Abstract
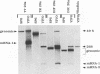
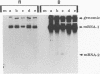
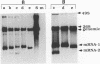
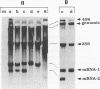
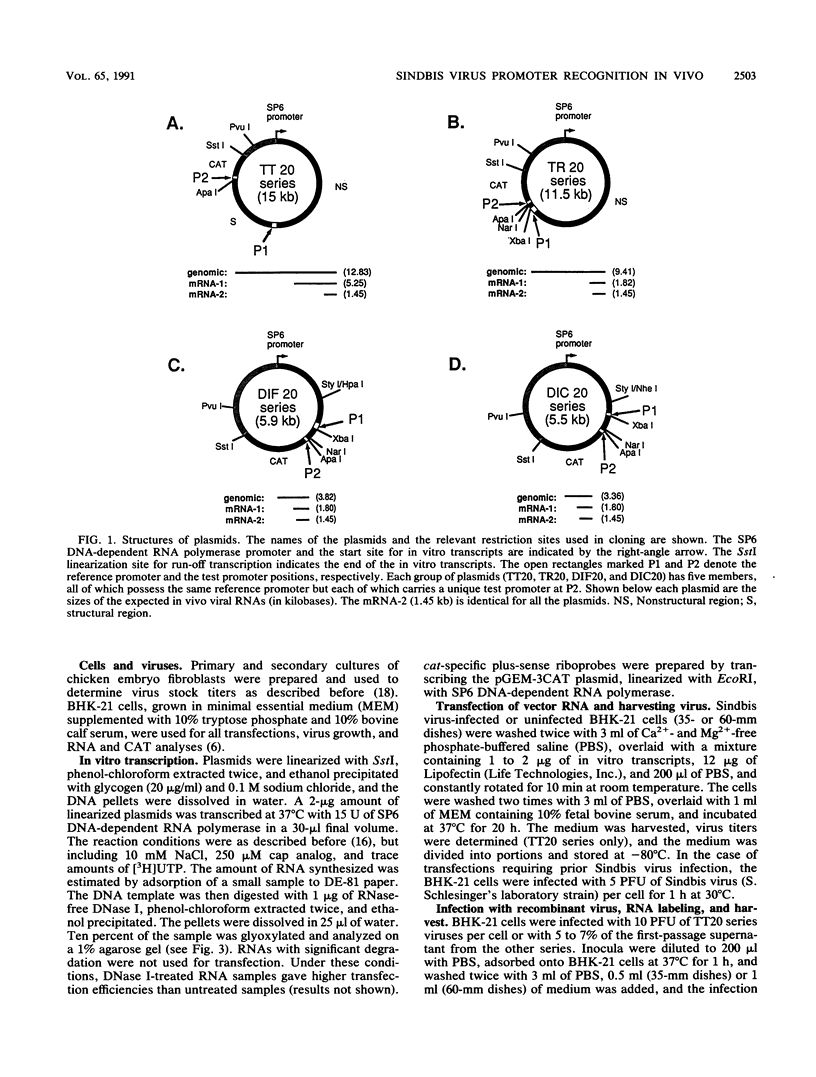
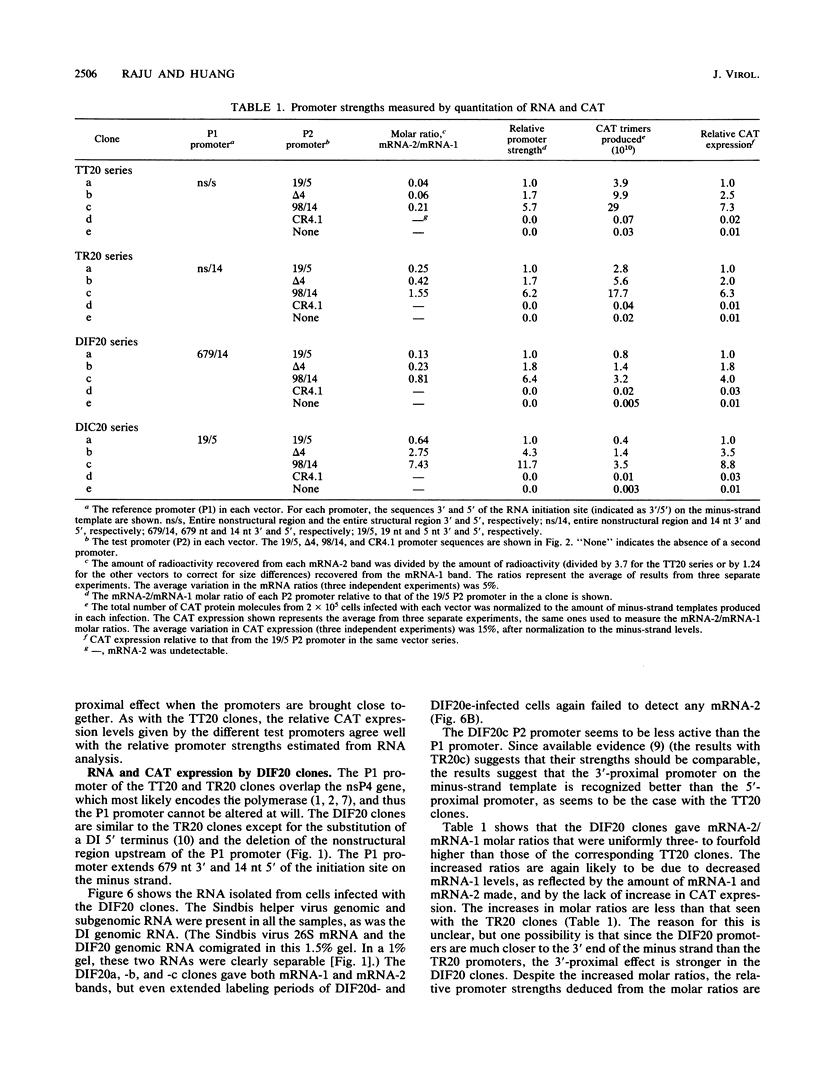
Four types of Sindbis virus vectors, each carrying two promoters for subgenomic mRNA synthesis, were designed to measure relative promoter strengths and to survey potential contextual effects on promoter strengths. One of the promoters in each vector was used as the reference promoter, while the other was the one being tested. We used these vectors to measure the relative strengths of four promoters: the minimal promoter, an extended sequence believed to have full promoter activity, and two mutant promoters, one with an inactivating 3-nucleotide insertion called CR4.1 and the other with a 4-nucleotide deletion called delta 4. The strengths of the promoters were measured by quantitating the RNA transcribed from each promoter in vivo and also by assaying for chloramphenicol acetyltransferase activity encoded by one of the two transcripts. We found that the relative strengths of the promoters were similar in different contexts. The complete promoter was 6-fold more active, the delta 4 promoter was (surprisingly) about twice as active, and the CR4.1 promoter was 100-fold less active than the minimal promoter. At least two contextual effects were identified that can alter the activity of one or both promoters in the vectors. One effect is that given identical promoters, the 3'-proximal promoter on the minus-strand template can be more active than the 5'-proximal promoter. This may be due to preferential association of one or more components of the transcription complex for the 3' end of the minus-strand template. A second effect is promoter competition, particularly when the promoters are closely spaced.
Full text
PDF






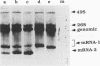
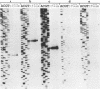
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barton D. J., Sawicki S. G., Sawicki D. L. Demonstration in vitro of temperature-sensitive elongation of RNA in Sindbis virus mutant ts6. J Virol. 1988 Oct;62(10):3597–3602. doi: 10.1128/jvi.62.10.3597-3602.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge B. W., Pfefferkorn E. R. Isolation and characterization of conditional-lethal mutants of Sindbis virus. Virology. 1966 Oct;30(2):204–213. doi: 10.1016/0042-6822(66)90096-1. [DOI] [PubMed] [Google Scholar]
- Carmichael G. G., McMaster G. K. The analysis of nucleic acids in gels using glyoxal and acridine orange. Methods Enzymol. 1980;65(1):380–391. doi: 10.1016/s0076-6879(80)65049-6. [DOI] [PubMed] [Google Scholar]
- French R., Ahlquist P. Characterization and engineering of sequences controlling in vivo synthesis of brome mosaic virus subgenomic RNA. J Virol. 1988 Jul;62(7):2411–2420. doi: 10.1128/jvi.62.7.2411-2420.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., Levis R., Raju R., Huang H. V., Rice C. M. A cis-acting mutation in the Sindbis virus junction region which affects subgenomic RNA synthesis. J Virol. 1989 Dec;63(12):5216–5227. doi: 10.1128/jvi.63.12.5216-5227.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn Y. S., Grakoui A., Rice C. M., Strauss E. G., Strauss J. H. Mapping of RNA- temperature-sensitive mutants of Sindbis virus: complementation group F mutants have lesions in nsP4. J Virol. 1989 Mar;63(3):1194–1202. doi: 10.1128/jvi.63.3.1194-1202.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Schlesinger S., Huang H. V. Promoter for Sindbis virus RNA-dependent subgenomic RNA transcription. J Virol. 1990 Apr;64(4):1726–1733. doi: 10.1128/jvi.64.4.1726-1733.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Weiss B. G., Tsiang M., Huang H., Schlesinger S. Deletion mapping of Sindbis virus DI RNAs derived from cDNAs defines the sequences essential for replication and packaging. Cell. 1986 Jan 17;44(1):137–145. doi: 10.1016/0092-8674(86)90492-7. [DOI] [PubMed] [Google Scholar]
- Ou J. H., Rice C. M., Dalgarno L., Strauss E. G., Strauss J. H. Sequence studies of several alphavirus genomic RNAs in the region containing the start of the subgenomic RNA. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5235–5239. doi: 10.1073/pnas.79.17.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju R., Kolakofsky D. Unusual transcripts in La Crosse virus-infected cells and the site for nucleocapsid assembly. J Virol. 1987 Mar;61(3):667–672. doi: 10.1128/jvi.61.3.667-672.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju R., Raju L., Hacker D., Garcin D., Compans R., Kolakofsky D. Nontemplated bases at the 5' ends of Tacaribe virus mRNAs. Virology. 1990 Jan;174(1):53–59. doi: 10.1016/0042-6822(90)90053-t. [DOI] [PubMed] [Google Scholar]
- Rice C. M., Levis R., Strauss J. H., Huang H. V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987 Dec;61(12):3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Strauss J. H. Nucleotide sequence of the 26S mRNA of Sindbis virus and deduced sequence of the encoded virus structural proteins. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2062–2066. doi: 10.1073/pnas.78.4.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss E. G., Lenches E. M., Strauss J. H. Mutants of sindbis virus. I. Isolation and partial characterization of 89 new temperature-sensitive mutants. Virology. 1976 Oct 1;74(1):154–168. doi: 10.1016/0042-6822(76)90137-9. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Rice C. M., Strauss J. H. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984 Feb;133(1):92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- Xiong C., Levis R., Shen P., Schlesinger S., Rice C. M., Huang H. V. Sindbis virus: an efficient, broad host range vector for gene expression in animal cells. Science. 1989 Mar 3;243(4895):1188–1191. doi: 10.1126/science.2922607. [DOI] [PubMed] [Google Scholar]