Abstract
Prostate-specific membrane antigen (PSMA) is a type 2 integral membrane glycoprotein that serves as an attractive target for cancer immunotherapy by virtue of its abundant and restricted expression on the surface of prostate carcinomas and the neovasculature of most other solid tumors. However, relatively little is known about the molecular structure of this target. Here, we report that PSMA is expressed on tumor cells as a noncovalent homodimer. A truncated PSMA protein, lacking transmembrane and cytoplasmic domains, also formed homodimers, indicating that the extracellular domain is sufficient for dimerization. PSMA dimers but not monomers displayed a native conformation and possessed high-level carboxypeptidase activity. A unique dimer-specific epitope was identified by using one of a panel of novel mAbs. When used to immunize animals, dimer but not monomer elicited antibodies that efficiently recognized PSMA-expressing tumor cells. These findings on PSMA structure and biology may have important implications for active and passive immunotherapy of prostate and other cancers.
According to the American Cancer Society, prostate cancer is the most prevalent cancer in American males and represents their second leading cause of cancer death (www.cancer.org/downloads/STT/CancerFacts&Figures.2002TM.pdf). Localized disease is generally treated with surgery or radiation, and recurrent disease can be controlled temporarily with hormonal therapy. However, almost all prostate carcinomas eventually become androgen-independent, and the use of chemotherapy (single agent or in combination) remains investigational. Because of the significant mortality and morbidity associated with disease progression, there is an urgent need for new and targeted treatments.
Recent advances in expression profiling have identified a number of candidate markers for prostate cancer (1–4). However, to serve as useful targets for immunotherapy, some suggest such markers should meet several additional criteria: (i) primarily restricted to the prostate, (ii) abundantly expressed as protein at all stages of disease, (iii) presented at the cell surface but not shed into the circulation, and (iv) associated with enzymatic or signaling activity. Prostate-specific membrane antigen (PSMA) remains among the few prostate cancer markers to fulfill these criteria and is overall the best validated one (5).
PSMA was originally discovered in the androgen-dependent LNCaP human prostatic adenocarcinoma cell line as the target for mAb 7E11 (6). A radiolabeled form of 7E11 is used clinically to image and stage prostate cancer (7), validating the utility of PSMA for tumor targeting. In normal prostate epithelia, PSMA is expressed primarily as a cytoplasmic protein termed PSM′ (8). In prostate carcinomas, however, differential mRNA splicing leads to expression of PSMA as a type 2 integral membrane glycoprotein possessing a 19-aa cytoplasmic fragment, a single 24-aa membrane-spanning domain, and a 707-aa extracellular region (9). PSMA expression generally increases with disease progression, becoming greatest in hormone-refractory and metastatic disease (8). PSMA also is expressed on the neovasculature of most other solid tumors but not in normal vasculature (10–14).
Despite the importance of PSMA as a marker for prostate cancer, relatively little is known about its biologic functions and its biophysical properties. PSMA possesses glutamate carboxypeptidase activity (EC 4.17.21), but the role of this activity in cellular transformation and metastasis is not understood. Although PSMA expression is highest in prostate, detectable levels of protein are also found in small intestine and brain. Intestinal PSMA may play a role in the metabolism of dietary γ-glutamated folate derivatives (15). Brain PSMA, also referred to N-acetylated α-linked acidic dipeptidase (NAALADase), may modulate glutamatergic neurotransmission (16), and a potent inhibitor of PSMA's NAALADase activity has shown activity in an animal model of ischemic stroke (17).
Here, we report that PSMA is expressed on prostate cancer cells as a noncovalently associated homodimer. Using a secreted form of the protein, we demonstrate that the extracellular domain is sufficient for dimerization and that dimerization is required for enzymatic activity. We also describe a dimer-specific mAb. Strikingly, when used as an immunogen, dimeric but not monomeric PSMA was capable of efficiently eliciting antibodies that recognize PSMA-expressing tumor cells. These findings provide insight into the structure and biology of an important target for cancer therapy.
Materials and Methods
Expression and Purification of the Extracellular Domain of PSMA (PSMAECTO). A fragment encoding the entire PSMAECTO (amino acids 44–750) was generated from the full-length PSMA cDNA plasmid p55A (9) by PCR. The resulting PCR product was cloned into mammalian expression vector PPI4 (18) downstream of the tissue plasminogen activator signal sequence to direct secretion of the protein. DNA sequencing was performed to confirm the fidelity of the construct. PPI4 encodes dihydrofolate reductase as a selectable marker and thus was transfected into dihydrofolate reductase-deficient Chinese hamster ovary DXB11 cells by using calcium phosphate. Transfectants were selected in nucleoside-free α-MEM containing dialyzed FBS (Life Technologies, Grand Island, NY) and then expression-amplified in stepwise increasing concentrations of methotrexate (Sigma). For protein expression, cells were cultured in the presence of serum-free Chinese hamster ovary cell media (JRH Biosciences, Lenexa, KS). PSMAECTO was purified from the culture supernatants by ion exchange, hydrophobic interaction, and hydroxyapatite chromatographies. A detailed description of the purification procedure is described in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org. Preparative size-exclusion chromatography (SEC) was used to obtain homogeneous monomeric and dimeric fractions of the protein. Purified PSMAECTO was stored in neutral PBS containing 1 mM Ca2+ and 0.5 mM Mg2+ (PBS+). Unless otherwise indicated, PSMA monomers represent spontaneously dissociated protein recovered over SEC rather than forcibly denatured material.
Antibodies. Mouse mAb MAB544 was obtained from Maine Biotechnology Services (Portland, ME). Additional mAbs to PSMA were generated by immunizing female mice s.c. at ≈3-week intervals with recombinant PSMA or 5 × 106 LNCaP cells adjuvanted with alum or Titermax (Sigma). Animals were administered a boost of antigen ≈3 days before death. Murine antibodies were raised by using BALB/c mice, and fully human antibodies were raised in XenoMice (Abgenix, Fremont, CA) in which the murine Ig gene locus had been inactivated and replaced with the human Ig gene locus (19). Hybridomas were generated by using standard methods by fusion of splenocytes with Sp2/0 or NS0 myeloma cells, selection in hypoxanthine-, aminopterin-, and thymidine-supplemented medium, and screening for reactivity with PSMA by ELISA. Hybridomas were cultured in CELLline flasks (Integra Biosciences, East Dundee, IL) in DMEM/F12 medium containing 10% ultra-low IgG FBS (Life Technologies). Antibodies were purified to >95% homogeneity by protein A-Sepharose Fast Flow chromatography (Amersham Biosciences), dialyzed into PBS, and stored at –80°C before use.
Protein Size Analyses. PSMA-3T3 cells (gift from M. Sadelain, Memorial Sloan–Kettering Cancer Center, New York) or LNCaP cells were lysed with M-PER reagent (Pierce) for 10 min and then centrifuged at 15,000 rpm for 30 min at 4°C. Full-length PSMA was recovered in the supernatant. For SDS/PAGE analysis, samples were suspended in Laemmli sample buffer in the presence (reducing) or absence (nonreducing) of 50 mM DTT. Blue Native (BN)/PAGE was performed according to published methods (20). Briefly, purified protein or cell lysates were diluted into an equal volume of pH 7.7 buffer containing 100 mM Mops, 100 mM Tris, 40% glycerol, and 0.1% Coomassie G-250. Samples were electrophoresed on 4–12% BisTris gels (Invitrogen) by using 50 mM Mops, 50 mM Tris, pH 7.7 as the running buffer. Gels were either stained with Coomassie blue or electroblotted onto nitrocellulose for Western blot analysis as described (20) by using MAB544 for detection. Analytical SEC was performed by using a TSK G3000SWXL (TosoHaas, Montgomeryville, PA) column equilibrated in PBS+. The column was calibrated by using BSA (67 kDa), IgG (150 kDa), ferritin (440 kDa), and thyroglobulin (670 kDa) as standards.
Enzyme Assays. Pteroyl γ-glutamyl carboxypeptidase (folate hydrolase) activity was determined by monitoring the cleavage of poly γ-glutamylated methotrexate as described (15) with the following exceptions. Di-γ-glutamylated methotrexate (MTX-glu2) was used as substrate and HPLC was used rather than capillary electrophoresis. At the completion of the incubation, 100 μl of 0.5 M Na2HPO4 was added to stop the reaction. Samples were loaded at a flow rate of 1.25 ml/min through a 50 × 4.6 mm, 3 μm PRISM reversed-phase column (Thermo Hypersil, Keystone Scientific, Bellefonte, PA) with a PRISM 10 × 4-mm guard column, eluted with 15% methanol in 85% 0.5 M K2HPO4, pH 7.0, and quantitated based on relative peak area observed at a wavelength of 313 nm. For NAALADase assays, PSMAECTO was incubated with N-acetyl-α-l-aspartyl-l-glutamate for 22 h at 37°C in the presence of 20 mM sodium phosphate, 50 mM NaCl, 10 mM ZnCl2, pH 7.1. Released l-glutamic acid was quantitated by using a commercial kit (R-Biopharm, Marshall, MI); 2-(phosphonomethyl)pentanedioic acid and Gly-Pro-7-amido-4-methylcoumarin were purchased from Sigma. Porcine kidney dipeptidyl peptidase IV (DPP IV) was used according to the manufacturer's instructions (Sigma).
Surface Plasmon Resonance Measurements. A Biacore 3000 instrument was used (Biacore, Piscataway, NJ). Antibodies were immobilized at ≈10,000 resonance units to CM5 sensor chips according to the manufacturer's instructions for amine coupling. A reference surface of isotype-matched antibody of irrelevant specificity was used as a background control. Binding experiments were performed at 25°C in PBS buffer with 0.005% (vol/vol) Surfactant P20. Purified PSMAECTO dimer (50 nM) or monomer (100 nM) was passed over control and test flow cells at a flow rate of 5 μl/min. The sensor surface was regenerated with two pulses of 20 mM HCl.
Immunogenicity Studies. BALB/c mice were immunized in groups of five with 5 μgofPSMAECTO on alum (250 μg per dose, Sigma) by s.c. injection at weeks 0, 1, 2, and 6. Serum was drawn 10 days after the final injection and analyzed by ELISA and flow cytometry. For ELISA, 96-well microtiter plates were coated with monomeric or dimeric PSMAECTO and blocked with a PBS/casein/Tween 20 buffer. Serially diluted immune or naïve mouse serum was added, and bound antibody was detected by using a goat anti-mouse IgG antibody conjugated to alkaline phosphatase, followed by the colorimetric substrate pNPP. The endpoint antibody titer was defined as the highest dilution that yielded an absorbance of 0.1 OD above background. For flow cytometry, cells were incubated on ice for 30 min with 200 μl of mouse serum diluted 1:50 in PBS+ with 0.1% sodium azide (PBSA). Cells were washed twice with PBSA and incubated for 30 min on ice with FITC-conjugated goat anti-mouse IgG. Cells were washed, resuspended in PBSA, and subjected to flow cytometry on a FACSCalibur (Becton Dickinson). Data are expressed in terms of mean fluorescence intensity (MFI) of staining.
Results
PSMA Is Expressed as a Noncovalent Homodimer, and Dimerization Is Mediated by Extracellular Residues. PSMAECTO was secreted from a stable Chinese hamster ovary cell line and purified to >95% homogeneity under nondenaturing conditions (Fig. 1A). The identity of the purified protein was confirmed by Western blot analysis (data not shown). The purified protein migrated at ≈100 kDa on denaturing SDS/PAGE gels under both reducing and nonreducing conditions, consistent with prior observations for full-length PSMA (9, 21, 22). Thus, like full-length PSMA, PSMAECTO is a monomer in the presence of denaturing detergents, and no disulfide or other covalent bonds are present to mediate oligomerization.
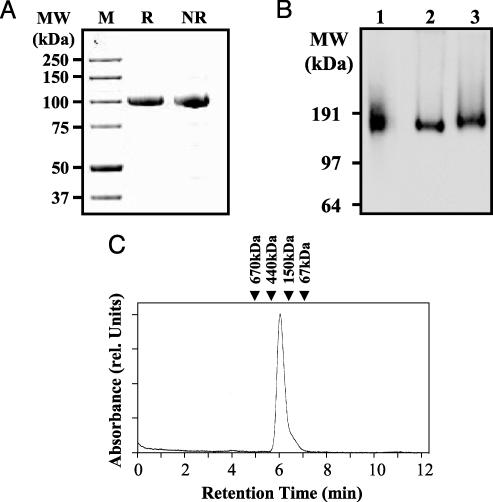
Fig. 1.
Full-length PSMA and PSMAECTO are expressed as noncovalent homodimers. (A) SDS/PAGE analysis of purified PSMAECTO under reducing (R) and nonreducing (NR) conditions. The protein (1 μg) was resolved on a 4–12% gradient gel and stained with Coomassie blue. Molecular mass markers (M) are indicated. Under denaturing conditions, PSMAECTO exhibits a molecular mass of 100 kDa, indicating the lack of covalent interactions between subunits. (B) BN/PAGE analysis of PSMAECTO (lane 1) and full-length PSMA extracted from PSMA-3T3 cells (lane 2) or LNCaP cells (lane 3). The apparent molecular masses indicate that both PSMAECTO and full-length PSMA are dimers under native conditions. (C) Analytical SEC of purified PSMAECTO in neutral PBS buffer. The arrows indicate the retention times of protein standards. The retention time of 260 kDa for PSMAECTO is consistent with that of a homodimer.
BN/PAGE was used to compare the oligomeric structure of PSMAECTO and full-length PMSA under native conditions. In BN/PAGE, proteins are sieved according to their mass by the progressively smaller pores of a polyacrylamide gradient gel run in the absence of denaturing detergents (23, 24). LNCaP and PSMA-3T3 cell membranes were solubilized by using nonionic detergents to preserve the native tertiary and quaternary structure of full-length PSMA. The whole-cell extracts or purified PSMAECTO were subjected to BN/PAGE and Western blotting. Both PSMAECTO and full-length PSMA migrated in BN/PAGE at just under 200 kDa, indicating that the proteins are noncovalently associated homodimers (Fig. 1B).
Compared with LNCaP-derived PSMA, PSMAECTO was slightly smaller, consistent with its lack of the ≈4.8-kDa transmembrane and cytoplasmic regions. A minor difference in size was observed between full-length PSMA extracted from LNCaP and PSMA-3T3 cells, with the 3T3-derived material being slightly smaller. The difference could be caused by differential glycosylation or other posttranslational modification of PSMA in the two cell lines.
PSMAECTO was subjected to analytical SEC as a second sizing method. When analyzed in neutral PBS+ buffer, purified PSMAECTO eluted as a single major peak with an apparent molecular mass of 260 kDa (Fig. 1C), slightly higher than expected. However, glycoproteins (such as PSMAECTO) are typically nonglobular in shape and run at higher apparent molecular mass than standard SEC calibration proteins (20). Therefore, an apparent molecular mass of 260 kDa is consistent with the proposed homodimeric structure of PSMAECTO. In contrast, purified monomeric PSMAECTO eluted with an apparent molecular mass of 130 kDa (data not shown).
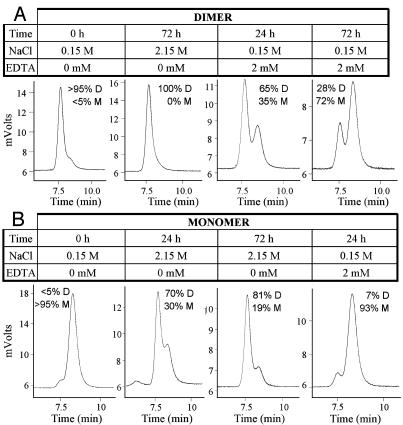
Monomer–Dimer Equilibrium. Purified dimeric and monomeric forms of PSMAECTO were resolved by preparative SEC in PBS+ buffer and collected in separate fractions. To assess whether dimer and monomer exist in a reversible equilibrium, the buffer conditions were perturbed, and the monomer–dimer ratio was analyzed by SEC. As indicated in Fig. 2A, a dimer preparation that contained ≈5% monomer initially was converted to 100% dimer upon incubation for 72 h at ambient temperature in PBS+ supplemented with 2M sodium chloride (Fig. 2 A). Conversely, the addition of 2 mM of the metal-chelating agent EDTA converted the dimer into monomer with a half-life of ≈2 days (Fig. 2 A), indicating that dimer stability is dependent upon the presence of metal ions, such as Zn2+ in the active site of PSMA. However, we cannot rule out that other metal ions (e.g., Ca2+) stabilize the dimer.
Fig. 2.
PSMA monomer–dimer equilibrium. Purified dimeric (A) and monomeric (B) PSMAECTO were subjected to varying buffer conditions and analyzed for size by analytical SEC. The percentages of monomer (M) and dimer (D) are indicated. The monomer and dimer were initially contained in PBS+ buffer at a concentration of 0.2 mg/ml. The buffer conditions were adjusted as indicated, and the proteins were incubated at ambient temperature for the indicated time periods before SEC analysis.
For a preparation that initially comprised >95% monomer, high salt similarly drove the equilibrium to mostly (81%) dimer within 72 h (Fig. 2B). EDTA had little influence on the oligomeric state of the monomer. Thus, regardless of the initial oligomeric state of the protein, high salt concentrations promote dimerization, whereas metal-chelating agents dissociate dimers into monomers.
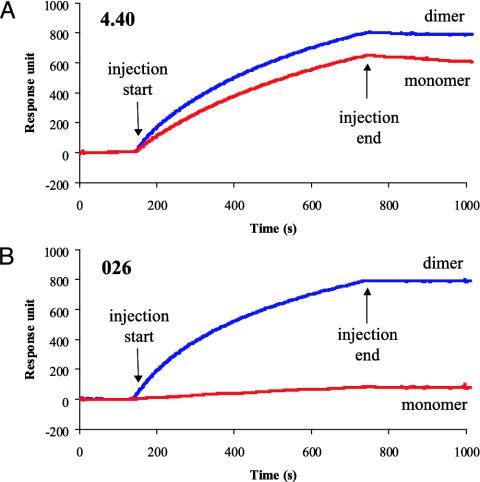
The Homodimer Represents the Native Conformation of PSMA. The conformations of cell-surface PSMA and PSMAECTO were explored by Biacore, flow cytometry, immunoprecipitation, and Western blot assays by using a panel of two mouse and two fully human mAbs against PSMA. The two human anti-PSMA mAbs, designated 026 and 4.40, specifically recognized full-length PSMA as expressed on the surface of LNCaP, C4-2, and PSMA-3T3 cells (data not shown). In addition, both mAbs recognized PSMAECTO dimer in Biacore (Fig. 3). However, whereas mAb 4.40 bound both monomeric and dimeric forms of PSMAECTO (Fig. 3A), mAb 026 was dimer-specific (Fig. 3B). Therefore, mAb 026 defines a dimer-specific epitope on PSMA.
Fig. 3.
Reactivity of monomeric and dimeric PSMAECTO with mAbs in Biacore. PSMA-specific or isotype-control antibodies (≈10,000 response units) were immobilized on CM5 sensor chip. PSMAECTO dimer (50 nM) or monomer (100 nM) was injected at 5 μl/min over control and test flow cells. Sensorgrams depict the net binding of PSMAECTO dimer and monomer to anti-PSMA mAbs. (A) mAb 4.40 recognizes monomeric and dimeric PSMAECTO with similar efficiency. (B) mAb 026 is specific for a dimer-specific epitope on PSMAECTO.
These studies were extended by using a broader panel of additional two human and four murine anti-PSMA mAbs. Uniformly, mAbs that recognized cell-surface PSMA in flow cytometry also reacted with PSMAECTO homodimer in Biacore. Most but not all bound PSMAECTO monomer. Dimer-specific mAbs such as 026 were inactive in Western blot analysis of PSMAECTO and full-length PSMA, highlighting the importance of tertiary and/or quaternary structure for recognition. In contrast, certain mAbs that recognized monomeric PSMA in Biacore were also active in Western blot. mAbs reactive in flow cytometry also efficiently immunoprecipitated PSMAECTO and full-length PSMA (data not shown). Taken together, these results indicate that the native conformation of PSMA is a homodimer and that the monomer possesses a partially denatured conformation or exposes epitopes located at the dimer interface that are not accessible in the dimer.
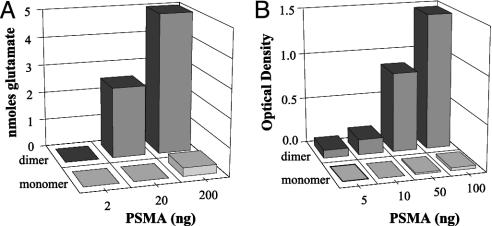
Homodimerization Is Required for Enzymatic Activity. PSMA has been reported to possess folate hydrolase, NAALADase, and DPP IV activities (15, 16, 25). The first two activities involve the hydrolysis of a carboxyl-terminal peptide bond to liberate a glutamic acid residue, whereas DPP IV cleaves downstream of an amino-terminal Aaa-Pro dipeptide sequence. We first evaluated the folate hydrolase activities of purified monomeric and dimeric forms of PSMAECTO. Whereas the dimer demonstrated high-level folate hydrolase activity, the monomer was essentially inactive (Fig. 4A). In fact, the residual activity of the monomer could be attributed to the residual amount (≈4%) of dimeric PSMAECTO present in the preparation. High-level folate hydrolase activity was also observed for LNCaP cell lysates (data not shown), consistent with prior observations (15). Similarly, dimeric but not monomeric forms of PSMAECTO possessed high-level NAALADase activity (Fig. 4B), which was abrogated by using 5 nM of the inhibitor 2-(phosphonomethyl)pentanedioic acid (data not shown).
Fig. 4.
Dimeric but not monomeric PSMAECTO is enzymatically active. Dimeric and monomeric PSMA were tested for folate hydrolase activity (A) and NAALADase activity (B). The background activity observed for PSMA monomer is consistent with the residual amount (≈4%) of dimer present in the preparation.
Lastly, neither monomer nor dimer demonstrated DPP IV activity under conditions where porcine DPP IV efficiently hydrolyzed the substrate Gly-Pro-7-amido-4-methylcoumarin (data not shown). Our result is consistent with that of Barinka et al. (26), who similarly failed to confirm the DPP IV activity previously reported for PSMA (25).
Dimeric but Not Monomeric Forms of PSMA Efficiently Elicit Antibodies Reactive Against PSMA-Expressing Tumor Cells. To comparatively evaluate the immunogenicity of different forms of PSMA, PSMAECTO monomers and dimers were formulated on the immunostimulatory adjuvant alum and used to immunize BALB/c mice. After the final immunization, sera were evaluated for reactivity with PSMA by ELISA and by flow cytometry to measure the humoral immune response.
When tested by ELISA, sera from both monomer- and dimer-immunized animals showed similar levels of anti-PSMA antibodies, indicating that each protein was immunogenic when formulated on alum. For dimer-immunized animals, the median endpoint titers were 1/6,400 (range 1/3,200 to 1/12,800) regardless of whether PSMAECTO monomer or dimer was used as the coating antigen. Similarly, monomer-immunized animals had median endpoint titers of 1/6,400 in both assay formats, although the range varied somewhat depending on whether the monomer (range <1/400 to 1/12,800) or dimer (range <1/400 to 1/6,400) was used for coating.
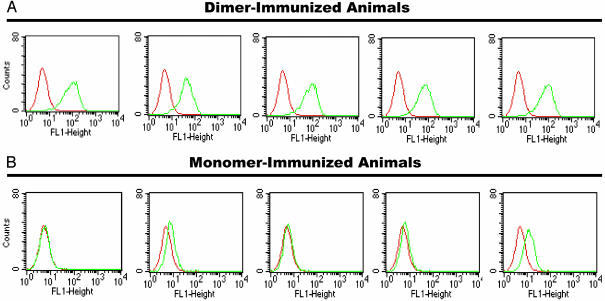
However, a sharp difference between sera was observed with cell-based flow cytometry (Fig. 5). Whereas each dimer-immunized animal elicited high-titered antibodies to PSMA-3T3 cells (median MFI = 74, range 41–84), such antibodies were weak in monomer-immunized animals (median MFI = 6, range = 5–12). The level of binding observed for monomer-immunized animals was comparable to that for naïve animals (data not shown). Similarly, background levels of binding to parental 3T3 cells were observed for all sera (median MFI <6 in all cases).
Fig. 5.
Dimeric but not monomeric PSMAECTO elicits antibodies that efficiently recognize cell-surface PSMA. Mice were immunized in groups of five with dimeric (A) and monomeric (B) PSMAECTO, and sera were tested at 1:50 dilution for reactivity with PSMA-3T3 cells by flow cytometry. Each panel represents a different animal. Sera from immunized animals (green) and a naïve animal (red) are depicted. Background levels of binding to parental 3T3 cells were observed for all sera (data not shown).
An identical pattern of reactivity was observed with human prostate cancer cell lines. Consistent, high-level reactivity with PSMA-expressing C4-2 cells was observed for sera from dimer-immunized animals (median MFI = 28.0, range 23.1–28.8) but not monomer-immunized (median MFI = 12.8, range 11.2–14.5) or control animals (median MFI = 12.3, range 8.6–16.0). Background levels of binding to PSMA-negative PC-3 cells were observed for all sera (median MFI <7 in all cases).
Thus, flow cytometry but not ELISA was able to reveal fundamental differences in the humoral immune responses elicited by monomeric and dimeric forms of PSMA. Chiefly, only antibodies elicited by dimeric PSMA cross-reacted with cell-surface PSMA. The inability of the ELISA to uncover such differences suggests that PSMAECTO adopts a partially denatured conformation upon adsorption to plastic.
Discussion
By virtue of its abundant and restricted surface expression on the neoplasms and neovasculature of prostate and other cancers, PSMA is an attractive target for active and passive immunotherapies of cancer. However, little is known about the molecular structure of this target. Here, we demonstrate that PSMA is natively expressed as a noncovalent homodimer and moreover that homodimerization is required for the enzymatic activity of PSMA. Although monomeric and dimeric PSMAECTO share certain antigenic epitopes, the dimer presents unique epitopes that can be discerned with a panel of mAbs. Remarkably, when used as immunogens, dimeric but not monomeric PSMAECTO was able to elicit antibodies that efficiently recognized PSMA-expressing tumor cells. These findings may have important implications for PSMA-targeted therapies of cancer and other diseases.
The homodimeric nature of PSMA was not predicted by prior experimental and theoretical analyses. One report described a minor, apparently covalently linked PSMA dimer (21). This minor dimeric species was observed on denaturing SDS/PAGE gels and thus is distinct from our native, noncovalent PSMA dimers, which represent the major species, dissociate in the presence of SDS, and are not detectable by SDS/PAGE.
Two reports have described topological models for PSMA (27, 28), but neither report proposes an oligomeric structure. The PSMA models are based on sequence homologies between its catalytic domain and that of zinc aminopeptidases from Vibrio proteolyticus and Streptomyces griseus. These enzymes belong to the M28 protease family that includes PSMA, and their x-ray crystal structures have been determined (29). Both the Vibrio and Streptomyces enzymes crystallize and are enzymatically active as monomers.
In addition, PSMA shares modest sequence and structural homology with human transferrin receptor (TfR), which contains a vestigial catalytic domain but lacks enzymatic activity. TfR is expressed as a type II membrane protein that forms a disulfide-linked homodimer, but the intermolecular disulfides are not required for dimerization (30). The high-resolution crystal structure of the TfR ectodomain reveals that the protein is organized into three distinct domains known as the protease-like, apical, and helical domains, with the last domain being principally responsible for dimerization (31). PSMA and TfR share 30%, 30%, and 24% sequence identity within these domains, respectively. Thus, despite their modest sequence identity, the helical dimerization domains of PSMA (residues 601–750) and TfR (residues 607–760) may have conserved function, and our experimental findings provide important guidance for future studies that probe the determinants of dimerization for this class of proteins.
The folate hydrolase and NAALADase activities of PSMA reside within its ectodomain and strictly depend on homodimerization. Both the enzymatic activity (32) and homodimerization (Fig. 2) of PSMA are disrupted by metal-chelating agents. Loss of the active site Zn2+ would abrogate enzymatic activity and may affect dimerization. However, additional metal-binding sites are contained within or contact the helical domain of TfR (31), and thus other metals (e.g., Ca2+) also may modulate the dimerization of PSMA. Intestinal PSMA is thought to hydrolyze γ-glutamyl linkages in pteroylpolyglutamate to facilitate absorption of folic acid, and brain PSMA is thought to hydrolyze N-acetyl-α-l-aspartyl-l-glutamate, the most abundant mammalian neuropeptide. The role of the enzymatic activities of PSMA in normal and diseased prostate remains unclear. The marked up-regulation of PSMA in malignant prostate cancer and its expression on tumor neovasculature raise the question as to whether the carboxypeptidase activity of the protein contributes to malignant transformation, metastasis, or angiogenesis. A PSMA active site inhibitor has demonstrated activity in an animal model of stroke (17), and our data suggest that similar activity may be observed for a class of agents that disrupts the dimer interface.
Our studies demonstrate that the PSMAECTO is sufficient for dimerization, and the similarities between PSMAECTO (amino acids 44–750) and PSM′ (amino acids 58–750) suggest that the latter protein is likely to dimerize as well. Consistent with this notion, Tiffany et al. (32) provide evidence that PSM′ possesses NAALADase activity, although with potentially altered active site kinetics. Although of moderate affinity in solution, the monomer–dimer equilibrium may be sufficient to maintain high-level dimerization in the more restricted, 2D space of the cell surface. In addition, the transmembrane and cytoplasmic regions may further contribute to stabilization of cell-surface dimer, as this issue is not addressed in our study.
There was a stark difference in the humoral immunity elicited by monomeric and dimeric forms of PSMAECTO. Whereas monomer and dimer generated similar overall levels of ELISA-reactive antibodies in mice, only the dimer efficiently elicited antibodies that recognized PSMA-expressing tumor cells, as would be required for effective antitumor immunity. The finding is surprising given that monomeric and cell-surface PSMAECTO share a number of antigenic epitopes. However, we are aware of the important differences between protein antigenicity and immunogenicity. For example, monomeric PSMAECTO may present one or more unique and immunodominant epitopes, or it may be more susceptible to limited proteolysis in vivo. The dichotomy observed for PSMAECTO monomer and dimer has important implications for PSMA-based vaccines that are intended to elicit humoral immunity.
We are not aware of another system with such sharply drawn differences between the functional immunogenicity of monomeric and oligomeric protein, but certain parallels can be drawn with the field of HIV-1 vaccine research. Monomeric forms of the viral envelope glycoproteins gp120 and gp41 elicit high-titer antibody responses, yet these antibodies do not recognize the native, fusogenic, trimeric form of these proteins as presented on the virion surface (reviewed in ref. 33). However, unlike our findings for PSMA, HIV-1 researchers have not yet succeeded in producing recombinant gp120/gp41 oligomers that reliably elicit antibodies to the native oligomer.
We also describe a mAb, designated 026, that specifically recognizes dimeric but not monomeric PSMAECTO. In addition, mAb 026 efficiently binds PSMA-expressing tumor cells, but not denatured PSMA, and thus defines an epitope unique to the quaternary structure of PSMA. Because of their exceptional selectivity, dimer-specific mAbs may be preferred for passive immunotherapy of PSMA-expressing tumors.
Several parallels can be drawn between PSMA and members of the epidermal growth factor receptor (EGFR, also known as HER and ErbB) superfamily of tyrosine kinases. Like PSMA, EGFRs are overexpressed in malignant disease, poor prognostic indicators, enzymatically active, and activated upon dimerization (reviewed in refs. 34 and 35). The anti-HER2 mAb Herceptin is indicated for therapy of breast cancers that overexpress this receptor. Interestingly, HER2 has no known ligands but rather seems to mediate cellular activation by serving as a dimerization partner for other members of the EGFR superfamily (36). The anti-HER1 mAb Erbitux is in advanced clinical development for colorectal cancer. Iressa is a small-molecule inhibitor of HER1 kinase activity and is approved for salvage therapy of nonsmall cell lung cancer. In addition, numerous other EGFR-targeted antibody, small-molecule, and vaccine therapies are in development (37), and such molecularly targeted agents are widely considered to be the vanguard of cancer therapy. Our findings on PSMA structure and function may foster the development of similar therapies based on this important tumor antigen.
In conclusion, we report that PSMA is expressed as a noncovalent homodimer. Dimerization is mediated by the extracellular domain of the molecule and is required for enzyme activity. Strikingly, PSMAECTO dimer but not monomer was capable of efficiently eliciting antibodies that recognize PSMA-expressing tumor cells, and a unique dimer-specific epitope was identified by using a mAb. Our findings provide insight into the structure and function of this important tumor antigen and thus may have significant implications for the development of effective immunotherapies for cancer.
Supplementary Material
Acknowledgments
We acknowledge the excellent technical assistance of Aimee Heyrman and thank M. Sadelain for the 3T3-PSMA cells and W. Goeckeler for critically reading the manuscript. PSMA Development Company, LLC, is a joint venture between Cytogen Corporation (Princeton) and Progenics Pharmaceuticals, Inc. The work was supported in part by National Institutes of Health Grants CA91746, CA92927, and CA96075 (to G.P.D.).
Abbreviations: PSMA, prostate-specific membrane antigen; PSMAECTO, extracellular domain of PSMA; NAALADase, N-acetylated α-linked acidic dipeptidase; BN, Blue Native; TfR, transferrin receptor; SEC, size-exclusion chromatography; DPP IV, dipeptidyl peptidase IV; MFI, mean fluorescence intensity.
W.A.H. is on the scientific advisory board of Progenics Pharmaceuticals, Inc.
References
- 1.Dhanasekaran, S. M., Barrette, T. R., Ghosh, D., Shah, R., Varambally, S., Kurachi, K., Pienta, K. J., Rubin, M. A. & Chinnaiyan, A. M. (2001) Nature 412 822–826. [DOI] [PubMed] [Google Scholar]
- 2.Welsh, J. B., Sapinoso, L. M., Su, A. I., Kern, S. G., Wang-Rodriguez, J., Moskaluk, C. A., Frierson, H. F., Jr., & Hampton, G. M. (2001) Cancer Res. 61, 5974–5978. [PubMed] [Google Scholar]
- 3.Magee, J. A., Araki, T., Patil, S., Ehrig, T., True, L., Humphrey, P. A., Catalona, W. J., Watson, M. A. & Milbrandt, J. (2001) Cancer Res. 61 5692–5696. [PubMed] [Google Scholar]
- 4.Luo, J., Duggan, D. J., Chen, Y., Sauvageot, J., Ewing, C. M., Bittner, M. L., Trent, J. M. & Isaacs, W. B. (2001) Cancer Res. 61 4683–4688. [PubMed] [Google Scholar]
- 5.Tasch, J., Gong, M., Sadelain, M. & Heston, W. D. (2001) Crit. Rev. Immunol. 21 249–261. [PubMed] [Google Scholar]
- 6.Horoszewicz, J. S., Kawinski, E. & Murphy, G. P. (1987) Anticancer Res. 7 927–935. [PubMed] [Google Scholar]
- 7.Freeman, L. M., Krynyckyi, B. R., Li, Y., Korupulu, G., Saleemi, K., Haseman, M. K. & Kahn, D. (2002) Q. J. Nucl. Med. 46 131–137. [PubMed] [Google Scholar]
- 8.Su, S. L., Huang, I. P., Fair, W. R., Powell, C. T. & Heston, W. D. (1995) Cancer Res. 55 1441–1443. [PubMed] [Google Scholar]
- 9.Israeli, R. S., Powell, C. T., Fair, W. R. & Heston, W. D. (1993) Cancer Res. 53 227–230. [PubMed] [Google Scholar]
- 10.Silver, D. A., Pellicer, I., Fair, W. R., Heston, W. D. & Cordon-Cardo, C. (1997) Clin. Cancer Res. 3 81–85. [PubMed] [Google Scholar]
- 11.Liu, H., Moy, P., Kim, S., Xia, Y., Rajasekaran, A., Navarro, V., Knudsen, B. & Bander, N. H. (1997) Cancer Res. 57 3629–3634. [PubMed] [Google Scholar]
- 12.Chang, S. S., O'Keefe, D. S., Bacich, D. J., Reuter, V. E., Heston, W. D. & Gaudin, P. B. (1999) Clin. Cancer Res. 5 2674–2681. [PubMed] [Google Scholar]
- 13.Chang, S. S., Reuter, V. E., Heston, W. D., Bander, N. H., Grauer, L. S. & Gaudin, P. B. (1999) Cancer Res. 59 3192–3198. [PubMed] [Google Scholar]
- 14.Chang, S. S., Reuter, V. E., Heston, W. D. & Gaudin, P. B. (2001) Urology 57 801–805. [DOI] [PubMed] [Google Scholar]
- 15.Pinto, J. T., Suffoletto, B. P., Berzin, T. M., Qiao, C. H., Lin, S., Tong, W. P., May, F., Mukherjee, B. & Heston, W. D. (1996) Clin. Cancer Res. 2 1445–1451. [PubMed] [Google Scholar]
- 16.Carter, R. E., Feldman, A. R. & Coyle, J. T. (1996) Proc. Natl. Acad. Sci. USA 93 749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slusher, B. S., Vornov, J. J., Thomas, A. G., Hurn, P. D., Harukuni, I., Bhardwaj, A., Traystman, R. J., Robinson, M. B., Britton, P., Lu, X. C., et al. (1999) Nat. Med. 5 1396–1402. [DOI] [PubMed] [Google Scholar]
- 18.Trkola, A., Dragic, T., Arthos, J., Binley, J. M., Olson, W. C., Allaway, G. P., Cheng-Mayer, C., Robinson, J., Maddon, P. J. & Moore, J. P. (1996) Nature 384 184–187. [DOI] [PubMed] [Google Scholar]
- 19.Mendez, M. J., Green, L. L., Corvalan, J. R., Jia, X. C., Maynard-Currie, C. E., Yang, X. D., Gallo, M. L., Louie, D. M., Lee, D. V., Erickson, K. L., et al. (1997) Nat. Genet. 15 146–156. [DOI] [PubMed] [Google Scholar]
- 20.Schülke, N., Vesanen, M., Sanders, R. W., Zhu, P., Anselma, D. J., Villa, A. R., Parren, P. W. H. I., Binley, J. M., Roux, K. H., Maddon, P. J., et al. (2002) J. Virol. 76 7760–7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Troyer, J. K., Beckett, M. L. & Wright, G. L., Jr. (1995) Int. J. Cancer 62 552–558. [DOI] [PubMed] [Google Scholar]
- 22.Israeli, R. S., Powell, C. T., Corr, J. G., Fair, W. R. & Heston, W. D. (1994) Cancer Res. 54 1807–1811. [PubMed] [Google Scholar]
- 23.Schägger, H. & von Jagow, G. (1991) Anal. Biochem. 199 223–231. [DOI] [PubMed] [Google Scholar]
- 24.Schägger, H., Cramer, W. A. & von Jagow, G. (1994) Anal. Biochem. 217 220–230. [DOI] [PubMed] [Google Scholar]
- 25.Pangalos, M. N., Neefs, J. M., Somers, M., Verhasselt, P., Bekkers, M., van der Helm, L., Fraiponts, E., Ashton, D. & Gordon, R. D. (1999) J. Biol. Chem. 274 8470–8483. [DOI] [PubMed] [Google Scholar]
- 26.Barinka, C., Rinnova, M., Sacha, P., Rojas, C., Majer, P., Slusher, B. S. & Konvalinka, J. (2002) J. Neurochem. 80 477–487. [DOI] [PubMed] [Google Scholar]
- 27.Rawlings, N. D. & Barrett, A. J. (1997) Biochim. Biophys. Acta 1339 247–252. [DOI] [PubMed] [Google Scholar]
- 28.Speno, H. S., Luthi-Carter, R., Macias, W. L., Valentine, S. L., Joshi, A. R. & Coyle, J. T. (1999) Mol. Pharmacol. 55 179–185. [DOI] [PubMed] [Google Scholar]
- 29.Chevrier, B., D'Orchymont, H., Schalk, C., Tarnus, C. & Moras, D. (1996) Eur. J. Biochem. 237 393–398. [DOI] [PubMed] [Google Scholar]
- 30.Alvarez, E., Girones, N. & Davis, R. J. (1989) EMBO J. 8 2231–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawrence, C. M., Ray, S., Babyonyshev, M., Galluser, R., Borhani, D. W. & Harrison, S. C. (1999) Science 286 779–782. [DOI] [PubMed] [Google Scholar]
- 32.Tiffany, C. W., Lapidus, R. G., Merion, A., Calvin, D. C. & Slusher, B. S. (1999) Prostate 39 28–35. [DOI] [PubMed] [Google Scholar]
- 33.Burton, D. R. & Moore, J. P. (1998) Nat. Med. 4 495–498. [DOI] [PubMed] [Google Scholar]
- 34.Schlessinger, J. (2000) Cell 103 211–225. [DOI] [PubMed] [Google Scholar]
- 35.Mendelsohn, J. (2002) J. Clin. Oncol. 20, 1S–13S. [PubMed] [Google Scholar]
- 36.Ranson, M. & Sliwkowski, M. X. (2002) Oncology 63 17–24. [DOI] [PubMed] [Google Scholar]
- 37.Sridhar, S. S., Seymour, L. & Shepherd, F. A. (2003) Lancet Oncol. 4 397–406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.