Summary:
Keratin 18 (K18) expression is a defining characteristic of internal epithelial cells of mammals. Here, we used the K18 gene and an internal ribosome entry site (IRES) to express green fluorescent protein, human placental alkaline phosphatase, and a modified Cre recombinase in an epithelial specific pattern in transgenic mice. The K18-driven alkaline phosphatase was expressed in liver, kidney, uterine endometrium, and other internal epithelia. The enzymatic activity of the Cre recombinase-mutant estrogen receptor fusion protein was dependent on tamoxifen administration and resulted in a mosaic pattern in internal epithelia, including bladder, uterus, liver, and kidney. This conditional Cre activity in internal epithelial organs should be valuable for strategies utilizing Cre for activation of gene expression. This study demonstrates that the tissue-specific, position-independent transcriptional activity of the K18 gene is not compromised by the use of an IRES element for the expression of a second protein from a bicistronic mRNA.
Keywords: keratin 18, Cre recombinase, transgenic mice, IRES, epithelium
INTRODUCTION
Successful targeting of gene expression to internal epithelia in transgenic mice has commonly utilized tissue-specific promoters which reflect the mature differentiated state of the epithelium in order to ensure specificity (Greenberg et al., 1994; Kim et al., 1993; Postic and Magnuson, 2000). However, the differentiated form of an epithelial tissue is not necessarily the best target for applications such as cancer modeling. Furthermore, the hormonal regulation of promoters such as those from mouse mammary tumor virus and probasin complicates investigations of hormone dependence in cancer progression in mouse models.
Keratin intermediate filaments are one of the defining characteristics of simple epithelia. Keratins 8 and 18 (K8, K18) are expressed in the first epithelial cell type to form during development and are persistently expressed in many adult carcinomas (Chu and Weiss, 2002). The regulation of the K18 gene is complex (Neznanov et al., 1997; Oshima et al., 1990; Rhodes and Oshima, 1998; Willoughby et al., 2000). However, the K18 gene is particularly well-behaved in transgenic mice, providing position-independent, copy number-dependent, and tissue-specific expression (Neznanov et al., 1993). Internal ribosome entry sites (IRES) permit the expression of additional reading frames from a chimeric mRNA and have been used successfully in gene trap applications, expression vectors for cell cultures, and transgenic mice (Bonaldo et al., 1998; Rees et al., 1996; Young et al., 1999). Here, we show that fusing the encephalomyocarditis virus (EMCV) IRES and a second coding region to the human K18 gene results in the expression of two reporter genes in an epithelium-specific pattern in transgenic mice. The utility of this strategy is further demonstrated by expressing a Cre recombinase-mutant estrogen receptor fusion (CreERT2) protein which results in tamoxifen-dependent Cre activity confined to epithelial tissues.
In order to determine if the EMCV IRES could be utilized within the context of the K18 gene, we generated transgenic mice from three different transgenes, shown in Figure 1. An enhanced green fluorescent protein (EGFP) cDNA, a human placental alkaline phosphatase (AP) cDNA, and a CreERT2 cDNA were placed downstream of the IRES sequence within the K18 gene. Four of 17 (23%) progeny injected with K18iresEGFP were identified as transgenic founders. Seven of 32 (22%) viable mice injected with K18iresCreERT2 carried the transgene. However, only one of 24 (4%) progeny was found to contain the K18iresAP gene. The low yield of K18iresAP transgenic mice is likely due to a deleterious effect of high-level AP expression on mouse development because the single viable K18iresAP line has few integrated copies and generates small litters (pup/litter = 4.0). In addition, the K18iresAP transgene was transmitted at only 60% of expected frequency. However, analysis of the AP expression patterns in additional transgenic embryos resulting directly from injection of the K18iresAP DNA revealed staining patterns very similar to the embryos of the established line (data not shown). Thus, the tissue-specific expression of AP in the established K18iresAP line appears representative of the gene.
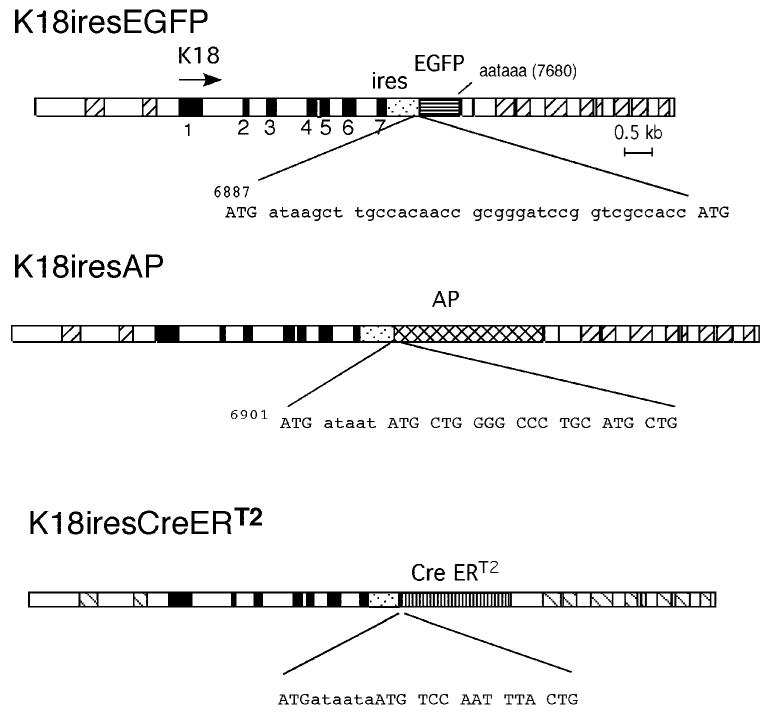
FIG. 1.
K18ires gene constructions. The structures of the K18iresEGFP, K18iresAP, and K18iresCreERT2 transgenes are shown. K18 exons are shown in filled boxes. The locations of Alu sequences are indicated by cross-hatching. The EMCV IRES sequence is indicated by light stippled rectangles. It is inserted downstream of the TGA of exon 7 before the polyadenylation signal (AATAAA), as indicated. The sequences between the last ATG of the IRES element and the initiating codons of the EGFP, AP and CreERT2 genes are shown below the maps.
Expression of the K18iresEGFP transgene was investigated by Northern blot analysis of adult organ RNAs. Figure 2 shows that all four founder lines expressed a major RNA form that hybridized with both the K18 and EGFP probes in liver and kidney. The size of the major transcript, approximately 2.8 kb, was larger than the normal 1.5 kb K18 mRNA (Fig. 2A; compare lane 2 and lanes 3-6), as expected for the fusion transcript, and was the same as that found after transient transfection of the transgene in cultured cells (Fig. 2A, lane 8). These results indicate that inclusion of the IRES and EGFP sequences generated the chimeric RNA primarily in epithelial tissues that express endogenous K18. Protein expression from the K18iresEGFP mRNA was confirmed by Western blotting of adult tissue extracts (data not shown). Human K18 and EGFP were detected only in the transgenic epithelial tissues. However, neither detection of the EGFP fluorescence nor EGFP immunostaining in organ sections was successful.
FIG. 2.
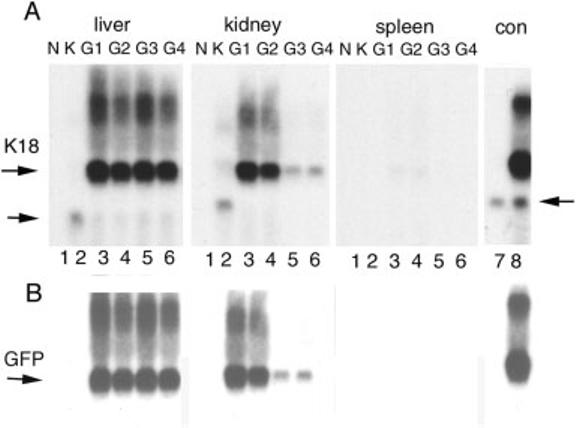
Northern blot analysis of K18iresEGFP transgenic tissues. Ten μg of total RNA was hybridized with radioactive probes for EGFP (B) and human K18 (A) sequentially. Samples for liver RNA were separated in a different gel than those for kidney and spleen. Samples from four founder lines are indicated by G1, G2, G3, and G4. N, nontransgenic mouse RNA; K, RNA from K18TG1, a low copy number human K18 transgenic mouse (Abe and Oshima, 1990); con, RNA from HR9 cells (lane 7) and the same cells transfected with the K18iresEGFP vector (lane 8). The position of the K18iresEGFP chimeric mRNA is indicated by the upper horizontal arrow. The lower arrow indicates the K18 mRNA from K18TG1. In lanes 7 and 8, the lower arrow indicates the cross-hybridization of the K18 probe with mouse K18. Overlaying the films of A and B revealed that the EGFP and major K18 signals were coincident.
The tissue-specific expression of AP activity from the K18iresAP transgene was compared with the pattern of endogenous mouse K18 (mK18) mRNA detected by whole-mount in situ hybridization (Fig. 3a,b). At embryonic day 10.5 (E10.5) mK18 RNA was detected in developing nasal cavity, gastrointestinal tract, liver, otic vesicle, and eye. The pattern of AP activity of the K18iresAP transgene at the same stage was nearly identical, with the exception of decreased eye expression and weak segmental staining in the body. Internal expression of K18 and AP was revealed in sections of E12.5 K18iresAP embryos (Fig. 3c,d). Human K18 protein was found in the liver, gastrointestinal tract, nasal cavity, developing lung, periderm, pituitary, and choroid plexus, consistent with the previously described pattern of human K18 gene expression in transgenic embryos (Thorey et al., 1993). AP activity was confirmed in the same tissues and additionally was seen in heart (Fig. 3d, arrow). Longer development of AP activity additionally detected activity in neopallial cortex and in punctuate vesicular structures throughout the body (Fig. 3f). The punctuate, vesicular staining pattern resembled developing blood vessels. Visualization of endothelial cells by staining with CD31 antibody (Fig. 3e) revealed a pattern strikingly similar to AP staining. This pattern of K18-driven AP expression was not paralleled by detection of K18 protein, most likely because of the lack of a complementary type II keratin to stabilize it. Thus, the segmental AP staining seen in the whole-mount embryo could be due to the transgene expression in developing vasculature.
FIG. 3.
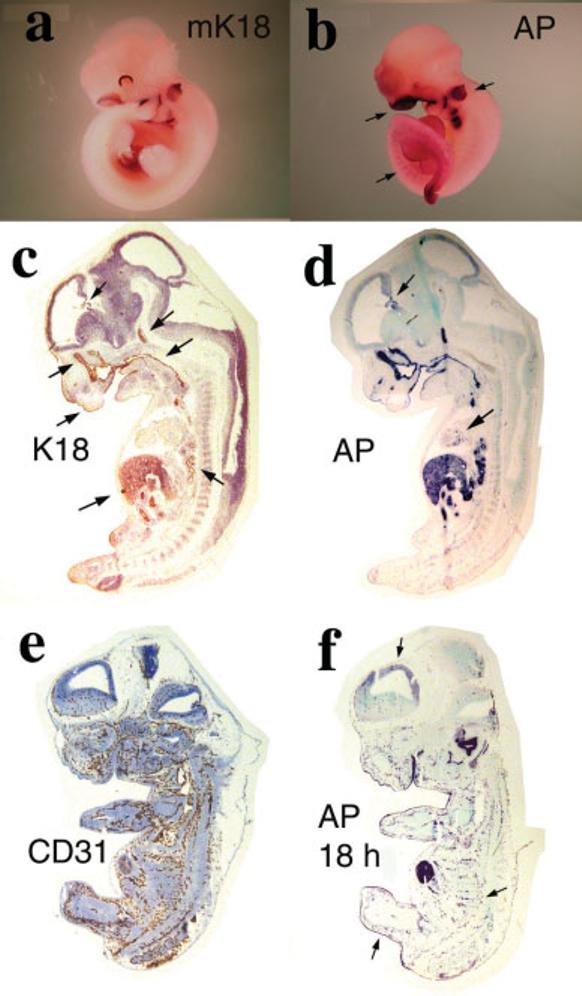
Human K18 and AP expression in K18iresAP embryos. a: ISH of E10.5 wild-type mouse embryo with mK18 riboprobe. b: AP staining of E10.5 K18iresAP embryo. Arrows point to strong staining in the otic vesicle and nasal area. Lower arrow points to weak, striated staining of the posterior trunk. c–f: Sections of E12.5 K18iresAP embryos. c: Human K18 antibody stain. Arrows point to staining of periderm, nasal epithelium, choroid plexus, pituitary, pharynx/esophagus, liver, and lung. d: Four-hour AP staining of adjacent section of c. Arrows point to staining in choroid plexus and heart. e: CD31 antibody localization of developing vasculature. f: 18-h AP staining of adjacent section to e. Arrows point to periderm, striated vascular staining, and weak neopallial cortex staining.
Human K18 and AP expression from adult K18iresAP mice was localized by both antibody staining of the proteins and AP enzymatic staining (Fig. 4). K18 was relatively uniformly expressed in hepatocytes and kidney tubules. AP protein was found in the same pattern. In skin, K18 protein and AP activity were colocalized to the hair follicle region. Additional AP activity staining of adult organs also revealed expression in other simple epithelia, including those of endometrium, prostate, pancreas (Fig. 4g-i), and mammary gland (not shown). Thus, AP expression paralleled human K18 expression in multiple adult tissues. However, unlike the embryonic pattern shown above, AP protein was low in gastrointestinal tract (data not shown).
FIG. 4.
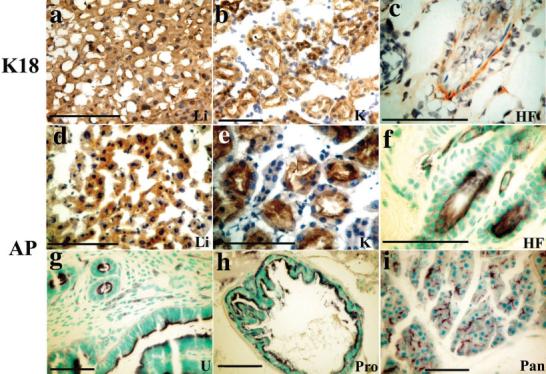
Histochemical localization of K18 and AP in adult K18iresAP mice. a–e: Immunostaining of frozen sections of mouse tissues for human K18 (a–c) or AP (d,e), counterstained with hematoxylin. f–i: Paraffin sections from whole-mount AP activity staining of mouse organs, counterstained with methylgreen. Li, liver; K, kidney, HF, hair follicle; U, uterus; Pro, prostate; Pan, pancreas. Both antibody staining and AP activity stain in presented tissues for wild-type animals had very low background (not shown). Scale bars = 50 μm.
We then examined the higher copy number K18iresCreERT2 founder lines for inducible Cre activity by mating the mice to the R26R Cre reporter line that expresses β-galactosidase (lacZ) when Cre is active (Soriano, 1999). Bigenic animals were treated with 4-hydroxy-tamoxifen (OHT) or vehicle only. High β-galactosidase (β-gal) activity was detected in brain, skin, kidney, liver, uterus, and bladder of the most active line, designated K18iresCreERT2-20 (Fig. 5). Lower but readily measured activity was found in skeletal muscle, spleen, lung, and mammary gland. β-Gal activity in control bigenic animals was negligible compared to that of OHT-treated mice. Thus, Cre activity is strictly OHT-dependent. Histochemical localization of the β-gal activity and human K18 protein revealed that Cre activity was expressed in subsets of cells that were K18-positive (Fig. 6). As expected, K18 was expressed in the liver, uterus, bladder, and other epithelial tissues (not shown). In addition, the epithelial covering of skeletal muscle expressed stable K18 (Fig. 6b), which may account for the β-gal activity detected by the sensitive luminescent assay. Cre activity was displayed in a mosaic pattern, even in tissues with relatively uniform K18 expression like liver and uterus. In skin and brain, both K18 protein and β-gal activity was localized in the hair follicle region and choroid plexus, respectively (data not shown). LacZ staining did not reveal the location of β-gal activity in muscle or spleen. Treatment of E11.5 embryos in utero with 20 mg/kg OHT (Sohal et al., 2001) resulted in similar mosaic expression in liver (data not shown). These results indicate that the OHT-regulated Cre activity was induced primarily in the epithelial tissues in a mosaic pattern, and therefore may be useful for applications not requiring Cre activity in every cell of a particular epithelium, such as oncogene activation.
FIG. 5.
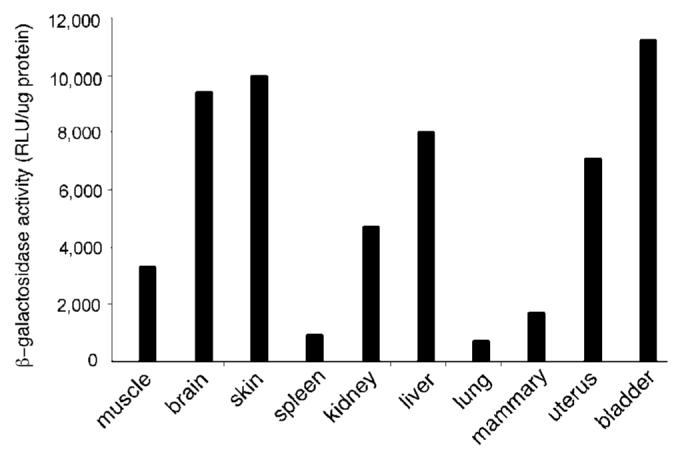
Inducible Cre-mediated β-gal activity in K18iresCreER; R26R mice. Bigenic animals were treated with vehicle or with 0.1 mg OHT for 5 days; 1–2 weeks after treatment, the indicated tissues were homogenized and assayed for Cre-mediated β-gal activity using a chemiluminescent assay as described. Enzymatic activities are displayed as average RLUs normalized to protein concentration. The background activity in control animals was negligible (<20 RLU/μg protein) after heat inactivation. The result shown here is representative of two independent experiments.
FIG. 6.
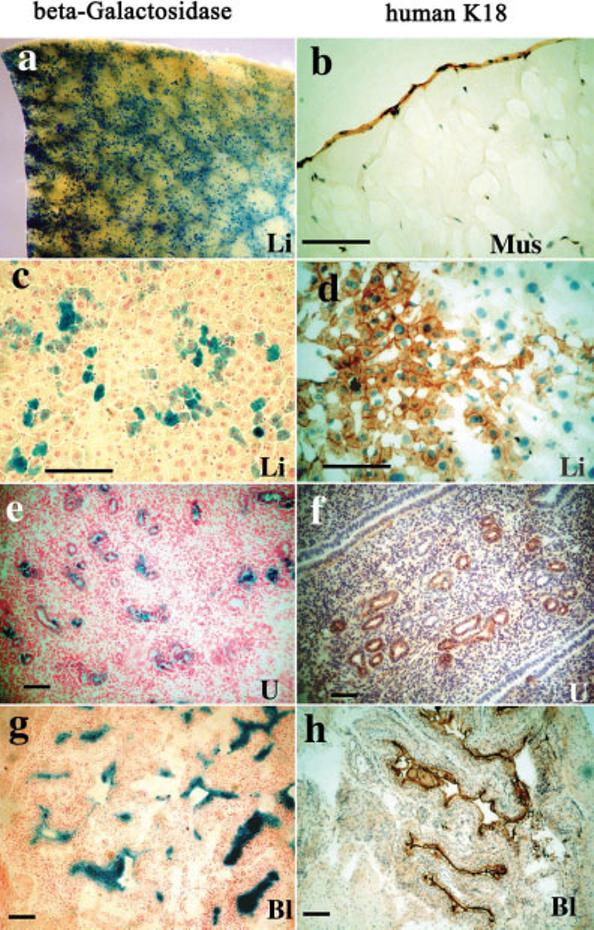
Immunohistochemical localization of K18 and β-gal activity of OHT-treated K18iresCreER;R26R mice. a: Whole-mount β-gal activity of liver (blue). b–h: Frozen sections of mouse tissues were stained for human K18 (b,d,f,h) or Cre-mediated β-gal activity (c,e,g). b: Localization of K18 in the epithelial covering of muscle. c,d: Hepatocytes stained for β-gal and K18 (brown). e,f: Uterine gland staining of the uterus. g,h: Transitional epithelium staining of the urinary bladder. Li, liver; Mus, muscle; U, uterus; Bl, bladder. Scale bars = 100 μm.
The EMCV IRES has been used to allow expression of a second reading frame in both cell culture and transgenic mice. While the IRES is utilized in many tissues (Creancier et al., 2001), the composition of the coding sequence can influence the efficiency of use (Hennecke et al., 2001), and others have found less efficient use of IRES in older mice (Shaw-Jackson and Michiels, 1999). The latter observation may contribute to the decreased AP activity we found in some adult K18iresAP tissues. The expression of K18 in early extraembryonic tissues is one restriction on the use of the K18ires vector for expression of genes with potent biological effects, which can be circumvented by conditional expression or activation strategies. The tamoxifen-inducible Cre activity we used here is one example of conditional strategies, which will be helpful when utilizing Cre to activate gene expression.
The fusion of Cre and mutant estrogen receptor has been used to regulate Cre activity in the epidermis (Indra et al., 1999; Vasioukhin et al., 1999) and even in embryos (Danielian et al., 1998). The Cre activity of CreERT2 is insensitive to estrogen but is half maximally activated by 6 nM OHT (Feil et al., 1997). This high specificity accounts for the low basal activation of the reporter gene in K18iresCreERT2;R26R mice. The mosaic pattern of the β-gal reporter represents the combination of IRES utilization, OHT availability, CreERT2 activity in the nucleus, and the expression and detection of β-gal. The sensitive chemiluminescent assay clearly detected β-gal activity in tissues which were not stained histochemically. The temporal control that CreERT2 provides should be useful in combination with a variety of recently described mouse tumor models (Andrechek et al., 2000; Jonkers et al., 2001). Furthermore, the K18ires vector should facilitate epithelial expression of cDNAs in transgenic mice. The integration site-independent expression of the K18 gene may also be useful for some gene therapy applications.
METHODS
DNA Constructions
K18iresEGFP was constructed from the K18 gene plasmid pGC1853 (Kulesh and Oshima, 1988). The EMCV IRES of the pCIN4 plasmid (Rees et al., 1996) was amplified by PCR and cloned into the SacII site of a polylinker inserted into the unique KpnI site of the K18 gene. The EGFP cDNA isolated from pEGFP (ClonTech, Palo Alto, CA) was cloned between the unique EcoRV and NotI sites of the K18ires vector polylinker. To make K18iresAP and K18iresCreERT2, an IRES-AP fused sequence and an IRES-CreERtam fusion were constructed by extending overlapping PCR products and replacing the 5′ end of the AP cDNA and an EcoRI fragment of the K14CreERtam gene (Vasioukhin et al., 1999), respectively. The CreERtam sequence was then converted to the CreERT2 (Indra et al., 1999) by replacing the AgeI/SacI fragment of the CreERtam sequence with the AgeI/SacI fragment of pCre-ERT2 containing the mutation of ER M543L544 to 543A544A. The fusion sequence was inserted between the SpeI and NotI sites of a synthetic polylinker. The sequence of the final plasmids is available on request.
Transgenic Mice
Transgenic mice were generated in the FVB/N strain as described previously (Abe and Oshima, 1990). Transgenic mice were identified by standard PCR analysis of tail lysates. The gene-specific primers used were for K18: ACAAGACAAACCAACTGCAAG and ACTTCACCCTGTTCCTTTATCC; for EGFP: AGGGCGAGGAGCTGTTCACCGG and GCTGGGGTAGCGGCTGAAGCAC; for AP: ATGACTACAGCCAAGGTGGG and TTCCGTATAGGAGGACCGTG, and for Cre: CTGGCATTTCTGGGGATTGC and ACGGAAATCCATCGCTCGAC. The R26R Cre reporter mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and genotyped following the instructions of the vendor.
Tamoxifen Treatment
Preparation of OHT (Sigma, St. Louis, MO) solutions has been described (Metzger and Chambon, 2001). Eight-week-bigenic mice were injected intraperitoneally once a day with vehicle or with 0.1 mg OHT for 5 consecutive days; 1–2 weeks after treatment, the animals were euthanized and tissues were collected for further analysis. The dosage was based on published descriptions (Indra et al., 1999) and preliminary trials in which higher dosages, up to 1 mg/day, did not increase the reporter activity but caused apparent toxicity (data not shown).
Northern Blot Analysis
RNA was prepared by the acidic phenol method using Tri Reagent (Molecular Research Center, Cincinnati, OH) and analyzed by Northern blotting (Neznanov et al., 1993) with a radioactive EGFP cDNA probe synthesized by PCR using primers AGGGCGAGGAGCTGTTCACCGG and GCTGGGGTAGCGGCTGAAGCAC. After appropriate exposure, the filters were stripped and rehybridized with a random-primed K18 cDNA probe. HR9 mouse parietal endodermal cells were maintained and transfected by the calcium phosphate precipitate method with the K18iresEGFP vector, as described previously (Oshima et al., 1990).
Immunohistochemistry
Acetone-fixed frozen sections were stained with CK5 mouse mAb (Sigma) against K18 (1:800) or an affinity-purified rabbit anti-AP antibody at 10 μg/ml. The Vector Laboratory (Burlingame, CA) ABC deluxe detection kit and DAB staining were used to visualize the appropriate secondary antibody or antibody complex (Animal Research Kit, Dako, Carpinteria, CA). Avidin/biotin blocking (Vector) was used on kidney and skin samples. Slides were counterstained with hematoxylin and photographed with a Nikon Coolpix 990 digital camera connected to a Lietz Dialux 20 microscope or an Olympus SZ40 dissection microscope. Digital images were processed with Adobe PhotoShop (Mountain View, CA) software. The background surrounding some embryo images was removed digitally for clarity. Whole-mount and frozen sections of adult tissues were X-gal stained as described previously (Lobe et al., 1999) except staining was carried out in 1 mg/ml X-Gal.
Chemiluminescent Assay of Tissue β-Gal Activity
Tissue β-gal activity was assessed using the Applied Biosystems (Foster City, CA) Dual-Light System. Tissue homogenates were incubated at 50°C for 30–60 min (Young et al., 1993) to inactivate the endogenous enzyme. Aliquots of 10 μl extract of each sample were used for the assay, following the instructions of the manufacturer. Protein concentrations were determined with the Bio-Rad (Hercules, CA) protein assay (Dye Reagent). LacZ activity was normalized to protein concentration to determine relative light units (RLU) per μg protein.
Whole-Mount In Situ Hybridization (ISH) and AP Staining
Using digoxygenin UTP (Roche, Nutley, NJ), the mK18 probe was prepared by transcription of a PCR amplified fragment (nt 460–1147) of the mK18 cDNA. ISH was performed as described previously (Wilkinson, 1998) with minor modifications. For whole-mount AP staining, adult organ slices or whole embryos were fixed in 4% paraformaldehyde / 0.2% glutaraldehyde in PBS for 30 min at 4°C with shaking, placed in PBS plus 10 mM MgCl2 and heated to 65°C for 45 min to inactivate the endogenous alkaline phosphatase. AP was stained as for ISH. The organ blocks were postfixed in 4% paraformaldehyde and sections from paraffin-embedded tissues were counterstained with methyl green.
ACKNOWLEDGMENTS
We thank Ms. Robbin Newlin, Ms. Jacqueline Avis, and Dr. Ling Wang for expert technical help with the generation of transgenic mice and immunohistochemical applications. We also thank Dr. Sonoko Narisawa for the gift of the affinity-purified AP antibody.
Contract grant sponsors: the National Cancer Institute (to R.G.O.), Cancer Center.
LITERATURE CITED
- Abe M, Oshima RG. A single human keratin 18 gene is expressed in diverse epithelial cells of transgenic mice. J Cell Biol. 1990;111:1197–1206. doi: 10.1083/jcb.111.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrechek ER, Hardy WR, Siegel PM, Rudnicki MA, Cardiff RD, Muller WJ. Amplification of the neu/erbB-2 oncogene in a mouse model of mammary tumorigenesis. Proc Natl Acad Sci USA. 2000;97:3444–3449. doi: 10.1073/pnas.050408497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaldo P, Chowdhury K, Stoykova A, Torres M, Gruss P. Efficient gene trap screening for novel developmental genes using IRES beta geo vector and in vitro preselection. Exp Cell Res. 1998;244:125–136. doi: 10.1006/excr.1998.4208. [DOI] [PubMed] [Google Scholar]
- Chu PG, Weiss LM. Keratin expression in human tissues and neoplasms. Histopathology. 2002;40:403–439. doi: 10.1046/j.1365-2559.2002.01387.x. [DOI] [PubMed] [Google Scholar]
- Creancier L, Mercier P, Prats AC, Morello D. c-myc internal ribosome entry site activity is developmentally controlled and subjected to a strong translational repression in adult transgenic mice. Mol Cell Biol. 2001;21:1833–1840. doi: 10.1128/MCB.21.5.1833-1840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- Greenberg NM, DeMayo FJ, Sheppard PC, Barrios R, Lebovitz R, Fine-gold M, Angelopoulou R, Dodd JG, Duckworth ML, Rosen JM, et al. The rat probasin gene promoter directs hormonally and developmentally regulated expression of a heterologous gene specifically to the prostate in transgenic mice. Mol Endocrinol. 1994;8:230–239. doi: 10.1210/mend.8.2.8170479. [DOI] [PubMed] [Google Scholar]
- Hennecke M, Kwissa M, Metzger K, Oumard A, Kroger A, Schirmbeck R, Reimann J, Hauser H. Composition and arrangement of genes define the strength of IRES-driven translation in bicistronic mRNAs. Nucleic Acids Res. 2001;29:3327–3334. doi: 10.1093/nar/29.16.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- Kim SH, Roth KA, Moser AR, Gordon JI. Transgenic mouse models that explore the multistep hypothesis of intestinal neoplasia. J Cell Biol. 1993;123:877–893. doi: 10.1083/jcb.123.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesh DA, Oshima RG. Cloning of the human keratin 18 gene and its expression in nonepithelial mouse cells. Mol Cell Biol. 1988;8:1540–1550. doi: 10.1128/mcb.8.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobe CG, Koop KE, Kreppner W, Lomeli H, Gertsenstein M, Nagy A. Z/AP, a double reporter for cre-mediated recombination. Dev Biol. 1999;208:281–292. doi: 10.1006/dbio.1999.9209. [DOI] [PubMed] [Google Scholar]
- Metzger D, Chambon P. Site- and time-specific gene targeting in the mouse. Methods. 2001;24:71–80. doi: 10.1006/meth.2001.1159. [DOI] [PubMed] [Google Scholar]
- Neznanov N, Thorey IS, Cecena G, Oshima RG. Transcriptional insulation of the human keratin 18 gene in transgenic mice. Mol Cell Biol. 1993;13:2214–2223. doi: 10.1128/mcb.13.4.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neznanov N, Umezawa A, Oshima RG. A regulatory element within a coding exon modulates keratin 18 gene expression in transgenic mice. J Biol Chem. 1997;272:27549–27557. doi: 10.1074/jbc.272.44.27549. [DOI] [PubMed] [Google Scholar]
- Oshima RG, Abrams L, Kulesh D. Activation of an intron enhancer within the keratin 18 gene by expression of c-fos and c-jun in undifferentiated F9 embryonal carcinoma cells. Genes Dev. 1990;4:835–848. doi: 10.1101/gad.4.5.835. [DOI] [PubMed] [Google Scholar]
- Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Rees S, Coote J, Stables J, Goodson S, Harris S, Lee MG. Bicistronic vector for the creation of stable mammalian cell lines that predisposes all antibiotic-resistant cells to express recombinant protein. Biotechniques. 1996;20:102–110. doi: 10.2144/96201st05. [DOI] [PubMed] [Google Scholar]
- Rhodes K, Oshima RG. A regulatory element of the human keratin 18 gene with AP-1-dependent promoter activity. J Biol Chem. 1998;273:26534–26542. doi: 10.1074/jbc.273.41.26534. [DOI] [PubMed] [Google Scholar]
- Shaw-Jackson C, Michiels T. Absence of internal ribosome entry site-mediated tissue specificity in the translation of a bicistronic transgene. J Virol. 1999;73:2729–2738. doi: 10.1128/jvi.73.4.2729-2738.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal DS, Nghiem M, Crackower MA, Witt SA, Kimball TR, Tymitz KM, Penninger JM, Molkentin JD. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89:20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Thorey IS, Meneses JJ, Neznanov N, Kulesh DA, Pedersen RA, Oshima RG. Embryonic expression of human keratin 18 and K18-beta-galactosidase fusion genes in transgenic mice. Dev Biol. 1993;160:519–534. doi: 10.1006/dbio.1993.1326. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci USA. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG. In situ hybridization: a practical approach. 2nd ed. Oxford University Press; New York: 1998. [Google Scholar]
- Willoughby DA, Vilalta A, Oshima RG. An Alu element from the K18 gene confers position-independent expression in transgenic mice. J Biol Chem. 2000;275:759–768. doi: 10.1074/jbc.275.2.759. [DOI] [PubMed] [Google Scholar]
- Young DC, Kingsley SD, Ryan KA, Dutko FJ. Selective inactivation of eukaryotic beta-galactosidase in assays for inhibitors of HIV-1 TAT using bacterial beta-galactosidase as a reporter enzyme. Anal Biochem. 1993;215:24–30. doi: 10.1006/abio.1993.1549. [DOI] [PubMed] [Google Scholar]
- Young WS, 3rd, Iacangelo A, Luo XZ, King C, Duncan K, Ginns EI. Transgenic expression of green fluorescent protein in mouse oxytocin neurones. J Neuroendocrinol. 1999;11:935–939. doi: 10.1046/j.1365-2826.1999.00410.x. [DOI] [PubMed] [Google Scholar]