Abstract
A pseudotriloop is formed by transloop base pairing between the first (5′) and the fifth nucleotide in a hexanucleotide RNA loop (“hexaloop”) to subtend a triloop of nucleotides 2–4. This structure has been found in hairpins involved in the regulation of iron metabolism in mammalian cells and in transcription of plant virus subgenomic RNA. Several hexaloop hairpins, including HIV-transactivation-responsive element and hepatitis B virus ε, potentially adopt a pseudotriloop conformation. Here we show that an RNA plant virus whose replication depends on a conventional triloop hairpin can be used to verify the existence of pseudotriloop structures in vivo. Our data suggest that the pseudotriloop may represent a common motif in RNA–protein recognition.
RNA–protein interactions play a fundamental role in various cellular processes and viral infections. Unlike DNA-binding proteins, which recognize sequence-specific motifs, proteins that interact with RNA often recognize structural features such as hairpin loops, bulges, and internal loops. The number of solved RNA–protein complexes has grown dramatically in recent years and has provided insight into the mechanisms of RNA recognition (1, 2). However, RNA motifs inferred from these studies are usually restricted to a specific class of related proteins and are not widely applicable.
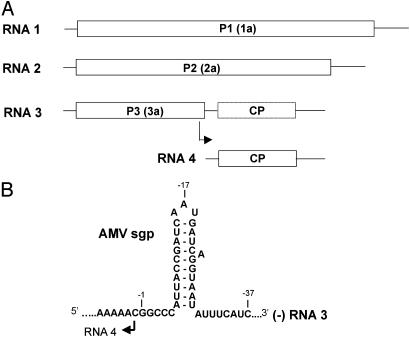
The family Bromoviridae comprises plant viruses with a tripartite RNA genome of messenger polarity (Fig. 1A). RNAs 1 and 2 encode the viral subunits of the RNA-dependent RNA polymerase (RdRp). The dicistronic RNA 3 encodes the movement protein (P3) and coat protein (CP). The latter is translated from a subgenomic (sg) mRNA 4. The best-studied members of this family are alfalfa mosaic virus (AMV) and brome mosaic virus (BMV) (3, 4).
Fig. 1.
(A) Schematic representation of the tripartite genome of viruses from the family Bromoviridae. RNA 1 encodes replicase subunit P1 with helicase and methyltransferase activities. RNA 2 encodes replicase subunit P2 with polymerase activity. RNA 3 encodes the P3 involved in cell-to-cell transport of the virus. RNA 4 is transcribed internally from the minus strand of RNA 3 and codes for the CP needed for cell-to-cell and long-distance transport of the virus. Identifiers for BMV proteins corresponding to AMV proteins P1, P2, and P3 are given in parentheses. (B) Structure of the minimal sgp region in AMV RNA 3 minus strands, as determined by in vitro mutational analysis (10). The marked nucleotides G-1, A-17, and C-37 serve as landmarks.
For BMV we found that in vitro sgRNA synthesis relies on the presence of a small hairpin with a CAUAGA loop, but that a conventional trinucleotide “triloop”-terminated hairpin could functionally replace this hexaloop-terminated hairpin structure (5). Using biochemical and biophysical techniques, we demonstrated that the CAUAGA loop adopts a triloop conformation by transloop base pairing of C1 and G5, creating a AUA triloop and bulging out A6. This conformation, which we have termed “pseudotriloop” (PTL), resembles the structure of iron-responsive elements (IREs) that regulate translation of mRNAs encoding proteins involved in iron uptake, storage, and utilization in mammalian cells. In the conserved loop motif CAGUGH (H = A, C, or U) of IREs, a similar transloop base pair between C1 and G5 is essential for recognition by iron-regulatory proteins (6–9). In contrast to the IRE loop, deletion of the bulge (A6) in the BMV loop was not detrimental for subgenomic promoter (sgp) activity. This finding suggested that the BMV RdRp is able to use both triloop and PTL hairpins for sg transcription (5).
We demonstrated (10) that AMV RdRp also recognizes a triloop hairpin to direct sgRNA synthesis in vitro (Fig. 1B). Given its relatedness to BMV, we anticipated that AMV RdRp would also accept PTL hairpins for sgRNA synthesis. To investigate this possibility, we have developed an in vivo replication system that critically depends on base pairing but not on sequence in the upper part of the AMV triloop hairpin and thus can be used to study the formation of PTL hairpins in heterologous RNAs. Several well known hexaloop hairpins were tested for their ability to replace the top 12 nucleotides of the viral triloop hairpin during a plant infection. The results indicate that hexaloops from various sources can adopt a PTL structure, suggesting that it may be a general motif in RNA–protein interactions.
Materials and Methods
Constructs. By means of PCR, EagI and NsiI restriction sites were introduced upstream and downstream of the hairpin, respectively, in a cDNA 3 plasmid that contains a premature stop codon after the codon encoding amino acid 255 of P3. The truncated P3 results in only slightly reduced movement relative to the full, 300-aa-residue movement protein (11). We also replaced the N-terminal amino acids of the CP ORF with the amino acids of a virulent strain that induces the formation of necrotic lesions on inoculated tobacco leaves (R.C.L.O., R. Miglino, and J.F.B., unpublished results). The desired mutations were obtained by cloning of complementary oligonucleotides into DNA 3 plasmids digested with EagI and NsiI. Synthesis of full-length RNA 3 transcripts with T7 RNA polymerase and inoculation of P12 tobacco plants, which expresses the AMV RdRp polypeptides, were performed as described (12).
Northern Blotting, RT-PCR, and Sequencing. Total RNA was extracted from Nicotiana tabacum P12 leaves 5 days after inoculation, separated by electrophoresis on a 1.5% agarose gel, and transferred to nylon membranes. Membranes were incubated with a 32P-labeled RNA probe that is complementary to RNA 3 and RNA 4 as described (13). Total RNA was used for reverse transcription with avian myeloblastosis virus reverse transcriptase and an oligonucleotide (36-BIO) that is complementary to nucleotides 1331–1355 of RNA 3. The resulting cDNA was amplified by PCR using 36-BIO and an oligonucleotide that is homologous to nucleotides 945–964 of RNA 3. PCR products were sequenced (BaseClear, Leiden, The Netherlands).
Results
Importance of Base Pairing in the AMV sgp Hairpin in Vivo. The AMV sgp is located on the minus strand of RNA 3 and constitutes an essential triloop hairpin (Fig. 1B). This hairpin partially overlaps with the P3 gene of plus-strand RNA 3; the 5′-A residue in the triloop corresponds to the U residue of the UGA stop codon. A truncated P3 (11) was engineered to permit a mutational analysis of sgp activity in plants without interfering significantly with the P3 function. RNA 3 transcripts with sgp mutations were inoculated onto transgenic tobacco plants (P12 plants) that express the AMV P1 and P2 replicase proteins. These plants support replication of RNA 3 in the absence of RNAs 1 and 2 (14). Because CP is required for cell-to-cell transport (15), accumulation of RNA 3 mutants in these plants depends on sgp activity. The infectivity of the mutants was monitored by the appearance of local lesions on inoculated leaves and by Northern blot hybridization.
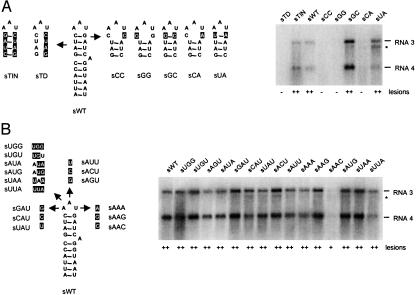
Fig. 2A shows the effects of mutations in the upper part of the sgp helix. Disruption of all four base pairs (sTD) abolished RNA 3 and 4 accumulation and lesion formation, whereas swapping these 4 base pairs (sTIN) led to wild-type levels of RNA accumulation and lesion formation. Previously, we concluded that the C·G base pair that closes the AAU triloop is critical for sgp activity in vitro (10). In vivo, disruption of this base pair was either detrimental to or strongly reduced RNA 4 synthesis (Fig. 2 A, sCC, sGG, and sCA). Restoring this base pair to G·C or U·A restored RNA 4 accumulation to wild-type level or higher and also led to high numbers of necrotic lesions on inoculated tobacco leaves (Fig. 2 A, sGC and sUA).
Fig. 2.
Mutational analysis of the AMV sgp hairpin in plants. (A Left) Mutations affecting base pairing in the upper half of the stem. The mutated nucleotides with respect to sWT are indicated in reversed print. For clarity, only the top part of the hairpin is shown for the mutants. (B Left) Mutation of the triloop nucleotides. (A and B Right) Northern blot analysis of AMV RNA 3 and 4 accumulation. * indicates RNA 3′, which is a degradation product of RNA 3 and is believed to lack the 5′-terminal 420 nucleotides of RNA 3 (25). Results of two independent lesion assays are summarized below each lane. In each assay, three different half-leaves of one plant were inoculated and monitored for 2 weeks. –, No necrotic lesions; +, 20–99; ++, 100 or more lesions developed per half-leaf.
Role of the Loop Nucleotides. A mutational analysis of the loop nucleotides of the sgp hairpin showed that mutation of A-16 into U or C and mutation of A-17 to C reduced sgp activity in vitro, whereas mutation of A-16 and A-17 to other nucleotides or mutation of U-18 to A, C, or G had little effect on sgp activity (10). Here, we analyzed the effect of these mutations on RNA 3 accumulation in P12 plants. In addition to these single-nucleotide mutants, a few loop mutants with double or triple mutations were analyzed (Fig. 2B). Local lesion numbers induced by mutant RNA 3 on P12 plants indicated that none of the loop mutations significantly affected infectivity of the virus. Northern blot analysis showed that the mutants accumulated RNAs 3 and 4 in wild-type ratios. Sequence analysis showed that the mutations were retained in the progenies of the mutants. Compared with the wild-type and other mutants, accumulation of RNAs 3 and 4 of loop mutant AAC was reduced (Fig. 2B), although this effect was not seen in vitro (10). Altogether, these data demonstrate that the secondary structure rather than the primary structure in the upper part of the hairpin is important for sgp activity in vivo.
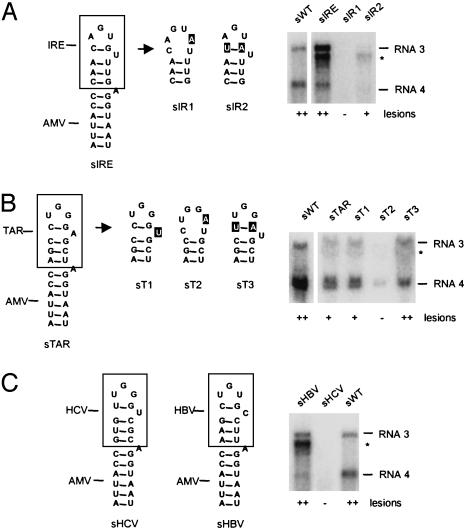
Chimeric sgp Hairpins Between AMV and IRE Are Active in Vivo. Given the low sequence requirements in the upper part of the AMV hairpin for sgp activity, we hypothesized that the system described here could be applied to investigate base pairing in heterologous sequences, e.g., those putatively forming a PTL hairpin. We started out by replacing the upper half of the sgp hairpin by the upper segment of a known PTL hairpin, an IRE from human ferritin mRNA (Fig. 3A). The resulting construct (sIRE) replicated in P12 plants as evidenced by the high number of necrotic lesions on inoculated leaves. Northern blots showed almost wild-type levels of RNA 3 and 4 accumulation (Fig. 3A). Analysis of the progeny RNA 3 by sequencing of RT-PCR products revealed that the IRE-derived sequence was maintained. The viability of the AMV-IRE chimera was strictly dependent on the presence of a base pair between N1 and N5 of the loop sequence. Conversion of the C1·G5 base pair to a C1–A5 mismatch, thereby creating a hexaloop structure, eliminated RNA accumulation and lesion formation (Fig. 3A, sIR1). The compensatory mutant U1·A5 was able to induce lesions again and replicated at a substantial level (Fig. 3A, sIR2). The lower accumulation of sIR2 as compared with sIRE is in accordance with a similarly lower binding capacity of the U1·A5 mutant of IRE to iron-regulatory protein 2 (16). These data indicate that the transloop C·G base pair in the IRE loop forms under in vivo circumstances and that the resulting PTL conformation is tolerated by the AMV RdRp.
Fig. 3.
Chimeras of AMV and proven or putative pseudotriloop hairpins. (A) Chimeric sgp hairpin between AMV and top part of an IRE of human ferritin (Fig. 4), and two mutants thereof. (B) Chimeric sgp hairpin of AMV and TAR loop from HIV-1 (Fig. 4) and three mutants thereof. (C) Chimeras of AMV with hairpin IIId of HCV and with ε sequence of hepatitis B virus (HBV)c (Fig. 4).
Chimeric Hairpins Between AMV and HIV Transactivation-Responsive Element (TAR) Show sgp Activity in Vivo. A well studied hexaloop structure in the HIV RNA is TAR, which is required for synthesis of viral transcripts from the integrated provirus (17). TAR interacts with the cyclin T1 (CycT1) subunit of the positive transcription elongation factor complex b and viral transcriptional activator protein (Tat) to activate elongation of RNA polymerase II transcription. The possibility of transloop base pairing in the TAR loop sequence CUGGGA has been proposed on the basis of enzymatic structure-probing data but could not be verified by NMR spectroscopy (18, 19). Recently, it was reported that CycT1–Tat binding to TAR in vitro depended on base pairing between C1 and G5 (20). This finding suggests that a PTL structure in the TAR loop is important for CycT1–Tat binding. Because our AMV system can be used to investigate PTL formation in vivo, we transplanted the top 12 nucleotides of HIV1-TAR onto the lower stem of the AMV sgp hairpin (Fig. 3B, sTAR). As judged by lesion formation and Northern blot analysis, sTAR was able to replicate in P12 plants, strongly suggesting the presence of a transloop base pair in the TAR loop. To test the importance of the C1·G5 base pair in sTAR without creating alternative tetraloop structures, we mutated A6 to a U, a natural variation in some lentiviruses. As expected, this mutation did not affect replication of this chimera (Fig. 3B, sT1). Disruption of the C·G base pair in the latter construct by a G-to-A change abolished lesion formation and reduced RNA 3 and 4 accumulation (Fig. 3B, sT2). Restoring the transloop base pair by the introduction of a compensatory C-to-U change also restored its ability to induce necrotic lesions and to support accumulation of RNAs 3 and 4 (Fig. 3B, sT3). These data support a PTL conformation of TAR.
Chimeras of AMV and Other PTL Hairpins. A rough survey of published hexaloop hairpins revealed that the PTL structure is putatively present in a variety of RNAs (Fig. 4). The hepatitis C virus (HCV) hairpin IIId has an unusual U–G transloop base pair, as determined by NMR spectroscopy (21, 22). Transplantation of the top of hairpin IIId onto the stem of the AMV hairpin did not yield an infectious virus (Fig. 3C, sHCV). The ε sequence of HBV folds into a large hairpin structure that serves both as encapsidation signal and as replication origin for synthesis of the first DNA strand (23). RNase protection assays revealed that in the CUGUGC hexaloop G3 is accessible to RNase T1 but that G5 is not (24), indicative of a putative C1·G5 transloop base pair. To verify this hypothesis we made an AMV-HBV chimeric hairpin (Fig. 3C, sHBV). This construct replicated to high levels in plants, as evidenced by local lesion formation and Northern blot analysis (Fig. 2C, sHBV). Note that the ratio between RNAs 3 and 4 is different than wild type (lane sWT). This effect was also visible with sIRE (Fig. 3A) and sUA (Fig. 2 A) and possibly suggests a correlation with loop stability that needs further investigation. Nonetheless, the in vivo data support the existence of a transloop base pair in the ε sequence.
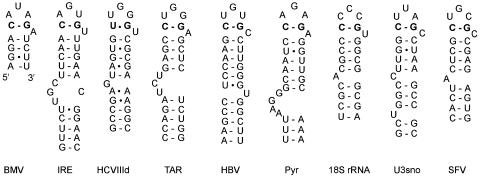
Fig. 4.
PTL structures formed by transloop C1·G5 or U1–G5 base pairing (in bold) in the hexanucleotide loops of several published RNA hairpins. For clarity, several hairpins are truncated at the bottom. BMV, brome mosaic virus sgp hairpin (5); IRE, iron-responsive element from human ferritin L-chain mRNA (26); HCVIIId, domain IIId of the internal ribosome entry site of HCV (21); TAR, transactivation-responsive element of HIV type 1 (17); HBV, encapsidation signal ε of hepatitis B virus (23); Pyr, PyrR-binding RNA hairpin BL2 from Bacillus subtilis (30); 18S rRNA, helix 11 of human 18S rRNA (31); U3sno, hairpin 4 of the U3 small nucleolar RNA from Saccharomyces cerevisiae (33); SFV, 5′-terminal hairpin of R–U5 of simian and human foamy virus (34).
Discussion
The in vivo data presented here corroborate our previous in vitro findings, which showed that AMV sgp activity depends on the presence of a 10-bp hairpin with a trinucleotide loop. In contrast to the in vitro data, we observed that mutations in the triloop sequence were less dramatic for sg transcription in vivo (10). This finding points to a difference in the specificity of template recognition by the native RdRp in vivo and the partially purified RdRp in vitro. The difference could be caused by protein factors or membrane structures that are present in native replication complexes but have been lost on purification of the enzyme. In vivo, sgp activity was found to be positively regulated by an enhancer element located between nucleotides –136 and –94, taking the start site for RNA 4 synthesis as +1 (25). This enhancer is absent in the core promoter fragments used in the in vitro studies and may contribute to sgp recognition in vivo.
In agreement with the in vitro data, we did find a major role of the top 4 base pairs in promoter activity in vivo. Disruption of these base pairs abolished infectivity of RNA 3 to P12 plants, whereas reversing them did not. Disruption of the C·G base pair that closes the triloop was by itself sufficient to abolish (Fig. 2 A, sCC and sGG) or reduce (sCA) infectivity. Replacement of this base pair by G·C or U·A base pairs resulted in almost wild-type levels of RNA accumulation. Thus, apart from having a structural function, the upper part of the sgp hairpin does not seem to harbor sequence-specific determinants for recognition by the AMV RdRp. What the exact sequence requirements are for sgp recognition remains to be determined. It is conceivable that these requirements turn out to be very minimal, because sgRNA synthesis is thought to take place in spherules where competition with other RNAs is almost absent (4).
The above-mentioned requirements for an active sgp allowed us to probe the formation of the transloop base pair in proven and putative PTL motifs. Chimeric RNAs between AMV and IRE, a proven PTL, were infectious to plants only when a transloop base pair at positions 1 and 5 of the hexanucleotide loop could form, indicating that the IRE PTL forms under in vivo circumstances. Recently, evidence for the structure of the ferritin IRE stem-loop in vivo was obtained by radical probing and protein footprinting in HeLa cells (26).
To support a PTL conformation in the TAR loop, chimeras between AMV and TAR were tested. It was observed that the sTAR series follows the same pattern of replication as the sIRE series. These results also show that the TAR loop shares several features with the IRE loop, as noted (16). A PTL conformation of the TAR loop is in fact also supported by phylogenetic comparison of HIV isolates (data not shown) and by data obtained from mutational analyses (27, 28). Final proof for the transloop base pair in TAR should come from in vivo transactivation assays or from HIV replication studies with the U1·A5 variant.
Finally, we tested chimeras between AMV and HCV or HBV. The AMV-HCV chimera was not infectious to plants. This lack of infectivity is possibly due to the U–G transloop base pair in the HCV loop that may be too weak and/or leads to alternative structures in the AMV system. A chimeric RNA between AMV and the ε sequence of HBV replicated efficiently in plants and therefore supports the existence of a transloop base pair in the ε sequence. Indeed, recent NMR data on ε show base pairing between C1 and G5 (29).
We have not tested the other proposed PTLs (Fig. 4), but data exist to support this conformation. The pyr hairpin is found in the pyrimidine nucleotide biosynthesis (pyr) operon of many eubacteria and is a substrate for regulatory protein PyrR. RNase protection assays have recently shown that the hexaloop “possesses some compact structure” and that mutation of C1 and G5, involved in the putative transloop base pair, was detrimental for PyrR binding (30). Structure probing of helix 11 of human 18S rRNA has shown that C1 and G5 are relatively inaccessible for single-strand-specific agents (31). The sequence variation of N2, N3, N4, and N6 in other 18S rRNAs is in agreement with a PTL structure (32). Structure-probing data on hairpin 4 of Saccharomyces cerevisiae U3 small nucleolar (sno) RNA are fully consistent with a PTL conformation, i.e., resistance of C1 and G5 but susceptibility of U2, U3, A4, and C6 to single-strand-specific agents (33). The SFV/HFV hairpin of simian and human foamy retroviruses is found at the 5′ terminus of the R-U5 region (34). Recent data suggest that this region has a function similar to that of the ε sequence of HBV (35).
A similar motif called the lone-pair triloop has recently been described (36). In lone-pair triloops the transloop base pair can consist of any non-Watson–Crick base pair and the bulge can be more than one nucleotide. So far, lone-pair triloops have been observed only in rRNA, and it is uncertain whether they are stable outside their natural context. To date, no experimental data exist to confirm their existence.
The structures shown in Fig. 4 are found in noncoding regions and, for those studied, perform essential regulatory functions in transcription, translation, or encapsidation. Because of their central function, PTL hairpins may be outstanding targets for drug design. We surmise that many more hexaloops with a CNNNGH consensus, and possibly also those with UNNNAH, GNNNCH, and ANNNUH, can adopt the PTL structure. Future experiments should point out whether the proposed PTLs are functionally relevant and whether proteins that interact with this motif share some common features.
Acknowledgments
We thank S. Barends, J. Beekwilder, J. van Duin, C. W. A. Pleij, M. H. de Smit, and S. H. E. van den Worm for editorial comments. This research was supported by The Netherlands Organization for Scientific Research, Earth and Life Sciences Research Council.
Abbreviations: PTL, pseudotriloop; sg, subgenomic; sgp, sg promoter; AMV, alfalfa mosaic virus; BMV, brome mosaic virus; IRE, iron-responsive element; RdRp, RNA-dependent RNA polymerase; CP, coat protein; TAR, transactivation-responsive element; HBV, hepatitis B virus; HCV, hepatitis C virus.
References
- 1.Cusack, S. (1999) Curr. Opin. Struct. Biol. 9 66–73. [DOI] [PubMed] [Google Scholar]
- 2.Frankel, A. D. (2000) Curr. Opin. Struct. Biol. 10 332–340. [DOI] [PubMed] [Google Scholar]
- 3.Bol, J. F. (1999) J. Gen. Virol. 80 1089–1102. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz, M. A., Chen, J., Janda, M., Sullivan, M., den Boon, J. & Ahlquist, P. (2002) Mol. Cell 9 505–514. [DOI] [PubMed] [Google Scholar]
- 5.Haasnoot, P. C. J., Olsthoorn, R. C. L. & Bol, J. F. (2002) RNA 8 110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sierzputowska-Gracz, H., McKenzie, R. A. & Theil, E. C. (1995) Nucleic Acids Res. 23 146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laing, L. G. & Hall, K. B. (1996) Biochemistry 35 13586–13596. [DOI] [PubMed] [Google Scholar]
- 8.Addess, K. J., Basilion, J. P., Klausner, R. D., Rouault, T. A. & Pardi, A. (1997) J. Mol. Biol. 274 72–83. [DOI] [PubMed] [Google Scholar]
- 9.Ke, Y., Sierzputowska-Gracz, H., Gdaniec, Z. & Theil, E. C. (2000) Biochemistry 39 6235–6242. [DOI] [PubMed] [Google Scholar]
- 10.Haasnoot, P. C. J., Brederode, F. T., Olsthoorn, R. C. L. & Bol, J. F. (2000) RNA 6 708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez-Navarro, J. A. & Bol, J. F. (2001) Mol. Plant–Microbe Interact. 14 1051–1062. [DOI] [PubMed] [Google Scholar]
- 12.Olsthoorn, R. C. L., Mertens, S., Brederode, F. T. & Bol, J. F. (1999) EMBO J. 18 4856–4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vlot, A. C., Menard, A. & Bol, J. F. (2002) J. Virol. 76 11321–11328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taschner, P. E., van der Kuyl, A. C., Neeleman, L. & Bol, J. F. (1991) Virology 181 445–450. [DOI] [PubMed] [Google Scholar]
- 15.van der Vossen, E. A. G., Neeleman, L. & Bol, J. F. (1994) Virology 202 891–903. [DOI] [PubMed] [Google Scholar]
- 16.Henderson, B. R., Menotti, E., Bonnard, C. & Kuhn, L. C. (1994) J. Biol. Chem. 269 17481–17489. [PubMed] [Google Scholar]
- 17.Berkhout, B. (1996) Prog. Nucleic Acids Res. Mol. Biol. 54 1–34. [DOI] [PubMed] [Google Scholar]
- 18.Colvin, R. A., White, S. W., Garcia-Blanco, M. A. & Hoffman, D. W. (1993) Biochemistry 32 1105–1112. [DOI] [PubMed] [Google Scholar]
- 19.Jaeger, J. A. & Tinoco, I. (1993) Biochemistry 32 12522–12530. [DOI] [PubMed] [Google Scholar]
- 20.Richter, S., Cao, H. & Rana, T. M. (2002) Biochemistry 41 6391–6397. [DOI] [PubMed] [Google Scholar]
- 21.Klinck, R., Westhof, E., Walker, S., Afshar, M., Collier, A. & Aboul-Ela, F. (2000) RNA 6 1423–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukavsky, P. J., Otto, G. A., Lancaster, A. M., Sarnow, P. & Puglisi, J. D. (2000) Nat. Struct. Biol. 7 1105–1110. [DOI] [PubMed] [Google Scholar]
- 23.Pollack, J. R. & Ganem, D. (1993) J. Virol. 67 3254–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knaus, T. & Nassal, M. (1993) Nucleic Acids Res. 21 3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Vossen, E. A. G., Notenboom, T. & Bol, J. F. (1995) Virology 212 663–672. [DOI] [PubMed] [Google Scholar]
- 26.Ke, Y. & Theil, E. C. (2002) J. Biol. Chem. 277 2373–2376. [DOI] [PubMed] [Google Scholar]
- 27.Feng, S. & Holland, E. C. (1988) Nature 334 165–167. [DOI] [PubMed] [Google Scholar]
- 28.Berkhout, B. & Jeang, K. T. (1989) J. Virol. 63 5501–5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flodell, S., Schleucher, J., Cromsigt, J., Ippel, H., Kidd-Ljunggren, K. & Wijmenga, S. (2002) Nucleic Acids Res. 30 4803–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonner, E. R., D'Elia, J. N., Billips, B. K. & Switzer, R. L. (2001) Nucleic Acids Res. 29 4851–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandiyan, V. & Boublik, M. (1990) Nucleic Acids Res. 18 7055–7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van de Peer, Y., De Rijk, P., Wuyts, J., Winkelmans, T. & De Wachter, R. (2000) Nucleic Acids Res. 28 175–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segault, V., Mougin, A., Gregoire, A., Banroques, J. & Branlant, C. (1992) Nucleic Acids Res. 20 3443–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park, J. & Mergia, A. (2000) Virology 274 203–212. [DOI] [PubMed] [Google Scholar]
- 35.Heinkelein, M., Leurs, C., Rammling, M., Peters, K., Hanenberg, H. & Rethwilm, A. (2002) J. Virol. 76 10069–10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, J. C., Cannone, J. J. & Gutell, R. R. (2003) J. Mol. Biol. 325 65–83. [DOI] [PubMed] [Google Scholar]