional associations. The fraction of positive controls (true positives) among the positive predictions drops quickly if the cut-offs are relaxed (figure 6), an observation that has been made before (e.g. Enault et al. 2003; von Mering et al. 2003a; Barker & Pagel 2005; Snitkin et al. 2006; Zhou et al. 2006) and can be explained by a number of technical causes, such as incorrect gene occurrence values due to misannotations of genomes or incorrect sets of positive and negative controls. One likely important cause for observing a low performance of phylogenetic profiling is the low number of only 26 genomes used here. Although it is of the same order of magnitude as in multiple other studies that used eukaryotic genomes (Barker & Pagel 2005; Snitkin et al. 2006; Barker et al. 2007), a recent study on the similarity/occurrence vector method of Date & Marcotte (2003) showed that this method yields significantly better results with increasing number of bacterial genomes (Sun et al. 2005). This probably applies also to the pure co-occurrence profiling methods benchmarked here. Nevertheless, also the studies on larger sets of genomes indicated a relatively limited coverage for gene co-occurrence (Huynen et al. 2003; von Mering et al. 2003a).
Figure 5.
(a) Phylogenetic profiling has low overall performance (b) but makes highly reliable predictions for the highest scoring 5% of orthologous group pairs. Plotted are the bootstrap medians of AUC and PPV0.05 estimated from a bootstrap sample (n=100) of positive and negative controls. (Results are based on a set of orthologous groups for 25 fungi and the microsporidium E. cuniculi (figure 1 of the electronic supplementary material) and functional associations for S. cerevisiae from the MIPS (Mewes et al. 2006) and KEGG (Kanehisa et al. 2004) databases. In the bootstrapping procedure, each MIPS complex (KEGG pathway) was given the same weight to account for the overrepresentation of some functional categories in the MIPS dataset, such as the large ribosomal subunit. The bootstraps for the PPV estimates were done such that positive and negative controls were sampled with the same probability as to produce an average P/(P+N) ratio of 0.5. Box plots of the ‘weighted’ as well as a ‘normal’, i.e. non-weighted, bootstrap distributions are shown in figure 2 of the electronic supplementary material.) Dashed line, performance of the random classifier; AUC, area under ROC curve; PPV0.05, positive predictive value, i.e. fraction of true positives among the 5% highest scoring predictions. Open circles, MIPS; open squares, KEGG.
Figure 6.
The positive predictive value drops quickly with increasing rate of positive predictions of the full MIPS dataset. The KEGG dataset produced similar results (data not shown). The dashed horizontal line indicates the performance of a random classifier, i.e. P/(P+N).
An alternative cause for the limited coverage of phylogenetic profiling is the discrepancy between the complexity of organisms and the severe simplifications required for a phylogenetic profiling analysis. A discussion of this discrepancy may suggest limitations of phylogenetic profiling and serves to introduce some methods that can be interpreted as extensions of, or at least closely related to phylogenetic profiling. For this discussion, it is helpful to consider ‘functional similarity’ as quantifiable. For example, a quantification can be based on similarity measures for controlled vocabularies that describe protein function, such as the function description in the clusters of orthologous groups (COG) database (Tatusov et al. 2000) or the gene ontology (Ashburner et al. 2000), e.g. the co-occurrence of terms associated with two proteins could be quantified by the Jaccard coefficient or the average Resnik similarity for ontology terms (Resnik 1999).
6.1 Functional divergence after gene duplication
Already during the process of profile construction, i.e. the mapping of the complexity of organisms into simple vectors of numbers, information is lost. The construction of phylogenetic profiles from groups of homologous genes is relatively easy by standard homology detection algorithms, but ignores that genome evolution is highly dynamic and driven by events such as de novo gene genesis, gene duplication, gene loss and HGT. Consequently, multiple homologous genes may coexist in the same genome. In particular, sub- or neo-functionalization of duplicated genes (reviewed in e.g. Roth et al. 2006) represents a major complication to any homology-based and genomic-context method, because they violate the assumption that homologous genes retain their function during evolution. For instance, if the homologue of ancestral function is not maintained but lost from one genome then, in the phylogenetic profile, the presence of homologous genes in different genomes will represent the presence of different functions.
Fortunately, the impact of functional divergence after duplication on the quality of genomic context-based predictions can be reduced by splitting homologous groups into orthologous groups. Two homologous genes are called orthologues if they were separated by a speciation. In contrast, if two genes have been separated by gene duplication they are called paralogues (Fitch 1970). Reliable orthology and paralogy relations between members of a gene family can be determined either by gene tree reconciliation, i.e. by comparing the tree of homologous genes to a trusted species tree or by examining the species overlap between subtrees to infer duplications (van der Heijden et al. 2007). Nevertheless, due to the practical complications of large-scale phylogenetic analyses, orthology is usually operationally based on some clustering approach or other heuristic that uses relative levels of sequence similarity (e.g. Tatusov et al. 1997; Remm et al. 2001). Alternative to constructing the orthologous groups and phylogenetic profiles by oneself, one can also download them from public databases such as RoundUp (Deluca et al. 2006) or COG (Tatusov et al. 2003).
By switching from homologous groups to orthologous groups, the impact of functional divergence after duplication can be reduced, but the situation is not principally solved. Despite the orthology relationship, still about 40% of the bacterial orthologous groups in the COG database contain paralogues that do not originate from species-specific family expansion (Tatusov et al. 2000). Furthermore, in a study of modularity based on various genomic-context methods, it was found that 40% of the genomic context-derived modules could not be unambiguously assigned to a metabolic pathway owing to the limited resolution of their orthologous groups (von Mering et al. 2003b).
6.2 Multifunctionality
Proteins often functionally interact with or depend on various other proteins. If all these associated proteins are functionally related with each other, then the functional context of the protein is homogeneous. An interesting situation arises for ‘multifunctional’ proteins whose functional context is heterogeneous, i.e. for those proteins that have ‘long-range’ dependencies bridging different processes. In this situation, it is not obvious whether it is more advantageous to maintain the gene or to lose it. If one part of the gene's functional context is lost, the loss of the gene itself may still affect the functioning of other parts of its context. Hence, multifunctionality probably influences the gene loss rate and thus the occurrence values in the phylogenetic profiles. There are numerous examples of multifunctional and ‘moonlighting’ proteins some of which are members or important cellular processes like glycolysis or tricarboxylic acid cycle (for a review see Jeffery 1999; Moore 2004). High-throughput protein interaction experiments indicate that many proteins are shared between different complexes (Gavin et al. 2002; Krause et al. 2004), which suggests that multifunctionality may be an abundant phenomenon. Various mechanisms allow a protein to act in different functional contexts, such as broad substrate specificity, differential expression and differential localization (Jeffery 1999). One of these mechanisms, the functional specialization of different domains of the protein, allows a protein to contribute to different processes by the activity of its different domains. It is amenable to what could be called ‘localized’ phylogenetic profiling methods. Both co-occurrence and similarity profiling methods have been extended in this direction.
Pagel et al. (2004) proposed a domain-based co-occurrence profiling method. For each PFAM and SCOP domain (Bateman et al. 2002; Andreeva et al. 2004), they constructed an occurrence profile and compared profile pairs with Hamming distance in order to predict functional relations between domains. Subsequently, proteins were considered as functionally related if they contained such functionally related domains. Thus, the approach taken by Pagel and co-workers compares proteins indirectly via their domain content. In contrast, the localized similarity profiling introduced by Kim & Subramaniam (2006) directly compares the sequences of proteins by local alignments using a variant of the phylogenetic profiling method of Date & Marcotte (2003). Each query protein (from a query species) is considered as a set of overlapping 120 amino acid segments that are individually compared to the database of proteins. The coevolution between two protein segments is quantified by mutual information, just as for full length proteins in the Date and Marcotte method. The correlation between two proteins was defined as the highest correlation between any of their segments (Kim & Subramaniam 2006). In order to be independent of pre-defined segments, Kim et al. (2006) further improved this method into residue-level profiles. Both local co-occurrence profiling and similarity profiling were found to be largely complementary to their global variants (Pagel et al. 2004; Kim & Subramaniam 2006). For example, the methods of Kim and Subramaniam and of Date and Marcotte had very similar performance but shared only about 36% of their predictions or less depending on the choice of cut-off (Kim & Subramaniam 2006; Kim et al. 2006). Localized phylogenetic profiling thus provides an additional source of evidence for functional relation and adds information when combined with global profiling methods.
6.3 Higher order functional relations
Most co-occurrence profiling methods make two simplifying assumptions about the dependencies of proteins: they only search for binary, i.e. pair-wise, and symmetric relationships. ‘Symmetric’ here means that if gene A is associated with gene B then the reverse, i.e. gene B is associated with gene A, is also true. For example, for Hamming distance, it is irrelevant if we calculate the distance dH(A, B) or dH(B, A). Consequently, co-occurrence profiling may frequently fail to detect asymmetric relationships. Barker & Pagel (2005) pointed out that their maximum likelihood method can be adapted to model contingent evolution of gene pairs, i.e. to model asymmetric, binary relationships between phylogenetic profiles. For example, asymmetric relationships could occur whenever a major system, such as a complex or pathway, is modified during evolution. While the main function of a complex is evolutionarily conserved, accessory proteins could be added or removed for fine tuning. Sometimes these modifications of systems happen in a gradual fashion over longer evolutionary time-scales, as has been observed for a variety of multi-protein complexes and metabolic pathways (Fothergill-Gilmore & Michels 1993; Petsko et al. 1993; Gabaldon et al. 2005; Tanaka et al. 2005). Asymmetric, binary relationships could be used to uncover these evolutionary trends, in particular, if more sequenced genomes become available and the resolution of phylogenetic trees is increased. Nevertheless, currently a systematic assessment of binary, asymmetric relationships between occurrence profiles is lacking. In contrast, ternary relationships, i.e. for triples of gene families, have been studied in the past. In the Boolean logic formalism proposed by Bowers et al. (2004), the profiles of orthologous groups are interpreted as vectors of truth values. For example, consider an enzyme C that uses substrates from both a pathway containing enzyme A and from a pathway containing enzyme B. The relation between the three enzymes can be expressed as the logic relation A∧B→C. Given such a relation, it can be concluded that both the activities of A and B are required for the activity of C. For example, for members A and B of two signalling cascades whose crosstalk involves the protein C. Bowers and co-workers identified eight possible ‘logic types’ that model different possible relationships between genes, excluding cases that could easily be modelled by binary relationships, and scored them using an entropy-related measure. Intriguingly, logic types that are easier to relate to our understanding of biological and evolutionary relationships tended to be more frequent than those that are difficult to interpret. Owing to the higher expressiveness of the ternary relation formalism, its results add to the results of classical, binary co-occurrence methods. Recently, the formalism has been used to model more complex relations of four genes (Zhang et al. 2006). It should, however, be noted that the identification of higher order relationships has practical limitations. The number of possible logic functions is exponential in the number of related genes. Each of these possible relations can be interpreted as an alternative hypothesis and the fitting of such a large number of hypotheses may not be possible with the currently available number of genomes. Furthermore, also the number of combinations of genes between which higher order relations are inferred grows quickly, which limits further extensions to small gene sets. Finally, with increasing complexity of the logic functions, a biological interpretation of the relationship between the involved genes becomes harder.
6.4 The evolution of functional context
If we consider binary functional dependencies and ignore that these dependencies can be asymmetric, we obtain a network of functional associations. Networks of functional associations between proteins can evolve on the level of edges, i.e. the interactions between proteins, and on the level of nodes, i.e. by gain and loss of protein-encoding genes. Obviously, both levels are linked because if a gene is lost from a genome also all functional links to it are lost. The edge-level evolution can be interpreted as a change of protein function; more specifically, by rewiring the network the functional context of a protein changes. The edge-level of the network is particularly relevant for phylogenetic profiling simply because phylogenetic profiling tries to predict the edges. However, it does this in a comparative way and thus disregards that the edges may have changed during evolution. Functional links between genes must have persisted sufficiently long during evolution to be detectable by phylogenetic profiling methods.
Unfortunately, no conclusive answer can yet be given on what is this rate of edge-level evolution, even for protein–protein interaction networks or networks derived from co-expression. The estimation of this rate is hampered by the high levels of noise in high-throughput data (Snel et al. 2004; Cesareni et al. 2005; Gandhi et al. 2006). For example, for the same dataset of tissue-specific expression in human and mouse, estimates of the fraction of gene pairs with conserved co-expression range between less than 10 and 84% (Liao & Zhang 2006; Tsaparas et al. 2006). In comparison, for gene pairs found in the relatively distantly related worm and budding yeast, the fraction of co-expressed gene pairs that share a transcription factor-binding site in yeast that is also co-expressed in worm was determined to be 76% (Snel et al. 2004), which indicates a high conservation of co-expression at least for conserved gene pairs.
A different aspect of the evolution of functional context is the ‘severity’ of change. A ‘severe’ change of functional context occurs if a protein is recruited to a new context that is unrelated to its original context. This recruitment may or may not involve a loss of the original functional links. Notably, if a protein maintains both its original and acquired functions, its functional context becomes heterogeneous, i.e. it can be considered multifunctional. However, if the original function is lost then the selective forces acting on the protein before and after the recruitment will be distinct. Recruitment appears to be a major mode in the evolution of metabolic pathways (Teichmann et al. 2001; Lecompte et al. 2002; Rison et al. 2002; Light & Kraulis 2004;). In contrast, ancestral and acquired functions can be related such that the ancestral and the divergent form of a gene may be subjected to similar selective constraints. For example, in the small-molecule metabolic network of Escherichia coli homologous genes separated by 11 or less reaction steps are overall rare but occur more frequently at very short distances (1–3), i.e. with relatively high functional similarity, than at larger distances (4–11; Rison et al. 2002). Similarly, duplicated physical complexes in most cases have similar functions (Pereira-Leal & Teichmann 2005).
6.5 Evolutionary modularity
A further abstraction from networks of functional links is to analyse the structure of organisms in terms of modularity. Usually, a module is defined as a group of proteins that have stronger associations among each other than to proteins outside of the module. The modular structure of biological networks is figured as hierarchical and overlapping (e.g. Ravasz et al. 2002; Palla et al. 2005): different modules are related to each other by combination into higher order modules or by coupling proteins. Certain topological properties of biological networks can be explained by high false positive rates in interaction data (Han et al. 2005) or by simple evolutionary models (Amoutzias et al. 2004; van Noort et al. 2004). Nevertheless, the modular organization is apparent and it remains an open question which selective forces or evolutionary mechanisms are responsible (e.g. Wagner 1996; Kashtan & Alon 2005). One would expect that the functional modularity of organisms constraints the evolutionary process, leading to a definition of evolutionary modularity that closely follows the general definition of modularity: an evolutionary module can be defined as a group of genes that have stronger co-evolved with each other than with genes outside of the module.
A number of genomic-context studies have included gene co-occurrence to find functional modules (Snel et al. 2002; Yanai & DeLisi 2002; von Mering et al. 2003b; Yamada et al. 2004, 2006; Spirin et al. 2006; Wu et al. 2006). Despite the technical and conceptual problems, genomic-context networks do indeed reflect functional modules, although the resulting modules are frequently distinct from traditional module definitions (Spirin et al. 2006; Yamada et al. 2006). The highest scoring pair-wise associations predicted by phylogenetic profiling can be considered as small co-gained and co-lost evolutionary modules and clearly contain information about function. Furthermore, also larger modular structures are reflected in the networks. Of particular value for their description are multifunctional network nodes that link different modules. For example, metabolic networks are usually modelled as graphs with nodes representing metabolites and the edges between them representing enzymes. The linkers in these networks are metabolites that are involved in diverse pathways. Indeed, in networks derived from genomic context, the evidence of a functional association between genes decreased with the degree of the metabolite node connecting them (Huynen & Snel 2003; von Mering et al. 2003b) and, consistently, for linear pathways the correspondence of the genomic-context network and the known metabolic network is highest (Spirin et al. 2006). A similar observation was made in a network based on gene order conservation. The linkers in these networks connect locally unconnected clusters and were enriched in multifunctional enzymes (Snel et al. 2002).
A lack of evolutionary modularity may be an important cause for the limited predictive coverage of phylogenetic profiling and other genomic-context methods. Studies that did not rely on genomic context to define their modules found a heterogeneous distribution of their evolutionary modularity; only about half of the functional classes, like metabolic pathways or protein complexes, evolved significantly modularly (Snel & Huynen 2004; Campillos et al. 2006). Consistent with this, genomic context is not equally informative about every cellular process (Campillos et al. 2006). For example, catabolic pathways were found to be less modular than biosynthetic pathways (Snel & Huynen 2004; Campillos et al. 2006) and showed a lower coverage in combined genomic-context networks (von Mering et al. 2003b). This suggests that the signal that genomic-context methods rely on may have only a limited coverage simply because there are many cellular processes whose evolution does not reflect a modular structure.
7. Concluding remarks
Phylogenetic profiling is a versatile method for the prediction of functional interactions, but its coverage is limited to those cellular systems that evolved in a modular fashion. Nevertheless, basically all experimental and computational methods have limits as they measure only one or few aspects of functional association. For instance, microarrays only capture co-expression on the level of mRNA abundance and thus ignore the effects of post-translational regulation. Numerous examples show that the evolutionary evidence for a functional interaction based on phylogenetic profiling as well as other genomic-context methods is frequently neither strong nor specific enough to pin down the exact function of a protein. Consequently, in virtually all examples, additional experimental evidence was used to stress the conclusions drawn from phylogenetic profiling and to understand protein function at a much higher level of detail.
The necessity to combine multiple lines of evidence extrapolates to phylogenetic profiling used as a broad-scale method that predicts functional interactions for a large number of gene pairs. Phylogenetic profiling has thus been integrated with other genomic-context methods. For prokaryotes, conservation of gene neighbourhood is usually found to have the highest coverage, followed by phylogenetic profiling and, finally, gene fusion/fission (Huynen et al. 2000; Manson McGuire & Church 2000; Yanai & DeLisi 2002; von Mering et al. 2003b). Genomic-context methods can have higher coverage than for example yeast two-hybrid at about the same accuracy (von Mering et al. 2002). Although such measurements depend somewhat on the benchmarking set (Lee et al. 2004), this shows that genomic-context methods can well compete with high-throughput experimental approaches. Frequently, one type of genomic evidence is clearly dominating (Huynen et al. 2000; Yanai & DeLisi 2002) and, thus, their contributions to the integrated network are complementary. It will be interesting to see if the predictions made by localized profiling approaches, ternary interactions and binary, asymmetric interactions will add further to the predictive coverage of phylogenetic profiling and of genomic-context methods in general.
8. Data and methods
8.1 Phylogenetic profiles
The protein sequences of the microsporidium Encephalitozoon cuniculi and of 25 fungi (Ustilago maydis, Cryptococcus neoformans, Phanerochaete chrysosporium, Stagonospora nodorum, Aspergillus nidulans, Aspergillus fumigatus, Magnaporthe grisea, Neurospora crassa, Trichoderma reesei, Fusarium graminearum, Schizosaccharomyces pombe, Yarrowia lipolytica, Debaromyces hansenii, Candida albicans, Candida glabrata, Ashbya gossypii, Kluyveromyces lactis, Kluyveromyces waltii, Saccharomyces kluyveri, Saccharomyces castellii, Saccharomyces bayanus, Saccharomyces kudriavzevii, Saccharomyces mikatae, Saccharomyces paradoxus, Saccharomyces cerevisiae) were downloaded from the corresponding sequencing projects. Similarity scores between the proteomes were computed using the Smith–Waterman P algorithm (Smith & Waterman 1981) on a TimeLogic DeCypher (matrix: Blosum62; e-value cut-off: 0.01; low-complexity filter on). The orthologous groups were constructed by an approach similar to that used for the COGs (Tatusov et al. 2000). Inparalogues specific to a clade usually are more similar to each other than to any gene outside the clade. This was used to construct species-specific inparalogous groups and inparalogous groups specific to the Saccharomyces sensu stricto clade (S. kudriavzevii, S. mikatae, S. bayanus, S. paradoxus, S. cerevisiae) whose members are closely related. Subsequently, triangles of mutual best hits between the inparalogous groups were merged if they shared two members (Tatusov et al. 2000). The resulting orthologous groups were further refined: we aligned the orthologous genes using Muscle v. 3.52 (Edgar 2004) with default parameters, calculated neighbour-joining trees with Bio-NJ (Gascuel 1997), inferred duplications with Loft (van der Heijden et al. 2007) and split orthologous groups according to ancient duplications. These refined orthologous groups were used to construct the phylogenetic profiles.
8.2 Control datasets
We retrieved the complex catalogue from the Munich Information Centre for Protein Sequences (MIPS; as of 14 November 2005; Mewes et al. 2006). From this hierarchy of categories, we removed those that had subcategories, contained the keywords ‘other’ or ‘complexes’, or referred to high-throughput studies (category 550). The procedure yielded a total of 1195 assignments of orthologues to 195 complexes. All pairs of orthologous groups that shared a MIPS complex were considered as positive controls. An alternative positive control dataset was constructed from pairs of orthologous groups with members in budding yeast that shared a KEGG map (as of 17 January 2006; 187 maps; Kanehisa et al. 2004) and are found in the same cellular location (Huh et al. 2003). We found that the latter requirement improved the benchmarking results (data not shown). The negative controls, which should be functionally unrelated, were constructed from the orthologous groups that occurred in the positive controls. To this aim, from each set of positive controls, we sampled pairs of orthologous groups that did not occur in any of the positive control datasets. Additionally, negative controls were not allowed to reside in the same cellular location in budding yeast (Huh et al. 2003). We took this approach to have the same distribution of profile entropies and loss rates in positive and negative controls because some methods, such as mutual information, are sensitive to the entropy of the profiles (see §8.4.8) or loss rate (Barker et al. 2007). Note that both control datasets only refer to a subset of the 5997 orthologous groups present in budding yeast. From all datasets, we excluded anti-correlating pairs (Pearson correlation r<0) because mutual information score gives high scores to both correlating and anti-correlating pairs and thus deviates from the other methods. Similarly, pan-orthologues, i.e. orthologues or profiles with presence values for all species in the dataset, were excluded because the maximum likelihood approach (Barker & Pagel 2005) and the Pearson correlation coefficient cannot be calculated for these. A summary of the data filtering is given in table 1 of the electronic supplementary material.
8.3 Fungal tree
The fungal tree was based on a concatenated alignment of selected sequences (Dutilh et al. 2007): only orthologous genes occurring in at least 25 of the 26 genomes were used. Thus, in order to have a maximum of residues for the tree construction, the unnecessary loss of orthologous groups that were only lacking from, for example, the degenerated genome of E. cuniculi was minimized. We started by merging inparalogous groups (see §8.1) into clusters based on the bidirectional best hits between them. Subsequently, species-specific expansions were filtered out: we aligned the genes in each cluster using Muscle (Edgar 2004) with default parameters, and calculated pair-wise protein distances with Tree-Puzzle v. 5.2 (Schmidt et al. 2002; approximate parameter estimates; parameter estimation uses neighbour-joining tree; JTT model of substitution; estimate amino acid frequencies from dataset; four gamma categories; alpha=1.00 (weak rate heterogeneity)). From the pair-wise distances, the neighbour-joining trees were constructed with Bio-NJ (Gascuel 1997) and after identification of species-specific duplications by Loft (van der Heijden et al. 2007) from each genome, only the duplicate with the shortest branch lengths to the root was retained. The underlying assumption is that the gene with the highest sequence conservation is most probably the one with the conserved function after duplication. This yielded 229 orthologous groups that contained no more than one gene in each of at least 25 species. For genes that were lacking from a genome, the concatenated Muscle alignments contained a gap. We used GBlocks (default parameters; Castresana 2000) to select blocks of unambiguously aligned amino acids, which resulted in a super-alignment of 132 409 conserved positions. Finally, we used PhyML to calculate the maximum likelihood tree depicted in figure 1 of the electronic supplementary material (JTT model of substitution; estimated proportion of invariable sites; four substitution rate categories; gamma fixed with alpha=1.00; Guindon & Gascuel 2003). This tree was used for the tree-guided and model-based profiling methods.
8.4 Phylogenetic profiling methods
8.4.1 Tree-kernel method
The C implementation of the tree-kernel method was downloaded from http://cg.ensmp.fr/∼vert/publi/ismb02/index.html and run with the default gain/loss and retention probabilities (Vert 2002). We used the distances in feature space as scores with small distances corresponding to high similarity of evolutionary history.
8.4.2 Maximum likelihood method
The likelihood ratios (LhRs) from the maximum likelihood method were calculated with a version of BayesMultiState program that allows fixing the state at the trees root (Barker & Pagel 2005), which was kindly provided by Andrew Meade. It is defined as , with denoting the maximum likelihood of the null model of independent evolution and the maximum likelihood of the alternative model of contingent evolution. The number of tries for finding the maximum likelihood of each model was set to 100, which deviates considerably from the default (10 tries) and was chosen as a suitable trade-off between computational costs and the variance of the estimated LhR as determined in five repeats of a sample of 100 profiles (data not shown). Despite the increased number of tries in few cases, we observed negative LhRs, which indicate a problem of the algorithm to find the global likelihood optimum. In these cases, we ran the algorithm another 100 tries, which reduced the number of orthologous group pairs with LhR less than 0 to 17.
8.4.3 Differential parsimony
The distance d(A, B) between the parsimonious reconstructions of two orthologous groups A and B was calculated as (D. Liberles 2006, personal communication)
Here, for instance, anc(ai) denotes the ancestral state for the branch i inferred for the orthologous group A and desc(ai) denotes the corresponding descendant's state. This gives no penalty to coordinate gains, coordinate losses or if no gain or loss of either orthologous group occurred, a penalty of 1 to independent gains or losses and a penalty of 2 for coordinate gain in one orthologous groups and loss in the other. In our implementation of differential parsimony, we use Dollo parsimony instead of Fitch parsimony (cf. Liberles et al. 2002) and we restricted the dataset to orthologous groups present in budding yeast. Consequently, a penalty of 2 did not occur. Notably, an alternative representation of the ancestral state reconstruction is as a vector with components corresponding to branches of the phylogeny and values corresponding to the ‘gain’ and ‘loss’ events as well as ‘no’ event. In the absence of penalty 2, differential parsimony is simply the Hamming distance between these gain/loss vectors. As an alternative similarity measure between the gain/loss vectors, we used a two-tailed Fisher's exact test to quantify the co-loss probability on the branches of the tree.
8.4.4 Tree-guided methods
Both mutual information and Jaccard coefficient used the tree-guided collapsing of subtrees that is described in §4.2. Furthermore, a single pseudocount for each of the four possible presence/absence combinations was added (von Mering et al. 2003a).
8.4.5 Jaccard coefficient
The Jaccard coefficient of two occurrence vectors A and B (Jaccard 1912) can be defined for co-occurrence of presences (a=1) as well as for co-occurrences of absences (a=0; see below). Here, we used the co-occurrence of presences (a=1).
8.4.6 Lp-norms
Lp-norms are defined as . Frequently used Lp-norms are the Manhattan distance or Hamming distance (p=1) and the Euclidean distance (p=2). For arbitrary values of p, the orthologous group pairs will have the same order when sorted by distance Lp and we thus use only the Hamming distance.
8.4.7 Pearson correlation coefficient
We used a standard linear correlation coefficient
8.4.8 Mutual information
Mutual information can be defined based on the Kullback entropy between two probability distributions {p0} and {p} as
The Kullback entropy quantifies the amount of information gained when substituting distribution {p0} by distribution {p}. The index i refers to the possible values drawn from the distributions. For the purpose of comparing two profiles A and B, the possible values correspond to the four combinations of occurrence values of two genes, i.e. we define pi≔p(a, b) and pi0≔p0(a, b), a∈A, b∈B. We furthermore assume that the occurrence values of the genes are statistically independent, i.e. p0(a, b)=p(a) p(b). Thus, the mutual information is defined as
The probabilities p are usually estimated by the frequencies of the occurrence values (for p(a) and p(b)) or combinations of occurrence values (for p(a, b)) in the compared profiles. Note that the maximally achievable mutual information is determined by the minimum entropy of the compared profiles (Steuer et al. 2002)
H(A) being the entropy of a profile defined as
8.5 Benchmarking
The positive predictive value (PPV) is defined as
Here, TP(x) and FP(x) are the numbers of true positives and false positives, respectively, in the test set that score better than the specified fraction or number x of positive prediction. If the fraction of positive predictions is set to x=1 then the positive predictive value will be completely determined by the ratio of positive and negative controls in the dataset, i.e. PPV=P/(P+N). This ratio is equivalent to the performance of the random classifier that selects positive predictions randomly from the controls. To be able to compare positive predictive values between datasets, it is important that the datasets have the same ratio of positive to negative controls. Alternatively, identical P/N ratios can be achieved by bootstrapping (see §8.5.1).
8.5.1 Bootstrapping
We used a weighted bootstrapping procedure that assigns equal weights to all complexes/pathways and generates a ratio of positive to negative controls of 1 : 1. The members of each complex/pathway were assigned the weight 1/(complex size). Proteins shared between complexes/pathways were assigned the average of the per complex/pathway weights. The negative controls were given the same weight as the sum of the weights of the positive controls.
Acknowledgments
We are grateful to David Liberles for giving us information on the differential parsimony approach and Andrew Meade for providing the newest version of the BayesMultiState software. We thank Fiona Nielsen for valuable discussion. We also thank the three anonymous referees who helped to improve this document. This work was funded by the Netherlands Bioinformatics Centre (NBIC), which is supported by the Netherlands Genomics Initiative (NGI) and by the European Union's Sixth Framework Programme EPISTEM (CT-2005-019067). The orthologous groups were constructed by V.N. The fungal tree was constructed by B.E.D.
Supplementary Material
Additional details on the analysis of co-occurrence profiling methods
References
- Altincicek B, Kollas A, Eberl M, Wiesner J, Sanderbrand S, Hintz M, Beck E, Jomaa H. LytB, a novel gene of the 2-c-methyl-d-erythritol 4-phosphate pathway of isoprenoid biosynthesis in Escherichia coli. FEBS Lett. 2001a;499:37–40. doi: 10.1016/S0014-5793(01)02516-9. [DOI] [PubMed] [Google Scholar]
- Altincicek B, Kollas A.K, Sanderbrand S, Wiesner J, Hintz M, Beck E, Jomaa H. GcpE is involved in the 2-c-methyl-d-erythritol 4-phosphate pathway of isoprenoid biosynthesis in Escherichia coli. J. Bacteriol. 2001b;183:2411–2416. doi: 10.1128/JB.183.8.2411-2416.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoutzias G.D, Robertson D.L, Oliver S.G, Bornberg-Bauer E. Convergent evolution of gene networks by single-gene duplications in higher eukaryotes. EMBO Rep. 2004;5:274–279. doi: 10.1038/sj.embor.7400096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J.O. Lateral gene transfer in eukaryotes. Cell. Mol. Life Sci. 2005;62:1182–1197. doi: 10.1007/s00018-005-4539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A, Howorth D, Brenner S.E, Hubbard T.J, Chothia C, Murzin A.G. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L. Guilt by association: contextual information in genome analysis. Genome Res. 2000;10:1074–1077. doi: 10.1101/gr.10.8.1074. [DOI] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader G.D, Hogue C.W.V. Analyzing yeast protein–protein interaction data obtained from different sources. Nat. Biotechnol. 2002;20:991–997. doi: 10.1038/nbt1002-991. [DOI] [PubMed] [Google Scholar]
- Barker D, Pagel M. Predicting functional gene links from phylogenetic–statistical analyses of whole genomes. PLoS Comput. Biol. 2005;1:e3. doi: 10.1371/journal.pcbi.0010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D, Meade A, Pagel M. Constrained models of evolution lead to improved prediction of functional linkage from correlated gain and loss of genes. Bioinformatics. 2007;23:14–20. doi: 10.1093/bioinformatics/btl558. [DOI] [PubMed] [Google Scholar]
- Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiko R.G, Harlow T.J, Ragan M.A. Highways of gene sharing in prokaryotes. Proc. Natl Acad. Sci. USA. 2005;102:14 332–14 337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann S, Ihmels J, Barkai N. Similarities and differences in genome-wide expression data of six organisms. PLoS Biol. 2004;2:E9. doi: 10.1371/journal.pbio.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovok I, Gorovitz B, Yanku M, Schreiber R, Gust B, Chater K, Aharonowitz Y, Cohen G. Alternative oxygen-dependent and oxygen-independent ribonucleotide reductases in Streptomyces: cross-regulation and physiological role in response to oxygen limitation. Mol. Microbiol. 2004;54:1022–1035. doi: 10.1111/j.1365-2958.2004.04325.x. [DOI] [PubMed] [Google Scholar]
- Bowers P.M, Cokus S.J, Elsenberg D, Yeates T.O. Use of logic relationships to decipher protein network organization. Science. 2004;306:2246–2249. doi: 10.1126/science.1103330. [DOI] [PubMed] [Google Scholar]
- Campillos M, von Mering C, Jensen L.J, Bork P. Identification and analysis of evolutionarily cohesive functional modules in protein networks. Genome Res. 2006;16:374–382. doi: 10.1101/gr.4336406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Cesareni G, Ceol A, Gavrila C, Palazzi L.M, Persico M, Schneider M.V. Comparative interactomics. FEBS Lett. 2005;579:1828–1833. doi: 10.1016/j.febslet.2005.01.064. [DOI] [PubMed] [Google Scholar]
- Cherry J.M, et al. SGD: Saccharomyces genome database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs D.J, Nagel R.J, Ares M, Jr, Stevens S.W. Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol. Cell Biol. 2006;26:523–534. doi: 10.1128/MCB.26.2.523-534.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley S.D. Evolution of a metabolic pathway for degradation of a toxic xenobiotic: the patchwork approach. Trends Biochem. Sci. 2000;25:261–265. doi: 10.1016/S0968-0004(00)01562-0. [DOI] [PubMed] [Google Scholar]
- Cunningham F.X, Jr, Lafond T.P, Gantt E. Evidence of a role for LytB in the nonmevalonate pathway of isoprenoid biosynthesis. J. Bacteriol. 2000;182:5841–5848. doi: 10.1128/JB.182.20.5841-5848.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. 1998;23:324–328. doi: 10.1016/S0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- Date S.V, Marcotte E.M. Discovery of uncharacterized cellular systems by genome-wide analysis of functional linkages. Nat. Biotechnol. 2003;21:1055–1062. doi: 10.1038/nbt861. [DOI] [PubMed] [Google Scholar]
- Date S.V, Marcotte E.M. Protein function prediction using the Protein Link EXplorer (PLEX) Bioinformatics. 2005;21:2558–2559. doi: 10.1093/bioinformatics/bti313. [DOI] [PubMed] [Google Scholar]
- Deluca T.F, Wu I.H, Pu J, Monaghan T, Peshkin L, Singh S, Wall D.P. Roundup: a multi-genome repository of orthologs and evolutionary distances. Bioinformatics. 2006;22:2044–2046. doi: 10.1093/bioinformatics/btl286. [DOI] [PubMed] [Google Scholar]
- Dutilh B.E, van Noort V, van der Heijden R.T, Boekhout T, Snel B, Huynen M.A. Assessment of phylogenomic and orthology approaches for phylogenetic inference. Bioinformatics. 2007;23:815–824. doi: 10.1093/bioinformatics/btm015. [DOI] [PubMed] [Google Scholar]
- Dynes J.L, Firtel R.A. Molecular complementation of a genetic marker in Dictyostelium using a genomic DNA library. Proc. Natl Acad. Sci. USA. 1989;86:7966–7970. doi: 10.1073/pnas.86.20.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enault F, Suhre K, Abergel C, Poirot O, Claverie J.M. Annotation of bacterial genomes using improved phylogenomic profiles. Bioinformatics. 2003;1(Suppl. 1):i105–i107. doi: 10.1093/bioinformatics/btg1013. [DOI] [PubMed] [Google Scholar]
- Enright A.J, Iliopoulos I, Kyrpides N.C, Ouzounis C.A. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- Farris J.S. Phylogenetic analysis under Dollos law. Syst. Zool. 1977;26:77–88. doi: 10.2307/2412867. [DOI] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am. Nat. 1985;125:1–15. doi: 10.1086/284325. [DOI] [PubMed] [Google Scholar]
- Fitch W.M. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. doi: 10.2307/2412448. [DOI] [PubMed] [Google Scholar]
- Fitch W.M. Toward defining course of evolution—minimum change for a specific tree topology. Syst. Zool. 1971;20:406–416. doi: 10.2307/2412116. [DOI] [Google Scholar]
- Fothergill-Gilmore L.A, Michels P.A. Evolution of glycolysis. Prog. Biophys. Mol. Biol. 1993;59:105–235. doi: 10.1016/0079-6107(93)90001-Z. [DOI] [PubMed] [Google Scholar]
- Gaasterland T, Ragan M.A. Microbial genescapes: phyletic and functional patterns of ORF distribution among prokaryotes. Microbiol. Comp. Genomics. 1998;3:199–217. doi: 10.1089/omi.1.1998.3.199. [DOI] [PubMed] [Google Scholar]
- Gabaldon T, Huynen M.A. Prediction of protein function and pathways in the genome era. Cell Mol. Life Sci. 2004;61:930–944. doi: 10.1007/s00018-003-3387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldon T, Rainey D, Huynen M.A. Tracing the evolution of a large protein complex in the eukaryotes, NADH: Ubiquinone oxidoreductase (Complex I) J. Mol. Biol. 2005;348:857–870. doi: 10.1016/j.jmb.2005.02.067. [DOI] [PubMed] [Google Scholar]
- Galperin M.Y, Koonin E.V. Who's your neighbor? New computational approaches for functional genomics. Nat. Biotechnol. 2000;18:609–613. doi: 10.1038/76443. [DOI] [PubMed] [Google Scholar]
- Gandhi T.K.B, et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat. Genet. 2006;38:285–293. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- Gascuel O. BioNJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- Gavin A.C, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gelfand M.S, Koonin E.V. Avoidance of palindromic words in bacterial and archaeal genomes: a close connection with restriction enzymes. Nucleic Acids Res. 1997;25:2430–2439. doi: 10.1093/nar/25.12.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J, Elfond G, Shustrova A, Weisinger M, Pellegrini M, Cokus S, Rothschild B. Inferring protein interactions from phylogenetic distance matrices. Bioinformatics. 2003;19:2039–2045. doi: 10.1093/bioinformatics/btg278. [DOI] [PubMed] [Google Scholar]
- Glazko G.V, Mushegian A.R. Detection of evolutionarily stable fragments of cellular pathways by hierarchical clustering of phyletic patterns. Genome Biol. 2004;5:R32. doi: 10.1186/gb-2004-5-5-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh C.S, Bogan A.A, Joachimiak M, Walther D, Cohen F.E. Co-evolution of proteins with their interaction partners. J. Mol. Biol. 2000;299:283–293. doi: 10.1006/jmbi.2000.3732. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Haft D.H, Paulsen I.T, Ward N, Selengut J.D. Exopolysaccharide-associated protein sorting in environmental organisms: the PEP–CTERM/EpsH system. Application of a novel phylogenetic profiling heuristic. BMC Biol. 2006;4:29. doi: 10.1186/1741-7007-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.D, Dupuy D, Bertin N, Cusick M.E, Vidal M. Effect of sampling on topology predictions of protein–protein interaction networks. Nat. Biotechnol. 2005;23:839–844. doi: 10.1038/nbt1116. [DOI] [PubMed] [Google Scholar]
- Huh W.K, Falvo J.V, Gerke L.C, Carroll A.S, Howson R.W, Weissman J.S, O'Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Huynen M.A, Bork P. Measuring genome evolution. Proc. Natl Acad. Sci. USA. 1998;95:5849–5856. doi: 10.1073/pnas.95.11.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen M.A, Snel B. Exploiting the variations in the genomic associations of genes to predict pathways and reconstruct their evolution. In: Galperin M.Y, Koonin E.V, editors. Frontiers in computational genomics. Caisters Academic Press; Norfolk, VA: 2003. pp. 145–166. [Google Scholar]
- Huynen M.A, Diaz-Lazcoz Y, Bork P. Differential genome display. Trends Genet. 1997;13:389–390. doi: 10.1016/S0168-9525(97)01255-9. [DOI] [PubMed] [Google Scholar]
- Huynen M, Snel B, Lathe W, Bork P. Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res. 2000;10:1204–1210. doi: 10.1101/gr.10.8.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen M.A, Snel B, Bork P, Gibson T.J. The phylogenetic distribution of frataxin indicates a role in iron–sulfur cluster protein assembly. Human Mol. Genet. 2001;10:2463–2468. doi: 10.1093/hmg/10.21.2463. [DOI] [PubMed] [Google Scholar]
- Huynen M.A, Snel B, von Mering C, Bork P. Function prediction and protein networks. Curr. Opin. Cell Biol. 2003;15:191–198. doi: 10.1016/S0955-0674(03)00009-7. [DOI] [PubMed] [Google Scholar]
- Jaccard P. The distribution of the flora of the alpine zone. New Phytol. 1912;11:37–50. doi: 10.1111/j.1469-8137.1912.tb05611.x. [DOI] [Google Scholar]
- Jeffery C.J. Moonlighting proteins. Trends Biochem. Sci. 1999;24:8–11. doi: 10.1016/S0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- Jim K, Parmar K, Singh M, Tavazoie S. A cross-genomic approach for systematic mapping of phenotypic traits to genes. Genome Res. 2004;14:109–115. doi: 10.1101/gr.1586704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai A.W, Susskind M.M, Sauer R.T. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashtan N, Alon U. Spontaneous evolution of modularity and network motifs. Proc. Natl Acad. Sci. USA. 2005;102:13 773–13 778. doi: 10.1073/pnas.0503610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Subramaniam S. Locally defined protein phylogenetic profiles reveal previously missed protein interactions and functional relationships. Proteins—Struct. Funct. Bioinform. 2006;62:1115–1124. doi: 10.1002/prot.20830. [DOI] [PubMed] [Google Scholar]
- Kim Y, Koyuturk M, Topkara U, Grama A, Subramaniam S. Inferring functional information from domain co-evolution. Bioinformatics. 2006;22:40–49. doi: 10.1093/bioinformatics/bti723. [DOI] [PubMed] [Google Scholar]
- Kispal G, et al. Biogenesis of cytosolic ribosomes requires the essential iron–sulphur protein Rli1p and mitochondria. EMBO J. 2005;24:589–598. doi: 10.1038/sj.emboj.7600541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V, Mushegian A.R, Bork P. Non-orthologous gene displacement. Trends Genet. 1996;12:334–336. doi: 10.1016/0168-9525(96)20010-1. [DOI] [PubMed] [Google Scholar]
- Korbel J.O, Jensen L.J, von Mering C, Bork P. Analysis of genomic context: prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nat. Biotechnol. 2004;22:911–917. doi: 10.1038/nbt988. [DOI] [PubMed] [Google Scholar]
- Korbel J.O, Doerks T, Jensen L.J, Perez-Iratxeta C, Kaczanowski S, Hooper S.D, Andrade M.A, Bork P. Systematic association of genes to phenotypes by genome and literature mining. PLoS Biol. 2005;3:815–825. doi: 10.1371/journal.pbio.0030134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause R, von Mering C, Bork P, Dandekar T. Shared components of protein complexes—versatile building blocks or biochemical artefacts? Bioessays. 2004;26:1333–1343. doi: 10.1002/bies.20141. [DOI] [PubMed] [Google Scholar]
- Krylov D.M, Wolf Y.I, Rogozin I.B, Koonin E.V. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 2003;13:2229–2235. doi: 10.1101/gr.1589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov G.V, Kumar R.A, Koc A, Sun Z, Gladyshev V.N. Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc. Natl Acad. Sci. USA. 2002;99:4245–4250. doi: 10.1073/pnas.072603099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn P, et al. Crystal structure of thy1, a thymidylate synthase complementing protein from Thermotoga maritima at 2.25 Å resolution. Proteins. 2002;49:142–145. doi: 10.1002/prot.10202. [DOI] [PubMed] [Google Scholar]
- Kunin V, Ouzounis C.A. The balance of driving forces during genome evolution in prokaryotes. Genome Res. 2003;13:1589–1594. doi: 10.1101/gr.1092603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecompte O, Ripp R, Thierry J.C, Moras D, Poch O. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 2002;30:5382–5390. doi: 10.1093/nar/gkf693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Date S.V, Adai A.T, Marcotte E.M. A probabilistic functional network of yeast genes. Science. 2004;306:1555–1558. doi: 10.1126/science.1099511. [DOI] [PubMed] [Google Scholar]
- Lee Y, Park B.C, Lee do H, Bae K.H, Cho S, Lee C.H, Lee J.S, Myung P.K, Park S.G. Mouse transthyretin-related protein is a hydrolase which degrades 5-hydroxyisourate, the end product of the uricase reaction. Mol. Cells. 2006;22:141–145. [PubMed] [Google Scholar]
- Li J.B, et al. Comparative and basal genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/S0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Liao B.Y, Zhang J.Z. Evolutionary conservation of expression profiles between human and mouse orthologous genes. Mol. Biol. Evol. 2006;23:530–540. doi: 10.1093/molbev/msj054. [DOI] [PubMed] [Google Scholar]
- Liberles D.A. Evaluation of methods for determination of a reconstructed history of gene sequence evolution. Mol. Biol. Evol. 2001;18:2040–2047. doi: 10.1093/oxfordjournals.molbev.a003745. [DOI] [PubMed] [Google Scholar]
- Liberles D.A, Thoren A, von Heijne G, Elofsson A. The use of phylogenetic profiles for gene prediction. Curr. Genomics. 2002:131–137. doi: 10.2174/1389202023350499. [DOI] [Google Scholar]
- Light S, Kraulis P. Network analysis of metabolic enzyme evolution in Escherichia coli. BMC Bioinform. 2004;5:15. doi: 10.1186/1471-2105-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li J, Sam L, Goh C.S, Gerstein M, Lussier Y.A. An integrative genomic approach to uncover molecular mechanisms of prokaryotic traits. PLoS Comput. Biol. 2006;2:e159. doi: 10.1371/journal.pcbi.0020159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttgen H, et al. Biosynthesis of terpenoids: YchB protein of Escherichia coli phosphorylates the 2-hydroxy group of 4-diphosphocytidyl-2c-methyl-d-erythritol. Proc. Natl Acad. Sci. USA. 2000;97:1062–1067. doi: 10.1073/pnas.97.3.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay D.J.C. Cambridge University Press; Cambridge, UK: 2005. Information theory, inference, and learning algorithms. [Google Scholar]
- Makarova K.S, Wolf Y.I, Koonin E.V. Potential genomic determinants of hyperthermophily. Trends Genet. 2003;19:172–176. doi: 10.1016/S0168-9525(03)00047-7. [DOI] [PubMed] [Google Scholar]
- Manson McGuire A, Church G.M. Predicting regulons and their cis-regulatory motifs by comparative genomics. Nucleic Acids Res. 2000;28:4523–4530. doi: 10.1093/nar/28.22.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte E.M. Computational genetics: finding protein function by nonhomology methods. Curr. Opin. Struct. Biol. 2000;10:359–365. doi: 10.1016/S0959-440X(00)00097-X. [DOI] [PubMed] [Google Scholar]
- Marcotte E.M, Pellegrini M, Ng H.L, Rice D.W, Yeates T.O, Eisenberg D. Detecting protein function and protein–protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- Mellor J.C, Yanai I, Clodfelter K.H, Mintseris J, DeLisi C. Predictome: a database of putative functional links between proteins. Nucleic Acids Res. 2002;30:306–309. doi: 10.1093/nar/30.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewes H.W, et al. MIPS: analysis and annotation of proteins from whole genomes in 2005. Nucleic Acids Res. 2006;34:D169–D172. doi: 10.1093/nar/gkj148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S, Galagan J.E, Mesirov J.P. Improving genome annotations using phylogenetic profile anomaly detection. Bioinformatics. 2005;21:464–470. doi: 10.1093/bioinformatics/bti027. [DOI] [PubMed] [Google Scholar]
- Mirkin B.G, Fenner T.I, Galperin M.Y, Koonin E.V. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol. Biol. 2003:3. doi: 10.1186/1471-2148-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar D, Bringel F, Schuren F.H, de Vos W.M, Siezen R.J, Kleerebezem M. Exploring Lactobacillus plantarum genome diversity by using microarrays. J. Bacteriol. 2005;187:6119–6127. doi: 10.1128/JB.187.17.6119-6127.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B. Bifunctional and moonlighting enzymes: lighting the way to regulatory control. Trends Plant Sci. 2004;9:221–228. doi: 10.1016/j.tplants.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Morett E, Korbel J.O, Rajan E, Saab-Rincon G, Olvera L, Olvera M, Schmidt S, Snel B, Bork P. Systematic discovery of analogous enzymes in thiamin biosynthesis. Nat. Biotechnol. 2003;21:790–795. doi: 10.1038/nbt834. [DOI] [PubMed] [Google Scholar]
- Muhlenhoff U, Richhardt N, Ristow M, Kispal G, Lill R. The yeast frataxin homolog Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Human Mol. Genet. 2002;11:2025–2036. doi: 10.1093/hmg/11.17.2025. [DOI] [PubMed] [Google Scholar]
- Myllykallio H, Lipowski G, Leduc D, Filee J, Forterre P, Liebl U. An alternative flavin-dependent mechanism for thymidylate synthesis. Science. 2002;297:105–107. doi: 10.1126/science.1072113. [DOI] [PubMed] [Google Scholar]
- Natale D.A, Galperin M.Y, Tatusov R.L, Koonin E.V. Using the COG database to improve gene recognition in complete genomes. Genetica. 2000;108:9–17. doi: 10.1023/A:1004031323748. [DOI] [PubMed] [Google Scholar]
- Ogilvie I, Kennaway N.G, Shoubridge E.A. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J. Clin. Invest. 2005;115:2784–2792. doi: 10.1172/JCI26020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omland K.E. The assumptions and challenges of ancestral state reconstructions. Syst. Biol. 1999;48:604–611. doi: 10.1080/106351599260175. [DOI] [Google Scholar]
- Overbeek R, Fonstein M, D'Souza M, Pusch G.D, Maltsev N. The use of gene clusters to infer functional coupling. Proc. Natl Acad. Sci. USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M. Detecting correlated evolution on phylogenies—a general-method for the comparative-analysis of discrete characters. Proc. R. Soc. B. 1994;255:37–45. doi: 10.1098/rspb.1994.0006. [DOI] [Google Scholar]
- Pagel P, Wong P, Frishman D. A domain interaction map based on phylogenetic profiling. J. Mol. Biol. 2004;344:1331–1346. doi: 10.1016/j.jmb.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Palla G, Derenyi I, Farkas I, Vicsek T. Uncovering the overlapping community structure of complex networks in nature and society. Nature. 2005;435:814–818. doi: 10.1038/nature03607. [DOI] [PubMed] [Google Scholar]
- Pazos F, Valencia A. Similarity of phylogenetic trees as indicator of protein–protein interaction. Protein Eng. 2001;14:609–614. doi: 10.1093/protein/14.9.609. [DOI] [PubMed] [Google Scholar]
- Pazos F, Ranea J.A, Juan D, Sternberg M.J. Assessing protein co-evolution in the context of the tree of life assists in the prediction of the interactome. J. Mol. Biol. 2005;352:1002–1015. doi: 10.1016/j.jmb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Pellegrini M, Marcotte E.M, Thompson M.J, Eisenberg D, Yeates T.O. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Leal J.B, Teichmann S.A. Novel specificities emerge by stepwise duplication of functional modules. Genome Res. 2005;15:552–559. doi: 10.1101/gr.3102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsko G.A, Kenyon G.L, Gerlt J.A, Ringe D, Kozarich J.W. On the origin of enzymatic species. Trends Biochem. Sci. 1993;18:372–376. doi: 10.1016/0968-0004(93)90091-Z. [DOI] [PubMed] [Google Scholar]
- Pretzer G, et al. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bacteriol. 2005;187:6128–6136. doi: 10.1128/JB.187.17.6128-6136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani A.K, Marcotte E.M. Exploiting the co-evolution of interacting proteins to discover interaction specificity. J. Mol. Biol. 2003;327:273–284. doi: 10.1016/S0022-2836(03)00114-1. [DOI] [PubMed] [Google Scholar]
- Ramazzina I, Folli C, Secchi A, Berni R, Percudani R. Completing the uric acid degradation pathway through phylogenetic comparison of whole genomes. Nat. Chem. Biol. 2006;2:144–148. doi: 10.1038/nchembio768. [DOI] [PubMed] [Google Scholar]
- Ravasz E, Somera A.L, Mongru D.A, Oltvai Z.N, Barabasi A.L. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- Reguly T, et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J. Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm M, Storm C.E, Sonnhammer E.L. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- Resnik P. Semantic similarity in a taxonomy: an information-based measure and its application to problems of ambiguity in natural language. J. Artif. Intell. Res. 1999;11:95–130. [Google Scholar]
- Rison S.C, Teichmann S.A, Thornton J.M. Homology, pathway distance and chromosomal localization of the small molecule metabolism enzymes in Escherichia coli. J. Mol. Biol. 2002;318:911–932. doi: 10.1016/S0022-2836(02)00140-7. [DOI] [PubMed] [Google Scholar]
- Rodionov D.A, Gelfand M.S. Identification of a bacterial regulatory system for ribonucleotide reductases by phylogenetic profiling. Trends Genet. 2005;21:385–389. doi: 10.1016/j.tig.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Rodionov D.A, Vitreschak A.G, Mironov A.A, Gelfand M.S. Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J. Biol. Chem. 2002;277:48 949–48 959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- Roth C, Rastogi S, Arvestad L, Dittmar K, Light S, Ekman D, Liberles D.A. Evolution after gene duplication: models, mechanisms, sequences, systems, and organisms. J. Exp. Zool. B Mol. Dev. Evol. 2006;308:58–73. doi: 10.1002/jez.b.21124. [DOI] [PubMed] [Google Scholar]
- Sato T, Imanaka H, Rashid N, Fukui T, Atomi H, Imanaka T. Genetic evidence identifying the true gluconeogenic fructose-1,6-bisphosphatase in Thermococcus kodakaraensis and other hyperthermophiles. J. Bacteriol. 2004;186:5799–5807. doi: 10.1128/JB.186.17.5799-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Yamanishi Y, Kanehisa M, Toh H. The inference of protein–protein interactions by co-evolutionary analysis is improved by excluding the information about the phylogenetic relationships. Bioinformatics. 2005;21:3482–3489. doi: 10.1093/bioinformatics/bti564. [DOI] [PubMed] [Google Scholar]
- Schluter D, Price T, Mooers A.O, Ludwig D. Likelihood of ancestor states in adaptive radiation. Evolution. 1997;51:1699–1711. doi: 10.2307/2410994. [DOI] [PubMed] [Google Scholar]
- Schmidt H.A, Strimmer K, Vingron M, von Haeseler A. Tree-Puzzle: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Slonim N, Elemento O, Tavazoie S. Ab initio genotype–phenotype association reveals intrinsic modularity in genetic networks. Mol. Syst. Biol. 2006;2:2006.0005. doi: 10.1038/msb4100047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.F, Waterman M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Snel B, Huynen M.A. Quantifying modularity in the evolution of biomolecular systems. Genome Res. 2004;14:391–397. doi: 10.1101/gr.1969504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen M.A. The identification of functional modules from the genomic association of genes. Proc. Natl Acad. Sci. USA. 2002;99:5890–5895. doi: 10.1073/pnas.092632599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B, van Noort V, Huynen M.A. Gene co-regulation is highly conserved in the evolution of eukaryotes and prokaryotes. Nucleic Acids Res. 2004;32:4725–4731. doi: 10.1093/nar/gkh815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snitkin E.S, Gustafson A.M, Mellor J, Wu J, DeLisi C. Comparative assessment of performance and genome dependence among phylogenetic profiling methods. BMC Bioinform. 2006;7:420. doi: 10.1186/1471-2105-7-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirin V, Gelfand M.S, Mironov A.A, Mirny L.A. A metabolic network in the evolutionary context: multiscale structure and modularity. Proc. Natl Acad. Sci. USA. 2006;103:8774–8779. doi: 10.1073/pnas.0510258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz B.J, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuer R, Kurths J, Daub C.O, Weise J, Selbig J. The mutual information: detecting and evaluating dependencies between variables. Bioinformatics. 2002;18:S231–S240. doi: 10.1093/bioinformatics/18.suppl_2.s231. [DOI] [PubMed] [Google Scholar]
- Stuart J.M, Segal E, Koller D, Kim S.K. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- Sun J, Xu J, Liu Z, Liu Q, Zhao A, Shi T, Li Y. Refined phylogenetic profiles method for predicting protein–protein interactions. Bioinformatics. 2005;21:3409–3415. doi: 10.1093/bioinformatics/bti532. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Tateno Y, Gojobori T. Evolution of vitamin B-6 (Pyridoxine) metabolism by gain and loss of genes. Mol. Biol. Evol. 2005;22:243–250. doi: 10.1093/molbev/msi011. [DOI] [PubMed] [Google Scholar]
- Tatusov R.L, Koonin E.V, Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Tatusov R.L, Galperin M.Y, Natale D.A, Koonin E.V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R.L, et al. The COG database: an updated version includes eukaryotes. BMC Bioinform. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann S.A, Rison S.C.G, Thornton J.M, Riley M, Gough J, Chothia C. Small-molecule metabolism: an enzyme mosaic. Trends Biotechnol. 2001;19:482–486. doi: 10.1016/S0167-7799(01)01813-3. [DOI] [PubMed] [Google Scholar]
- Ternes P, Sperling P, Albrecht S, Franke S, Cregg J.M, Warnecke D, Heinz E. Identification of fungal sphingolipid C9-methyltransferases by phylogenetic profiling. J. Biol. Chem. 2006;281:5582–5592. doi: 10.1074/jbc.M512864200. [DOI] [PubMed] [Google Scholar]
- Tringe S.G, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- Tsaparas P, Marino-Ramirez L, Bodenreider O, Koonin E.V, Jordan I.K. Global similarity and local divergence in human and mouse gene co-expression networks. BMC Evol. Biol. 2006;6:70. doi: 10.1186/1471-2148-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson G.W, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Urbonavicius J, Skouloubris S, Myllykallio H, Grosjean H. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria—evolutionary implications. Nucleic Acids Res. 2005;33:3955–3964. doi: 10.1093/nar/gki703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden R.T, Snel B, van Noort V, Huynen M.A. Orthology prediction at scalable resolution through automated analysis of phylogenetic trees. BMC Bioinform. 2007;8:83. doi: 10.1186/1471-2105-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noort V, Snel B, Huynen M.A. Predicting gene function by conserved co-expression. Trends Genet. 2003;19:238–242. doi: 10.1016/S0168-9525(03)00056-8. [DOI] [PubMed] [Google Scholar]
- van Noort V, Snel B, Huynen M.A. The yeast coexpression network has a small-world, scale-free architecture and can be explained by a simple model. EMBO Rep. 2004;5:280–284. doi: 10.1038/sj.embor.7400090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter J.C, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Vert J.P. A tree kernel to analyse phylogenetic profiles. Bioinformatics. 2002;1(Suppl. 1):S276–S284. doi: 10.1093/bioinformatics/18.suppl_1.s276. [DOI] [PubMed] [Google Scholar]
- von Mering C, Krause R, Snel B, Cornell M, Oliver S.G, Fields S, Bork P. Comparative assessment of large-scale data sets of protein–protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. String: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003a;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Zdobnov E.M, Tsoka S, Ciccarelli F.D, Pereira-Leal J.B, Ouzounis C.A, Bork P. Genome evolution reveals biochemical networks and functional modules. Proc. Natl Acad. Sci. USA. 2003b;100:15 428–15 433. doi: 10.1073/pnas.2136809100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Jensen L.J, Kuhn M, Chaffron S, Doerks T, Kruger B, Snel B, Bork P. String 7-recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G.P. Homologues, natural kinds and the evolution of modularity. Am. Zool. 1996;36:36–43. [Google Scholar]
- Wu J, Kasif S, DeLisi C. Identification of functional links between genes using phylogenetic profiles. Bioinformatics. 2003;19:1524–1530. doi: 10.1093/bioinformatics/btg187. [DOI] [PubMed] [Google Scholar]
- Wu J, Hu Z, DeLisi C. Gene annotation and network inference by phylogenetic profiling. BMC Bioinform. 2006;7:80. doi: 10.1186/1471-2105-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Goto S, Kanehisa M. Extraction of phylogenetic network modules from prokayrote metabolic pathways. Genome Inform. Ser. Workshop Genome Inform. 2004;15:249–258. [PubMed] [Google Scholar]
- Yamada T, Kanehisa M, Goto S. Extraction of phylogenetic network modules from the metabolic network. BMC Bioinform. 2006;7:130. doi: 10.1186/1471-2105-7-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, DeLisi C. The society of genes: networks of functional links between genes from comparative genomics. Genome Biol. 2002;3:research0064. doi: 10.1186/gb-2002-3-11-research0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Derti A, DeLisi C. Genes linked by fusion events are generally of the same functional category: a systematic analysis of 30 microbial genomes. Proc. Natl Acad. Sci. USA. 2001;98:7940–7945. doi: 10.1073/pnas.141236298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Mellor J.C, DeLisi C. Identifying functional links between genes using conserved chromosomal proximity. Trends Genet. 2002;18:176–179. doi: 10.1016/S0168-9525(01)02621-X. [DOI] [PubMed] [Google Scholar]
- Yarunin A, Panse V.G, Petfalski E, Dez C, Tollervey D, Hurt E.C. Functional link between ribosome formation and biogenesis of iron–sulfur proteins. EMBO J. 2005;24:580–588. doi: 10.1038/sj.emboj.7600540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Kim S, Wang T, Baral C. Joint learning of logic relationships for studying protein function using phylogenetic profiles and the Rosetta Stone method. IEEE Trans. Signal Process. 2006;54:2427–2435. doi: 10.1109/TSP.2006.873718. [DOI] [Google Scholar]
- Zhou Y, Wang R, Li L, Xia X.F, Sun Z.R. Inferring functional linkages between proteins from evolutionary scenarios. J. Mol. Biol. 2006;359:1150–1159. doi: 10.1016/j.jmb.2006.04.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional details on the analysis of co-occurrence profiling methods