Abstract
Many adult regenerative cells divide infrequently but have high proliferative capacity. We developed a strategy to fluorescently label slow-cycling cells in a cell type–specific fashion. We used this method to purify the label-retaining cells (LRCs) that mark the skin stem cell (SC) niche. We found that these cells rarely divide within their niche but change properties abruptly when stimulated to exit. We determined their transcriptional profile, which, when compared to progeny and other SCs, defines the niche. Many of the >100 messenger RNAs preferentially expressed in the niche encode surface receptors and secreted proteins, enabling LRCs to signal and respond to their environment.
Epidermis and its appendages undergo continuous renewal and maintain reservoirs of multipotent SCs whose descendants are organized spatially and temporally. The epidermal basal layer (BL) contains putative SCs in addition to the transiently amplifying (TA) cells, which give rise to terminally differentiating suprabasal layers (1-3). The BL and the hair follicle outer root sheath (ORS) are contiguous and biochemically similar (fig. S1A). In the hair bulb, the dermal papilla (DP) maintains contact with matrix TA cells until they differentiate to form the inner root sheath (IRS) and hair shaft. Follicles periodically undergo cycles of growth (anagen), destruction (catagen), and rest (telogen). The zone between noncycling and cycling segments is a SC niche, the ORS “bulge” (4, 5).
Multipotent epithelial SCs with high proliferative potential reside in the bulge (6, 7). The bulge contains the majority of infrequently cycling, label-retaining cells (LRCs), which can respond to anagen DP signals to regenerate the follicle. After wounding or transplantation, bulge cells give rise to epidermis, follicles, and sebaceous glands. Additionally, when dissected from rat whiskers and cultured, bulge cells yield more colonies than other follicle segments (7).
It is not known what features define this specialized SC niche, what its interactions with bulge LRCs are, and whether all LRCs are SCs. To begin to address these issues, we devised a strategy based on the prediction that bulge SCs are uniquely both slow-cycling and active for a keratinocyte-specific promoter. With this strategy, we purify and characterize bulge LRCs and related keratinocyte progeny in the BL and ORS. Analyses of their transcriptional profiles reveal the skin LRC mRNAs; some of these mRNAs are found in SCs of other tissues, whereas others specify the unique environment of the skin SC niche.
To mark infrequently cycling cells of adult skin epithelium, we engineered transgenic mice to express histone H2B–green fluorescent protein (GFP) (8) controlled by a tetracycline-responsive regulatory element (TRE). A tightly regulated TRE-mCMV-H2B-GFP founder animal was crossed with mice harboring a keratin 5 (K5) promoter–driven tet repressor–VP16 transgene (9), and offspring were selected for doxycycline (Tet)–controlled regulation restricted to skin epithelium (Fig. 1A). Without Tet, backskin epithelial cells exhibited ∼103 to 104 units of GFP fluorescence (9, 10) (Fig. 1B). After feeding 4-week-old mice Tet for 4 weeks to 4 months (chase), only bulge cells (<1% total) retained fluorescence at ≥103 units (Fig. 1B) (fig. S1). Independent of cell surface markers, our approach marks putative bulge SCs on the basis of their slow-cycling properties. Although we used keratinocyte-specific TetoffVP16 mice, TRE-mCMV-H2B-GFP animals could be used with mice expressing other promoter/enhancer-driven, Tet-regulatable activators/repressors to isolate LRCs from other tissues.
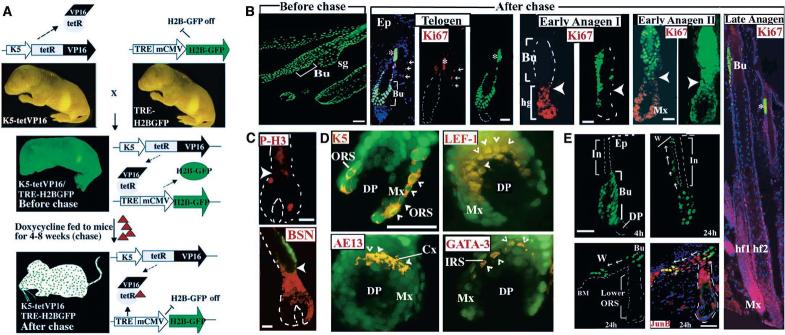
Fig. 1.
System for marking slow-cycling SCs in vivo and monitoring their fate. (A) Strategy. (B to D) Skin sections of mice before and after 4-week chase. Shown are epifluorescence of H2B-GFP (green) and 4′,6′-diamidino-2-phenylindole (DAPI) (blue), and indirect immunofluorescence with antibodies (Abs) indicated (Texas Red). The hair cycle stage is indicated on each set of “after chase” frames (see also fig. S1, B to D, and fig. S2). Arrows (B) denote Ki67+ sebaceous gland cells in telogen. Arrowheads [(B) and (C)] denote transition zone between bulge and newly generated follicle downgrowth. Late anagen (Ki67 in red): GFP-bright cells are retained in the bulge; their progeny rapidly divide, diluting H2B-GFP. (D) Early anagen II bulb overexposed for GFP and double-labeled (small arrowheads) with Abs against each differentiation cell type. (E) Mice “after chase” were scratch-wounded and analyzed by immunofluorescence. Arrows denote likely directions of movements of GFP-positive LRCs and progeny. Abbreviations: Bu, bulge; DP, dermal papilla; Mx, matrix; hg, hair germ; Ep, epidermis; asterisk, hair shaft (autofluorescent); hf, hair follicle; Cx, cortex; ORS/IRS, outer/inner root sheaths; BM, basement membrane; In, infundibulum; W, wound. Scale bars, 50 μm.
To track LRC fate during hair cycling, we monitored GFP fluorescence intensities relative to the proliferation-associated markers Ki67, phosphorylated histone H3 (P-H3; G2/M) and basonuclin (BSN) (Fig. 1, B and C). Throughout the cycle, most LRCs remained in the bulge as GFP-bright and Ki67−, P-H3−, and BSNlow cells (Fig. 1, B and C). During anagen, an abrupt switch in proliferation markers and H2B-GFP intensity occurred at the transition between bulge and new follicle downgrowth (large arrowheads). Overexposure verified that this downgrowth was largely GFP-positive deriving from bulge LRCs. Newly created GFP-positive populations included ORS (K5+), matrix (Lef1+/K5−), hair (AE13+/K5−), and IRS (GATA-3+/K5−) (Fig. 1D). We conclude that only a few bulge LRCs initiate each new follicle, and that upon exit their progeny rapidly proliferate, change biochemistry, and regenerate all differentiated cell types.
To determine whether bulge LRCs can react to injury, we scratch-wounded 8-week-old chased mice. Within 24 to 48 hours, GFPhigh cells were detected outside the bulge (Fig. 1E). These were not “scattered” bulge cells, because they localized to infundibulum and displayed underlying basement membrane (anti–laminin 5 immunoreactivity; not shown). Not seen in unwounded skin, GFP-positive cells within infundibulum and epidermis expressed nuclear junB, a stress-response protein (Fig. 1E). Thus, in response to wound stimuli, LRCs change their biochemistry, exit the bulge, migrate, and proliferate to repopulate infundibulum and epidermis. The ability of LRCs to regenerate hair follicles and epidermis is a feature characteristic of bulge SCs (6, 7).
Immunofluorescence microscopy revealed that the zone harboring keratinocyte-specific, H2B-GFP–bright LRCs was more restricted than that defined by known bulge-preferred markers, including K15, K19, α6-integrin, β1-integrin, CD34, S100A4, and S100A6 (11-14) (fig. S3). To further analyze the properties of LRCs, we prepared single-cell suspensions from 8-week-old, chased transgenic skins and performed fluorescence-activated cell sorting (FACS). Of this population, 12% displayed 10 to 104 units of GFP fluorescence, with 1 to 2% exhibiting 103 to 104 units, relative to background (Fig. 2A).
Fig. 2.
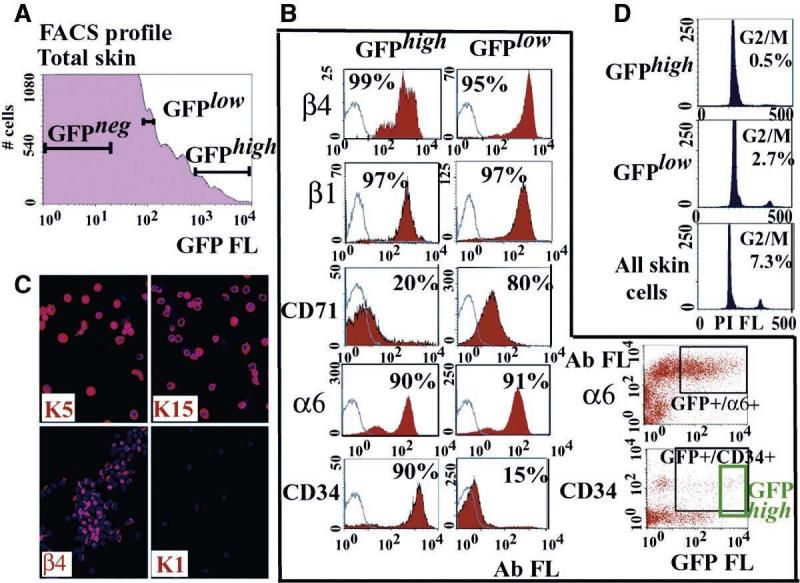
Isolation and preliminary characterization of bulge LRCs and progeny. Animals were Tet-fed for 4 weeks beginning at t = 4 weeks. (A) FACS analyses of single-cell suspensions of skins. GFP fluorescence (FL) is in arbitrary units. (B) Two-color FACS analyses for GFP and five surface markers. α6/CD34 data illustrate that GFPhigh LRCs represent only ∼30% of α6/CD34/K5-H2B-GFP–positive cells. White, without primary Abs; red, with Abs. Percentages of total cells scoring positive are indicated. (C) GFPhigh FACS population analyzed by immunofluorescence to illustrate homogeneity. Figure S4B provides quantification of all three fractions screened for six markers. (D) Propidium iodide (PI)–FACS cell cycle profiles by DNA content: G0/G1 (n = 1), G2/M (n = 2), and S (n = 1 or 2). Percentage of total cells in G2/M is indicated.
Populations gated at 103 to 104 units (GFPhigh) and 50 to 100 units (GFPlow) excluded propidium iodide and exhibited surface β4-, β1-, and α6-integrins, typical of BL/ORS cells (Fig. 2B). GFPhigh cells were enriched in CD34 (14), whereas GFPlow cells had more CD71 (down-regulated in bulge) (13). Two-color analyses indicated that only ∼30% of GFP+/CD34+ (also α6+) cells were GFPhigh (Fig. 2B) (fig. S4B). Semiquantitative fluorescence documented that GFPhigh cells corresponded in fluorescence intensity to bulge cells, whereas GFPlow fluorescence placed them outside the niche (fig. S4A).
Although GFPhigh and GFPlow cells differed in CD34/CD71 expression, they both expressed BL/ORS keratins K5, K14, and K15 (1, 12), but not differentiation-specific K1 (1) (Fig. 2C). FACS by surface-β4 yielded a larger pool of cells that were similar to GFPlow but with lower fluorescence (fig. S4B). Immunofluorescence with six markers indicated that these three FACS populations were >90% homogeneous (fig. S4B), and semiquantitative reverse transcription polymerase chain reactions (RT-PCRs) documented their distinctive characteristics (fig. S4C). Cell cycle profiles showed that only 0.5% GFPhigh LRCs were in G2/M (Fig. 2D). Finally, although existing methods did not permit long-term culturing of adult murine bulge cells (7, 14), GFPhigh LRCs were highly enriched (>10× to 15×) for cells forming colonies, some of which were >500 cells (fig. S4D), consistent with the high proliferative capacity documented for rat whisker bulge (7).
Using microarray analyses, we obtained transcriptional profiles for the three populations, and high-stringency analyses uncovered the distinguishing features of LRCs (table S2 and Materials and Methods); ∼4800 of 12,000 mRNAs were scored as present in each population. When bulge LRCs were compared with SC databases from hematopoietic (HSC), embryonic (ESC), and neuronal (NSC) tissues (15, 16), SCs were found to express 68% of mRNAs present in LRCs and ∼40% of mRNAs up-regulated in LRCs relative to BL/ORS (Fig. 3A). Moreover, overlap existed between HSCs and LRCs relative to their respective progenies (table S1).
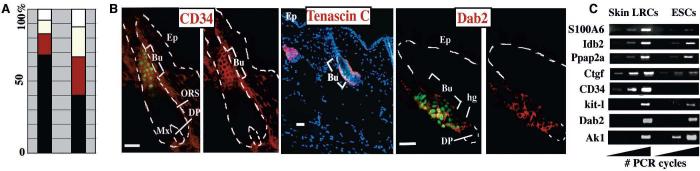
Fig. 3.
Transcriptional profiling of bulge LRCs relative to other SCs. Duplicate mRNAs of bulge LRCs and their two progeny populations were amplified and hybridized to Affymetrix oligonucleotide chips. Files were analyzed by Microarray Suite (MAS5.0) Affymetrix software followed by public database searches, functional annotation, and comparison with similar databases from embryonic (ESC), neural (NSC), and hematopoietic (HSC) SCs (15, 16). (A) Bar graphs: Percentage of mRNAs called present in skin LRCs (4839 total; left) and of mRNAs increased in LRCs versus BL/ORS (154 total; right) and also called present in three other SCs (black), two other SCs (red), one other SC (yellow), and no other SCs (white). (B) GFP epifluorescence and immunofluorescence of skin sections from 8-week-old mice after 4-week chase. Abs are against known SC markers found up-regulated in bulge LRCs. Abbreviations are as in Fig. 1 legend; scale bars, 50 μm. Lower magnification for tenascin-C illustrates marker-specificity. (C) Semiquantitative RT-PCR. Probes are for SC markers, up-regulated in skin LRCs. Mouse ESC mRNAs are shown for comparison. PCR was run for 29, 32, and 35 cycles.
The complete database with the raw Affymetrix data files is available at www.rockefeller.edu/labheads/fuchs/database.php. Up-regulated skin LRC mRNAs included known SC markers such as stem cell factor (kit ligand), Dab2, ephrin tyrosine kinase receptors (Ephs), tenascin C ( Tnc), interleukin-11 receptor, Id binding protein–2 (Idb-2), four-and-a-half lim domains (Fhl1), CD34, S100A6, and growth arrest–specific (Gas) proteins (17-21). Immunofluorescence and/or RT-PCR confirmed their bulge-preferred location (Fig. 3, B and C). Candidates to be involved in SC maintenance and/or activation, the shared LRC SC factors encompassed proteins regulating cell growth and survival; receptors able to sense and respond to growth factors, hormones, and extra-cellular matrix; and transcription factors. This newfound relation between bulge LRCs and other SC populations (15, 16) opens important avenues for future investigation.
As shown in table S2, 154 mRNAs were up-regulated by a factor of ≥2 in all four comparisons of bulge LRCs to GFPlow and β4-positive BL/ORS progeny; Table 1 shows functional classifications for a subset of these mRNAs. With 25 primer sets, RT-PCR verified up-regulation of 24 putative LRC mRNAs relative to at least one of the two progeny (fig. S5). Many known bulge markers surfaced as up-regulated LRC transcripts, including CD34 (9×), S100A4 (5×) (22), S100A6 (3×) (22), Barx2 (2×) (23), and Tcf3 (3×) (24) ( Table 1). Immunofluorescence confirmed their bulge-preferred location relative to BL/ORS (fig. S3). Most LRC mRNAs were specifically expressed in bulge relative to upper ORS and BL. Some were exclusive for the bulge within the skin. Some were present not only in bulge, but also other skin cells not analyzed here. An example is tektin2 (14×), a putative microtubule-binding protein, increased in bulge relative to BL/ORS and also present in arrector pili muscles (Fig. 4A).
Table 1.
Transcriptional profiling of bulge LRCs relative to their BL/ORS progeny. Functional classification is shown for 66 mRNAs scored as increased in bulge LRCs relative to BL/ORS progeny (full list in table S2). Average relative increase across the four comparisons is in parentheses; an mRNA that is present but not increased is denoted P.
| Category | mRNAs |
|---|---|
| Known bulge factors | Cd34 (9×)*, S100a4 (5×)*, S100a6 (3×)*, Tcf3 (3×)*, β1-integrin (P)*, β4-integrin (P)*, α6-integrin (P)*, Barx2 (2×)* |
| Cell cycle | Gas1 (growth arrest specific 1) (4×), Ltbp1 (latent TGFβ binding protein) (8×)*, Ltpb2 (10×), Ltbp3 (3×), TGFβ2 (3×)*, Inhbb (3×), Ak1 (3×) |
| TGFβ-induced factors | Idb1 (2×), Idb2 (8×), Idb3 (2×), Idb4 (4×), Ctgf (8×)*, Ltbp1 (8×)*, Ltbp2 (10×), Ltbp3 (3×), Igfbp5 (6×),* Igfbp7 (2×)*, Timp2 (5×), β6-integrin (6×)*, Tnc (3×)*, EfnB1 (2×) |
| Wnt signaling | Sfrp1 (7×), Dab2 (9×), Dkk3 (5×), Fzd2 (5×), Fzd3 (3×), Fzd7 (4×), Ctbp2 (2×), Fts (2×), Tcf3 (3×)* |
| Other signal transduction/ cell-cell communication factors |
Stem cell factor (Kit-l) (2×), EfnA4 (2×), EfnB1 (2×), EfnB2 (2×), Bdnf (8×)*, Tpst1 (3×), Ptprk (3×), Ppap2a (8×), Gkap42-pending (6×), Igfbp5 (6×) |
| Extracellular matrix/basement membrane proteins |
Tnc (3×)*, Col6a1 (3×)*, Col18a1 (2×)*, Mtn2 (2×), Timp2 (4×)*, Bgn (2×), Agn (2×), Sdc1 (2×)*, syndecan bp (2×) |
| Nuclear proteins/ transcription factors |
Idb1 (2×), Idb2 (8×), Idb3 (2×), Idb4 (4×), Nfatc1 (2×), Osf (5×), Pbx3 (4×), Nfib (3×)*, Hist1h2bc (3×), vdr (2×)*, Mad4 (2×)*, Max (2×)*, LIM domain only 1 (2×), Cited2 (2×) |
| Cytoskeletal/cell adhesion factors |
Macf1 (ACF7) (3×)*, Tekt2 (14×), β6-integrin (6×)*, Pdlim3 (15×), Actn1 (4×), Myo1b (4×), Pfn2 (3×), Krt2-6a (7×)*, Ndn (3×) |
mRNAs previously reported in skin, irrespective of location.
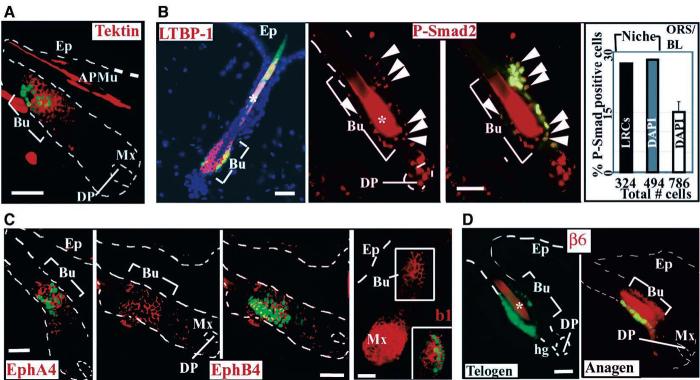
Fig. 4.
Implementation of array analyses to examine characteristics and dynamics of the skin SC niche. GFP (green) and immunofluorescence (red) of skin sections from 8-week-old mice (4-week chase). Examples shown: (A) An mRNA up-regulated >2× in LRCs relative to epidermis/ORS. (B) Activated (nuclear phospho-Smad2; arrowheads) or up-regulated (LTBP-1) LRC factors involved in TGFβ signaling. Quantification is at right (graph). (C) Tissue polarity proteins expressed in the SC niche. EphA4, EphB4, and EfnB1 (right of EphB4); boxed bulge in frame is also shown with GFP colabeling. (D) Dynamics of the niche during cycles of SC activation (telogen/anagen transition). Abbreviations are as in Fig. 1 legend; APMu, arrector pili muscle; asterisk denotes hair shaft autofluorescence. Scale bars, 50 μm.
Table 1 groups mRNAs into categories useful in considering the properties of LRCs relative to progeny cells. Several points are worthy of special mention. Consistent with their slow-cycling properties, skin LRCs expressed elevated transcripts encoding cell cycle regulatory proteins, and in particular, keratinocyte growth inhibitors implicated in transforming growth factor–β ( TGFβ) signaling (25). One of these, LTBP-1, is necessary for latent TGFβ activation (26) and was strongly and specifically localized to bulge (Fig. 4B). Indicative of TGFβ receptor activation, nuclear phospho-Smad2 immunoreactivity was more prevalent in bulge than progeny (Fig. 4B). Activated TGFβ/phospho-Smad target genes and/or Smad interacting proteins were also included in up-regulated LRC mRNAs ( Table 1) (fig. S5) (27). Conversely, transcripts down-regulated (163 total) encoded many proliferation-associated proteins, including Ki67 (3×) (Fig. 1, B and C), Cdc25C (2×), and N-myc “downstream-regulated-like” (2×).
Additional up-regulated LRC mRNAs encoded members of the Wnt pathway, essential for follicle morphogenesis and hair cycle activation (28, 29). These were inhibitors, including Srfp1, Dab2, Dkk3, Ctbp2, Tcf3, and Tle1, and the Wnt receptors Fzd3, F2d7, and Fzd2 (Table 1) (fig. S5). Conversely, Wnt3a and Wnt3 were down-regulated >3× in LRCs. Consistent with Tcf3's repressor function (24), a Wnt-inhibited niche would explain why at most stages in the hair cycle, the Wnt reporter gene TOPGAL is silent in the bulge (29).
Many bulge LRC–up-regulated mRNAs (43%) encoded secretory or integral membrane proteins (Fig. 5), which suggests the ability of skin LRCs to organize their niche, communicate with neighboring cells, and respond to their special environment. A case in point may be ephrin receptors (Ephs) and their membrane-bound ligands (Efns), which signal bidirectionally in cell-cell communication and tissue boundary formation (21). Although not specific, EfnB1, EphA4, and EphB4 were expressed in LRCs (Fig. 4C). EfnB1 was up-regulated further in matrix, a compartment not analyzed here, and EphA4 and EphB4 were also in a subset of lower ORS cells in full anagen (Fig. 4C) (30).
Fig. 5.
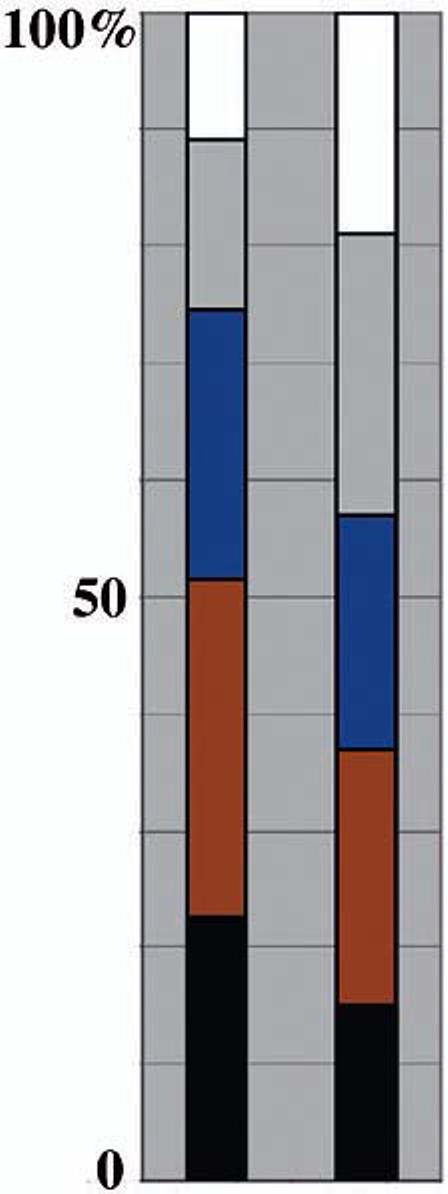
Comparison of cellular localization of bulge LRC mRNAs increased relative to BL/ORS and mRNA present in LRCs. Left, increased in LRCs relative to BL/ORS (154 total); right, present in skin LRCs (three pools of 150 mRNAs each of the 4839 present were analyzed). Black, expressed sequence tags; red, intracellular/cytosolic; blue, nuclear; gray, integral to membrane; white, secreted.
Although most markers labeled bulge LRCs irrespective of whether follicles were in anagen or telogen, Bdnf and TGFβ2 changed LRC expression with the hair cycle (31, 32). Transient stimuli mediated by DP may be particularly important in influencing signaling pathways within the niche. Another example is β6-integrin, present only in early anagen and not telogen (Fig. 4D). αvβ6 makes an attractive candidate for SC activation/migration, as it uses tenascin-C as ligand and is activated during skin wounding and tumorigenesis. Similarly, although not yet formally tested, DP-induced changes in Wnt signaling could explain why TOPGAL is transiently activated in the bulge at early anagen (29). If signaling pathways (e.g., Wnts) are generally important in SC self-renewal, as they are in hematopoietic SCs (33, 34), then differences in their status could have an impact on rates at which SCs divide and are mobilized from their niche.
In summary, we have uncovered a constellation of distinguishing features of bulge LRCs relative to related keratinocyte progeny, which, together with their localization, likely accounts for their special properties. Our findings suggest that the bulge SC niche is a growth and differentiation–restricted environment. The LRC-related changes that have thus far surfaced are already suggestive of a broader interaction between environmental stimuli and the SC niche.
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/1092436/DC1 Materials and Methods Figs. S1 to S5 Tables S1 and S2
References and Notes
- 1.Fuchs E, Raghavan S. Nature Rev. Genet. 2002;3(199) doi: 10.1038/nrg758. [DOI] [PubMed] [Google Scholar]
- 2.Potten CS, Morris RJ. J. Cell Sci. Suppl. 1988;10(45) doi: 10.1242/jcs.1988.supplement_10.4. [DOI] [PubMed] [Google Scholar]
- 3.Mackenzie IC. J. Invest. Dermatol. 1997;109(377) doi: 10.1111/1523-1747.ep12336255. [DOI] [PubMed] [Google Scholar]
- 4.Cotsarelis G, Sun TT, Lavker RM. Cell. 1990;61(1329) doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 5.Morris RJ, Potten CS. J. Invest. Dermatol. 1999;112(470) doi: 10.1046/j.1523-1747.1999.00537.x. [DOI] [PubMed] [Google Scholar]
- 6.Taylor G, Lehrer MS, Jensen PJ, Sun TT, Lavker RM. Cell. 2000;102(451) doi: 10.1016/s0092-8674(00)00050-7. [DOI] [PubMed] [Google Scholar]
- 7.Oshima H, Rochat A, Kedzia C, Kobayashi K, Barrandon Y. Cell. 2001;104(233) doi: 10.1016/s0092-8674(01)00208-2. [DOI] [PubMed] [Google Scholar]
- 8.Kanda T, Sullivan KF, Wahl GM. Curr. Biol. 1998;8(377) doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 9.Diamond I, Owolabi T, Marco M, Lam C, Glick A. J. Invest. Dermatol. 2000;115(788) doi: 10.1046/j.1523-1747.2000.00144.x. [DOI] [PubMed] [Google Scholar]
- 10.Vasioukhin V, Degenstein L, Wise B, Fuchs E. Proc. Natl. Acad. Sci. U.S.A. 1999;96(8551) doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones PH, Harper S, Watt FM. Cell. 1995;80(83) doi: 10.1016/0092-8674(95)90453-0. [DOI] [PubMed] [Google Scholar]
- 12.Lyle S, et al. J. Investig. Dermatol. Symp. Proc. 1999;4(296) doi: 10.1038/sj.jidsp.5640233. [DOI] [PubMed] [Google Scholar]
- 13.Tani H, Morris RJ, Kaur P. Proc. Natl. Acad. Sci. U.S.A. 2000;97(10960) doi: 10.1073/pnas.97.20.10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trempus CS, et al. J. Invest. Dermatol. 2003;120(501) doi: 10.1046/j.1523-1747.2003.12088.x. [DOI] [PubMed] [Google Scholar]
- 15.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. Science. 2002;298(597) doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 16.Ivanova NB, et al. Science. 2002;298(601) doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 17.Sette C, Dolci S, Geremia R, Rossi P. Int. J. Dev. Biol. 2000;44(599) [PubMed] [Google Scholar]
- 18.Garcion E, Faissner A, French-Constant C. Development. 2001;128(2485) doi: 10.1242/dev.128.13.2485. [DOI] [PubMed] [Google Scholar]
- 19.Hocevar BA, et al. EMBO J. 2003;22(3084) doi: 10.1093/emboj/cdg286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molofsky AV, et al. Nature. 2003;425(962) doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kullander K, Klein R. Nature Rev. Mol. Cell Biol. 2002;3(475) doi: 10.1038/nrm856. [DOI] [PubMed] [Google Scholar]
- 22.Ito M, Kizawa K. J. Invest. Dermatol. 2001;116(956) doi: 10.1046/j.0022-202x.2001.01369.x. [DOI] [PubMed] [Google Scholar]
- 23.Sander G, et al. J. Invest. Dermatol. 2000;115(753) doi: 10.1046/j.1523-1747.2000.00122.x. [DOI] [PubMed] [Google Scholar]
- 24.Merrill BJ, Gat U, DasGupta R, Fuchs E. Genes Dev. 2001;15(1688) doi: 10.1101/gad.891401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iavarone A, Massague J. Mol. Cell. Biol. 1999;19(916) doi: 10.1128/mcb.19.1.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isogai Z, et al. J. Biol. Chem. 2003;278(2750) doi: 10.1074/jbc.M209256200. [DOI] [PubMed] [Google Scholar]
- 27.Yang YC, et al. Proc. Natl. Acad. Sci. U.S.A. 2003;100(10269) [Google Scholar]
- 28.Andl T, Reddy ST, Gaddapara T, Millar SE. Dev. Cell. 2002;2(643) doi: 10.1016/s1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- 29.Alonso L, Fuchs E. Genes Dev. 2003;17(1189) doi: 10.1101/gad.1086903. [DOI] [PubMed] [Google Scholar]
- 30.Tumbar T, et al. data not shown. [Google Scholar]
- 31.Botchkarev VA, Yaar M, Gilchrest BA, Paus R. J. Invest. Dermatol. 2003;120(168) doi: 10.1046/j.1523-1747.2003.12003.x. [DOI] [PubMed] [Google Scholar]
- 32.Soma Y, et al. Arch. Dermatol. Res. 2002;293(609) doi: 10.1007/s00403-001-0279-6. [DOI] [PubMed] [Google Scholar]
- 33.Willert K, et al. Nature. 2003;423(448) doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 34.Reya T, et al. Nature. 2003;423(409) doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 35.We thank L. Degenstein, J. Fan, and L. Polak for help with mice; A. Glick for the K5Tetoff mice; S. Mazel and T. Shengelia for flow cytometry; and all who assisted with reagents (see supporting online material). Affymetrix hybridizations were conducted by the HHMI Stanford Microarray facility. Supported by postdoctoral fellowships from the Life Sciences Foundation (T.T.), Human Frontier Science Program (G.G., C.B.), EMBO (V.G.), Austrian Science Foundation (M.R.), BAEF and NATO (C.B.), and NIH (W.E.L.). E.F. is an Investigator of the Howard Hughes Medical Institute.