Abstract
MutY, like many DNA base excision repair enzymes, contains a [4Fe4S]2+ cluster of undetermined function. Electrochemical studies of MutY bound to a DNA-modified gold electrode demonstrate that the [4Fe4S] cluster of MutY can be accessed in a DNA-mediated redox reaction. Although not detectable without DNA, the redox potential of DNA-bound MutY is ≈275 mV versus NHE, which is characteristic of HiPiP iron proteins. Binding to DNA is thus associated with a change in [4Fe4S]3+/2+ potential, activating the cluster toward oxidation. Given that DNA charge transport chemistry is exquisitely sensitive to perturbations in base pair structure, such as mismatches, we propose that this redox process of MutY bound to DNA exploits DNA charge transport and provides a DNA signaling mechanism to scan for mismatches and lesions in vivo.
DNA repair proteins that contain a FeS redox cofactor are ubiquitous (1–7), yet a role for these factors has been lacking. Two examples, highly homologous (8), are MutY (1) and endonuclease III (Endo III) (2, 9), base excision repair enzymes from Escherichia coli (10). MutY, containing 350 residues, acts as a glycosylase to remove adenine from G:A (11–13) and 7,8-dihydro-8-oxo-2-deoxyguanonsine:A mismatches (14–21); Endo III removes pyrimidines damaged by ring saturation, contraction, or fragmentation (22–29). Although MutY and Endo III have dramatically different substrate recognition features, they both contain a [4Fe4S]2+ cluster (1, 2, 9) within a Cys-X6-Cys-X2-Cys-X5-Cys loop located near the protein C terminus (1, 9, 30). Based on sequence alignment, a loop defined by the first two ligating cysteines, called the FCL, is proposed to be a common element of DNA repair proteins (30), present throughout phylogeny (31–37). The function, if any, for these clusters remains undetermined, although the FCL has been proposed as a structural element, aiding in DNA binding (9, 30, 38). Interestingly, however, MutY is capable of folding without the cluster; the [4Fe4S]2+ cluster adds no stability to the enzyme, but it is critical for substrate binding and catalysis (39). The solvent-accessible [4Fe4S]2+ cluster of Endo III undergoes decomposition with ferricyanide and is resistant to reduction, with an estimated midpoint potential of <–600 mV for the [4Fe4S]2+/1+ couple (2, 38).
Here, we consider whether the [4Fe4S]2+ cluster in MutY can function in DNA-mediated charge transport (CT). Many laboratories have probed DNA-mediated CT chemistry (40–42). Using biochemical, spectroscopic, and electrochemical methods, we have shown that CT through DNA can proceed over long molecular distances in a reaction that is remarkably sensitive to intervening dynamical base pair structure (43–50). DNA-mediated CT has been shown to yield oxidative DNA damage from a distance within nucleosome core particles (47) and HeLa cell nuclei (51). DNA binding proteins and peptides have also been shown to modulate and participate in long-range CT chemistry (48, 49), raising the question of the physiological relevance of DNA CT.
Methods
DNA-Modified Electrodes. Thiol-modified oligonucleotides were prepared by using phosphoramidite synthesis as described (50, 52–57). Thiol-terminated linkers were attached to single-stranded oligonucleotides, HPLC-purified, and hybridized to their complements. Self-assembly of thiol-modified duplexes was carried out (100 μM duplex, 5 mM phosphate, 50 mM NaCl, pH 7, ambient temperature) without added Mg2+, to create loosely packed DNA films (52). After assembly of the duplexes, the remaining exposed surface was passivated with mercaptohexanol (100 mM). After backfilling, the electrode was rinsed with MutY dilution buffer (20 mM sodium phosphate, pH 7.5/1 mM Na2EDTA/20% glycerol/500 mM NaCl) and used for protein binding and electrochemical experiments. MutY (800 μM) was added to the DNA-modified electrode and allowed to incubate for 10 min at ambient temperature before investigation by electrochemistry.
Protein Samples. Proteins fused to maltose-binding protein (MBP) were used for the electrochemical studies owing to the need for high protein concentrations. The MBP-MutY fusion protein has similar activity as the WT with G:A and 7,8-dihydro-8-oxo-2-deoxyguanonsine:A-containing duplexes but is stable at much higher protein concentrations. The WT MutY or C199H MutY gene was ligated into a pMAL-c2x expression vector (New England Biolabs) to produce an N-terminal MBP fusion protein. Both the mutant and WT MBP-MutY proteins were expressed in E. coli and purified as described (58) with the exception of an amylose column replacing the cation exchange column in the chromatography step. Protein samples were diluted to the desired concentrations by using a buffer of 20 mM sodium phosphate, pH 7.5/1 mM Na2EDTA/20% glycerol/500 mM NaCl.
Cyclic Voltammetry (CV). CV was carried out on 0.2-cm2 Au (111) electrodes on mica by using a BAS (West Lafayette, IN) model CV-50W electrochemical analyzer equipped with a low-current module. Buffer and electrolyte conditions were 20 mM sodium phosphate (pH 7.5), 1 mM Na2EDTA, 20% glycerol, 500 mM NaCl, ambient temperature (±800 μM MutY). A scan rate of 50 mV/sec was used in all experiments. A Ag wire reference electrode and a Pt wire auxiliary electrode were used, and potentials were recorded versus Ag wire (52). Ag wire was used rather than a saturated calomel electrode because of the low volume constraints in the protein experiments.
Results and Discussion
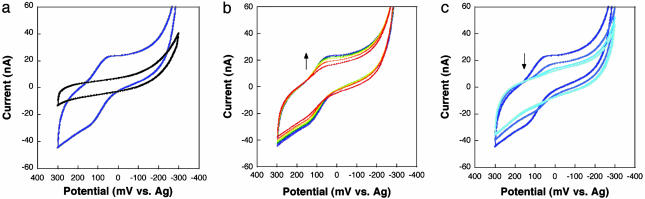
MutY Electrochemistry on DNA Films. The redox characteristics of MutY-bound DNA were probed electrochemically on loosely packed DNA films (Fig. 1). Protein electrochemistry has benefited from experiments conducted on monolayers of adsorbed species (59–63), and in our laboratory, electrochemistry on loosely packed DNA-modified surfaces has been a particularly useful method to probe protein/DNA interactions (52). A thiol-terminated duplex SH-5′-AGTACAGTCATCGCG [nonspecific DNA binding site, Kd (MutY) ≈250 nM (64)] was prepared and used in modifying gold electrode surfaces (50, 52). After passivation of the remaining gold surface with mercaptohexanol, the electrodes were incubated for 10 min at ambient temperature with 800 μM MutY, expressed as a fusion protein with MBP (see Methods). As evident in Fig. 2, CV of MutY at the DNA-modified surface yields a pronounced electrochemical signal at ≈275 mV versus NHE. This signal corresponds to ≈2 pmol/cm2; the density of the DNA surfaces is ≈12 pmol/cm2 as measured by radioactive tagging (52, 53). The peak shape suggests slow diffusive kinetics associated with this redox reaction; the scan rate data yield a slow diffusion constant of ≈10–11 cm2/sec. Similar CV data have been reported for cytochrome c on an electrode modified with a substrate mediator (59, 60). The electrochemical signal for MutY grows in over the 10-min incubation time, and the signal persists as the MutY solution is exchanged for MutY storage buffer; three solution exchanges are needed for bound MutY to diffuse away, consistent with MutY being bound to the DNA film. Moreover, MutY (800 μM) at a surface without DNA but coated with mercaptohexanol yields no electrochemical signal. The DNA is therefore required to reduce the protein electrochemically.
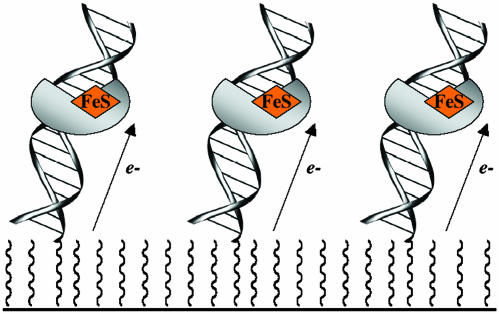
Fig. 1.
Schematic illustration of DNA-modified electrodes used in these studies to determine the DNA-bound redox potential of MutY electrochemically.
Fig. 2.
CV of 800 μM MutY at a gold surface modified with the thiol-terminated duplex SH-5′-AGTACAGTCATCGCG and passivated with mercaptohexanol. (a) The redox potential of MutY measured in these experiments requires DNA binding. The CV of MutY at a surface modified with mercaptohexanol (MCH) only (black) shows no redox process, whereas at a DNA/MCH-modified Au surface (blue), the DNA-bound redox potential of MutY is ≈75 mV vs. Ag (≈275 mV vs. NHE). (b and c) This electrochemical signal grows in with DNA-binding time (CVs for incubation from 1–10 min) (b) and persists as the MutY solution is exchanged for pure buffer (20 mM sodium phosphate, pH 7.5/1 mM Na2EDTA/20% glycerol/500 mM NaCl) (c), indicating that MutY is bound to the DNA-modified surface.
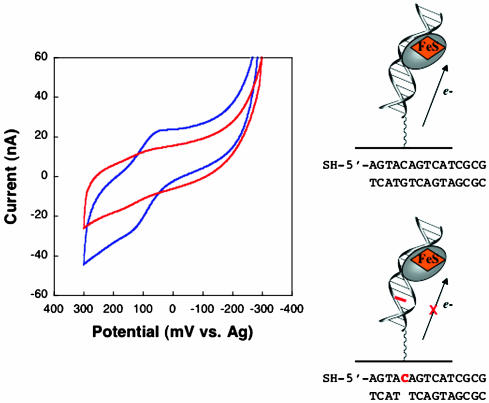
That the redox reaction of MutY is DNA-mediated rather than that the DNA serves to locally concentrate MutY at the gold surface is established in experiments where the intervening DNA structure is perturbed (Fig. 3). No electrochemical signal for MutY is observed at an electrode modified with the duplex SH-5′-AGTACAGTCATCGCG hybridized to a complement incorporating an abasic site opposite the underlined cytosine (64). It has been established that the presence of an intervening abasic site or other DNA base-stacking perturbation attenuates the reduction of intercalators bound to DNA films (50, 52, 57). Given the attenuation in signal seen here on DNA surfaces containing an abasic site, it is clear that the abasic site must intervene between MutY and the gold electrode. It is not, then, the case that MutY is simply delivered to the gold surface by DNA binding. Indeed the MutY redox process is DNA-mediated.
Fig. 3.
The redox chemistry of MutY is DNA-mediated. CV of 800 μM MutY at a surface modified with the thiol-terminated duplex SH-5′-AGTACAGTCATCGCG hybridized to a fully base-paired complement (blue) versus a complement with an abasic site opposite the red C is shown.
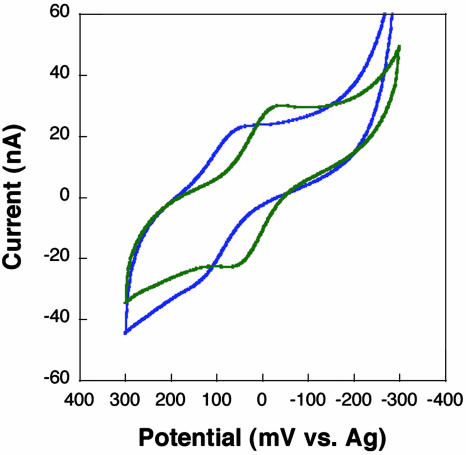
Is the redox reaction at the [4Fe4S]2+ cluster of MutY? To probe this question, we examined a mutant protein, C199H, also expressed as a fusion with MBP, in which Cys-199 is mutated to a histidine. C199H has similar activity, substrate specificity, and structure as the WT, except for histidine ligation of the cluster at position 199 (17, 65, 66). EPR and crystallography indicate that coordination to the histidine is somewhat destabilized, leading to loss of Fe with the partial formation of [3Fe4S] clusters. With C199H MutY bound to a DNA-modified electrode, we again observe a reversible cyclic voltammogram, but the potential is shifted ≈70 mV negative (Fig. 4). This shift in potential indicates that we are accessing the cluster of MutY in these electrochemistry experiments; in all other respects, C199H is electrochemically identical to the WT protein.
Fig. 4.
CV of 800 μM WT (blue) and C199H mutant (green) MutY at a gold electrode modified with the thiol-terminated sequence SH-5′-AGTACAGTCATCGCG hybridized to a fully base-paired complement. A –70 mV potential shift is observed with the cluster mutant.
Redox Properties of MutY and HiPiP Iron Proteins. The redox potentials for both the WT [4Fe4S] cluster and mutant bound to DNA are consistent with those found in HiPiP ferredoxins and small models (67, 68). Ferredoxins that contain [4Fe4S] clusters generally use the 2+/1+ couple; HiPiP iron proteins instead shuttle between the 3+/2+ forms. The electrochemical couple we observe on the DNA electrodes is in fact in the range found typically for the 3+/2+ couple of [4Fe4S] clusters of HiPiP proteins (67–74). Although redox studies of Endo III suggested that reduction of the [4Fe4S]2+ must occur at potentials <–600 mV versus NHE and oxidation could not be accomplished without decomposition (2, 38), these studies were conducted in the absence of DNA, where the FCL is accessible to solvent.
Because the [4Fe4S] cluster of MutY is in the 2+ form in the absence of DNA, owing to the change in DNA-bound redox potential, a shift in equilibrium toward a mixture of clusters in the 2+ and 3+ form is therefore expected in the presence of DNA. UV-visible spectroscopy of MutY in solution shows a small, but significant, increase in absorption at 410 nm that grows in upon addition of DNA, consistent with formation of [4Fe4S]3+ (data not shown). Low-temperature EPR also yields a signal consistent with [4Fe4S]3+ in the presence of a DNA-bound oxidant (E. Yavin, A. Boal, and J.K.B., unpublished results). Furthermore, in the electrochemistry experiments on DNA-modified electrodes, CV experiments in which the initial potential is varied indicate that both oxidized and reduced species are present initially. Chronocoulometry also reveals that the CV signal is not completely reversible; there is somewhat more electrochemical reduction than oxidation, consistent with MutY in the 3+ form having somewhat higher affinity for DNA. These data all are consistent with the environment around the [4Fe4S] cluster being altered upon DNA binding so as to favor oxidation of the cluster.
Because [4Fe4S] cluster potentials are well known to be extremely sensitive to environment beyond the ligating atoms, it is reasonable that binding of the DNA polyanion would alter the potential (67–74). Both biochemical and recent structural studies indicate that the FCL resides along the DNA-bound interface (9, 30, 38, 75, 76). Electrostatic arguments would suggest that binding of the DNA polyanion would stabilize an oxidized form of the cluster. It is also noteworthy that the many aromatic residues surrounding the cluster in MutY (1) are typically found in HiPiP proteins (69–74); DNA binding would be expected to further shield the cluster from solution and associated oxidative decomposition in the 3+ form. The change in potential on histidine ligation is also consistent with a neutral nitrogen donor stabilizing Fe(II); variations in potential of similar magnitude have been observed in ferredoxin mutants where the coordinating residue is changed (77). Moreover, if a significant population of DNA-bound protein had lost an Fe (66), a shift to more negative potentials would also be expected (78).
The [4Fe4S] cluster of these base excision repair enzymes, containing a unique ligating peptide sequence, is thus well designed for its function: robust in solution as [4Fe4S]2+, stable to oxidizing and reducing conditions of the cell, but, once buried within DNA, activated to carry out redox chemistry.
A Role for DNA CT in DNA Repair. Importantly, our electrochemical data indicate that DNA-mediated CT involving the [4Fe4S] cluster of MutY is feasible. The primary lesson learned from our many studies of DNA CT chemistry is that DNA CT is exquisitely sensitive to perturbations in base pair structure (43–50). Indeed we have shown DNA CT electrochemistry as a diagnostic tool for mutations and lesions in DNA (50, 57). Might DNA repair proteins, MutY in particular, similarly exploit DNA CT to detect mismatches and lesions in DNA?
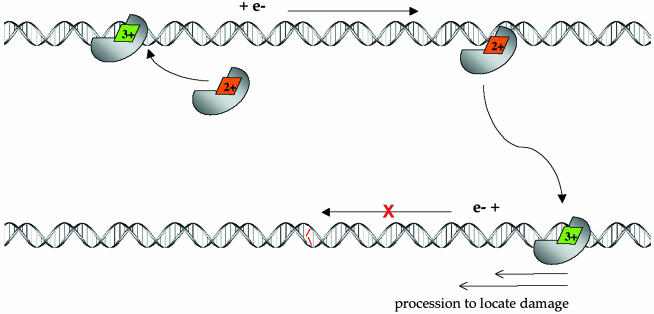
Fig. 5 illustrates a model in which DNA-mediated CT is used by the repair protein in scanning DNA for mismatches and lesions. Based on results of others, within the cell, MutY contains a [4Fe4S]2+ cluster (2, 38, 39). On binding DNA nonspecifically, as our results indicate, MutY undergoes a shift in redox potential, and the 3+/2+ couple becomes accessible, so that the cluster may become oxidized and release an electron in a DNA-mediated reaction. Upon oxidation, MutY could then serve, through DNA-mediated CT, to reduce an alternate DNA repair protein, perhaps another MutY, bound at a distant site along the duplex. The electrochemistry data indicate that such a DNA-mediated process is possible. Moreover, association of two MutY equivalents on the DNA template has been proposed based on kinetic experiments (79). In the reduced form, the DNA affinity of MutY should be diminished, facilitating dissociation of the protein from its DNA site. We propose that this process, as described, constitutes a scan of this region of the genome. Significantly, the region must be well stacked and contain no mismatches or lesions for CT to occur. Also shown is association of MutY to a region containing a mismatch, where the associated stacking perturbation would not permit DNA-mediated CT to occur. Here, the protein would remain associated with the DNA, processively diffusing to the mismatch site on a slower time scale. Data in support of this slower processive mechanism for MutY (80) as well as descriptive models (81, 82) have also been reported. The CT scanning mechanism does not obviate these schemes. Instead, the DNA CT scanning strategy confines the search to a manageable regime within the genome. Furthermore, although the specificity ratio, 10–1,000, of these repair proteins for their target site versus well-matched DNA is too low to explain preferential recognition of mismatches within the genome, this small level of specificity is sufficient for target binding within the subset of mismatch-containing strands.
Fig. 5.
Schematic model of long-range scanning for mismatches by MutY through DNA-mediated CT chemistry. (Upper) Nonspecific binding of MutY to DNA, where binding is associated with a shift in redox potential of the [4Fe4S]2+ cluster, leading to oxidation to the 3+ form. Associated with cluster oxidation is DNA-mediated CT to an alternately bound MutY, where reduction to the 2+ cluster promotes dissociation of the protein. Because this CT process proceeds without interruption by an intervening mismatch, the process constitutes a scan of this region of the genome. (Lower) Association of MutY to a region containing a mismatch (red), where the associated stacking perturbation does not permit DNA-mediated CT to occur. Here, the protein is shown processively diffusing to the mismatch site.
Overall, this process provides a rationale for the widespread distribution of redox cofactors in DNA repair proteins (1–7, 31–37). Through this route, the rapid scanning of the genome can be accomplished by using few copies of repair protein. Significantly, this DNA-mediated signaling strategy takes advantage of the unique chemical characteristics of DNA CT. This chemistry now requires consideration more generally with respect to mechanisms of DNA repair and signaling in vivo.
Acknowledgments
We thank D. Ceres for technical assistance, M.-P. Golinelli for preparation of MutY mutants, and M. G. Hill and E. D. A. Stemp for helpful discussions. We thank the National Institutes of Health and the National Foundation for Cancer Research for their financial support of this research.
Abbreviations: Endo III, endonuclease III; CT, charge transport; MBP, maltose-binding protein; CV, cyclic voltammetry.
References
- 1.Guan, Y., Manuel, R. C., Arvai, A. S., Parikh, S. S., Mol, C. D., Miller, J. H., Lloyd, S. & Tainer, J. A. (1998) Nat. Struct. Biol. 5 1058–1064. [DOI] [PubMed] [Google Scholar]
- 2.Cunningham, R. P., Asahara, H., Bank, J. F., Scholes, C. P., Salerno, J. C., Surerus, K., Munck, E., McCracken, J., Peisach, J. & Emptage, M. H. (1989) Biochemistry 28 4450–4455. [DOI] [PubMed] [Google Scholar]
- 3.Pomposiello, P. J. & Demple, B. (2001) Trends Biotechnol. 19 109–114. [DOI] [PubMed] [Google Scholar]
- 4.Unden, G. & Bongaerts, J. (1997) Biochim. Biophys. Acta 1320 217–222. [DOI] [PubMed] [Google Scholar]
- 5.Rebeil, R., Sun, Y., Chooback, L., Pedraza-Reyes, M., Kinsland, C., Begley, T. P. & Nicholson, W. L. (1998) J. Bacteriol. 180 4879–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinks, J. A., Evans, M. C. W., de Miguel, Y., Sartori, A. A., Jiricny, J. & Pearl, L. H. (2002) J. Biol. Chem. 277 16936–16940. [DOI] [PubMed] [Google Scholar]
- 7.Lee, C. H., Kim, S. H., Choi, J. I., Choi, J. Y., Lee, C. E. & Kim, J. (2002) Mol. Cells 13 154–164. [PubMed] [Google Scholar]
- 8.Michaels, M. L., Pham, L., Nghiem, Y., Cruz, C. & Miller, J. H. (1990) Nucleic Acids Res. 18 3841–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kou, C.-F., McRee, D. E., Fischer, C. L., O'Handley, S. F., Cunningham, R. P. & Tainer, J. A. (1992) Science 258 434–440. [DOI] [PubMed] [Google Scholar]
- 10.Williams, S. D. & David, S. S. (1998) Chem. Rev. 98 1221–1232. [DOI] [PubMed] [Google Scholar]
- 11.Au, K. G., Cabrera, M., Miller, J. H. & Modrich, P. (1988) Proc. Natl. Acad. Sci. USA 85 9163–9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Au, K. G., Clark, S., Miller, J. H. & Modrich, P. (1989) Proc. Natl. Acad. Sci. USA 86 8877–8881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu, A.-L. & Chang, D. (1988) Cell 54 805–809. [DOI] [PubMed] [Google Scholar]
- 14.Michaels, M. L. & Miller, J. H. (1992) J. Bacteriol. 174 6321–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michaels, M. L., Tchou, J., Grollman, A. P. & Miller, J. H. (1992) Biochemistry 31 10964–10968. [DOI] [PubMed] [Google Scholar]
- 16.Michaels, M. L., Cruz, C., Grollman, A. P. & Miller, J. H. (1992) Proc. Natl. Acad. Sci. USA 89 7022–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porello, S. L., Leyes, A. E. & David, S. S. (1998) Biochemistry 37 14756–14764. [DOI] [PubMed] [Google Scholar]
- 18.Lu, A. L., Tsai-Wu, J. J. & Cillo, J. (1995) J. Biol. Chem. 270 23582–23589. [DOI] [PubMed] [Google Scholar]
- 19.Manuel, R. C. & Lloyd, R. S. (1997) Biochemistry 36 11140–11152. [DOI] [PubMed] [Google Scholar]
- 20.Gogos, A., Cillo, J., Clarke, N. D. & Lu, A. L. (1996) Biochemistry 35 16665–16671. [DOI] [PubMed] [Google Scholar]
- 21.Manuel, R. C., Czerwinski, E. W. & Lloyd, R. S. (1996) J. Biol. Chem. 271 16218–16226. [DOI] [PubMed] [Google Scholar]
- 22.Breimer, L. & Lindahl, T. (1980) Nucleic Acids Res. 8 6199–6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katcher, H. L. & Wallace, S. S. (1983) Biochemistry 22 4072–4081. [DOI] [PubMed] [Google Scholar]
- 24.Demple, B. & Linn, S. (1980) Nature 287 203–208. [DOI] [PubMed] [Google Scholar]
- 25.Boorstein, R. J., Hilbert, T. P., Cadet, J., Cunningham, R. P. & Teebor, G. W. (1989) Biochemistry 28 6164–6169. [DOI] [PubMed] [Google Scholar]
- 26.Breimer, L. H. & Lindahl, T. (1984) J. Biol. Chem. 259 5543–5548. [PubMed] [Google Scholar]
- 27.Dizdaroglu, M., Laval, J. & Boiteux, S. (1993) Biochemistry 32 12105–12111. [DOI] [PubMed] [Google Scholar]
- 28.Wagner, J. R., Blount, B. C. & Weinfeld, M. (1996) Anal. Biochem. 233 76–81. [DOI] [PubMed] [Google Scholar]
- 29.Breimer, L. H. & Lindahl, T. (1985) Biochemistry 24 4018–4022. [DOI] [PubMed] [Google Scholar]
- 30.Thayer, M. M., Ahern, H., Xing, D., Cunningham, R. P. & Tainer, J. A. (1995) EMBO J. 14 4108–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cunningham, R. P. (1997) Mut. Res. 383 189–196. [DOI] [PubMed] [Google Scholar]
- 32.Begley, T. J., Haas, B. J., Noel, J., Shektman, A., Williams, W. A. & Cunningham, R. P. (1999) Curr. Biol. 9 653–659. [DOI] [PubMed] [Google Scholar]
- 33.Desiraju, V., Shanabruch, W. & Lu, A. (1993) J. Bacteriol. 175 541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nolling, J., van Eeden, F. J. M., Eggen, R. I. L. & de Vos, W. M. (1992) Nucleic Acids Res. 20 6501–6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piersen, C. E., Prince, M. A., Augustine, M. L., Dodson, M. L. & Lloyd, R. S. (1995) J. Biol. Chem. 270 23475–23484. [DOI] [PubMed] [Google Scholar]
- 36.Hilbert, T. P., Boorstein, R. J., Kung, H. C., Bolton, P. H., Xing, D., Cunningham, R. P. & Teerbor, G. W. (1996) Biochemistry 35 2505–2511. [DOI] [PubMed] [Google Scholar]
- 37.McGoldrick, J. P., Yeh, Y. C., Solomon, M., Essigmann, J. M. & Lu, A. L. (1995) Mol. Cell. Biol. 15 989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu, W., O'Handley, S., Cunningham, R. P. & Johnson, M. K. (1992) J. Biol. Chem. 267 16135–16140. [PubMed] [Google Scholar]
- 39.Porello, S. L., Cannon, M. J. & David, S. S. (1998) Biochemistry 37 6465–6475. [DOI] [PubMed] [Google Scholar]
- 40.Boon, E. M. & Barton, J. K. (2002) Curr. Opin. Struct. Biol. 12 320–329. [DOI] [PubMed] [Google Scholar]
- 41.Schuster, G. B. (2000) Acc. Chem. Res. 33 253–258. [DOI] [PubMed] [Google Scholar]
- 42.Giese, B. (2002) Annu. Rev. Biochem. 71 51–70. [DOI] [PubMed] [Google Scholar]
- 43.Hall, D. B., Holmlin, R. E. & Barton, J. K. (1996) Nature 382 731–735. [DOI] [PubMed] [Google Scholar]
- 44.Nunez, M. E., Hall, D. B. & Barton, J. K. (1999) Chem. Biol. 6 85–97. [DOI] [PubMed] [Google Scholar]
- 45.Bhattacharya, P. K. & Barton, J. K. (2001) J. Am. Chem. Soc. 123 8949–8956. [DOI] [PubMed] [Google Scholar]
- 46.Kelley, S. O. & Barton, J. K. (1999) Science 283 375–381. [DOI] [PubMed] [Google Scholar]
- 47.Nunez, M. E., Noyes, K. T. & Barton, J. K. (2002) Chem. Biol. 9 403–415. [DOI] [PubMed] [Google Scholar]
- 48.Rajski, S. R., Kumar, S., Roberts, R. J. & Barton, J. K. (1999) J. Am. Chem. Soc. 121 5615–5616. [Google Scholar]
- 49.Wagenknecht, H.-A., Stemp, E. D. A. & Barton, J. K. (2000) J. Am. Chem. Soc. 122 1–7. [Google Scholar]
- 50.Boon, E. M., Ceres, D. M., Drummond, T. G., Hill, M. G. & Barton, J. K. (2000) Nat. Biotechnol. 18 1096–1100. [DOI] [PubMed] [Google Scholar]
- 51.Nunez, M. E., Holmquist, G. P. & Barton, J. K. (2001) Biochemistry. 40 12465–12471. [DOI] [PubMed] [Google Scholar]
- 52.Boon, E. M., Salas, J. E. & Barton, J. K. (2001) Nat. Biotechnol. 20 282–286. [DOI] [PubMed] [Google Scholar]
- 53.Kelley, S. O., Barton, J. K., Jackson, N. M. & Hill, M. G. (1997) Bioconjugate Chem. 8 31–36. [DOI] [PubMed] [Google Scholar]
- 54.Kelley, S. O., Barton, J. K., Jackson, N. M., McPherson, L. D., Potter, A. B., Spain, E. M., Allen, M. J. & Hill, M. G. (1998) Langmuir 14 6781–6786. [Google Scholar]
- 55.Sam, M., Boon, E. M., Barton, J. K., Hill, M. G. & Spain, E. M. (2001) Langmuir 17 5727–5730. [Google Scholar]
- 56.Kelley, S. O., Jackson, N. M., Hill, M. G. & Barton, J. K. (199) Angew. Chem. Int. Ed. Engl. 38 941–945. [DOI] [PubMed] [Google Scholar]
- 57.Kelley, S. O., Boon, E. M., Barton, J. K., Jackson, N. M. & Hill, M. G. (1999) Nucleic Acids Res. 27 4830–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chmiel, N. H., Golinelli, M. P., Francis, A. W. & David, S. S. (2001) Nucleic Acids Res. 29 553–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eddowes, M. J. & Hill, H. A. O. (1979) J. Am. Chem. Soc. 701 4461–4462. [Google Scholar]
- 60.Hill, H. A. O., Page, D. J. & Walton, N. J. (1987) J. Electroanal. Chem. 217 129–140. [Google Scholar]
- 61.Guo, L. H. & Hill, H. A. O. (1991) Adv. Inorg. Chem. 36 341–375. [Google Scholar]
- 62.Armstrong, F. A., Heering, H. A. & Hirst, J. (1997) Chem. Soc. Rev. 26 169–188. [Google Scholar]
- 63.Liu, H. H., Lu, J. L., Zhang, M., Pang, D. W. & Abruna, H. D. (2003) J. Electroanal. Chem. 544 93–100. [Google Scholar]
- 64.Porello, S. L., Williams, S. D., Kuhn, H., Michaels, M. L. & David, S. S. (1996) J. Am. Chem. Soc. 118 10684–10692. [Google Scholar]
- 65.Golinelli, M. P., Chmiel, N. H. & David, S. S. (1999) Biochemistry 38 6997–7007. [DOI] [PubMed] [Google Scholar]
- 66.Messick, T. E., Chmiel, N. H., Golinelli, M. P., Langer, M. R., Joshua-Tor, L. & David, S. S. (2002) Biochemistry 41 3931–3942. [DOI] [PubMed] [Google Scholar]
- 67.Beinert, H., Holm, R. H. & Munck, E. (1997) Science 277 653–659. [DOI] [PubMed] [Google Scholar]
- 68.Carter, C. W., Jr., Kraut, J., Freer, S. T., Alden, R. A., Sieker, L. C., Adman, E. & Jensen, L. H. (1972) Proc. Natl. Acad. Sci. USA 69 3526–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Glaser, T., Bertini, I., Moura, J. J. G., Hedman, B., Hodgson, K. O. & Solomon, E. I. (2001) J. Am. Chem. Soc. 123 4859–4860. [DOI] [PubMed] [Google Scholar]
- 70.Adman, E., Watenpaugh, K. D. & Jensen, L. H. (1975) Proc. Natl. Acad. Sci. USA 72 4854–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kodaka, M., Tomohiro, T. & Okuno, H. (1991) J. Phys. Chem. 95 6741–6744. [Google Scholar]
- 72.Stephens, P. J., Jollie, D. R. & Warshel, A. (1996) Chem. Rev. 96 2491–2500. [DOI] [PubMed] [Google Scholar]
- 73.Banci, L., Bertini, I., Savellini, G. G. & Luchinat, C. (1996) Inorg. Chem. 35 4248–4253. [DOI] [PubMed] [Google Scholar]
- 74.Cowan, J. A. & Lui, S. M. (1998) Adv. Inorg. Chem. 45 313–350. [Google Scholar]
- 75.Chepanoske, C. L., Golinelli, M. P., Williams, S. D. & David, S. S. (2000) Arch. Biochem. Biophys. 380 11–19. [DOI] [PubMed] [Google Scholar]
- 76.Fromme, J. C. & Verdine, G. L. (2003) EMBO J. 22 3461–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen, K. S., Bonagura, C. A., Tilley, G. J., McEvoy, J. P., Jung, Y. S., Armstrong, F. A., Stout, C. D. & Burgess, B. K. (2002) Nat. Struct. Biol. 9 188–192. [DOI] [PubMed] [Google Scholar]
- 78.Zhou, J., Hu, Z., Munck, E. & Holm, R. H. (1996) J. Am. Chem. Soc. 118 1966–1967. [Google Scholar]
- 79.Wong, I., Bernards, A. S., Miller, J. K. & Wirz, J. A. (2003) J. Biol. Chem. 278 2411–2418. [DOI] [PubMed] [Google Scholar]
- 80.Francis, A. W. & David, S. S. (2003) Biochemistry 42 801–810. [DOI] [PubMed] [Google Scholar]
- 81.Verdine, G. L. & Bruner, S. D. (1997) Chem. Biol. 4 329–334. [DOI] [PubMed] [Google Scholar]
- 82.Chen, L. W., Haushalter, K. A., Lieber, C. M. & Verdine, G. L. (2002) Chem. Biol. 9 345–350. [DOI] [PubMed] [Google Scholar]