Abstract
The structural analysis of an enzymatic reaction intermediate affords a unique opportunity to study a catalytic mechanism in extraordinary detail. Here we present the structure of a tetrahedral intermediate in the catalytic cycle of aspartate-β-semialdehyde dehydrogenase (ASADH) from Haemophilus influenzae at 2.0-Å resolution. ASADH is not found in humans, yet its catalytic activity is required for the biosynthesis of essential amino acids in plants and microorganisms. Diaminopimelic acid, also formed by this enzymatic pathway, is an integral component of bacterial cell walls, thus making ASADH an attractive target for the development of new antibiotics. This enzyme is able to capture the substrates aspartate-β-semialdehyde and phosphate as an active complex that does not complete the catalytic cycle in the absence of NADP. A distinctive binding pocket in which the hemithioacetal oxygen of the bound substrate is stabilized by interaction with a backbone amide group dictates the R stereochemistry of the tetrahedral intermediate. This pocket, reminiscent of the oxyanion hole found in serine proteases, is completed through hydrogen bonding to the bound phosphate substrate.
The activation of the β-carboxyl group of l-aspartic acid is the commitment step to the aspartate biosynthetic pathway (1, 2). l-aspartate-β-semialdehyde (ASA) dehydrogenase (ASADH, EC 1.2.1.11) catalyzes the subsequent reductive dephosphorylation of β-aspartylphosphate (βAP) to ASA, leading to the first branch point in this pathway. At this point, ASA is either converted to homoserine, a common intermediate in the biosynthesis of threonine, isoleucine, and methionine, or is condensed with pyruvate leading to the production of diaminopimelic acid (DAP). This metabolite is a cross-linking component in the peptidoglycan layer of cell walls in Gram-negative bacteria and serves as the direct precursor of lysine (3). The aspartate biosynthetic pathway is not found in humans or other eukaryotes, making these amino acids essential nutrients and implicating ASADH as a potential target for the development of new antimicrobial compounds. Perturbations to the asd gene have been shown to be lethal to the microorganism, and bacterial strains with this gene deletion are auxotrophic for DAP (4). Because of the importance of this enzyme in amino acid biosynthesis, there is an ongoing interest in the development of effective microbial ASADH inhibitors (5–7).
The structures of ASADHs from Escherichia coli (8, 9) and from Vibrio cholerae (10) were recently solved, both as the apoenzyme and as a ternary complex with NADP and a covalently bound active site inhibitor, S-methyl-l-cysteine sulfoxide (SMCS). Each of these structures has further clarified the role of several active site amino acids identified earlier by site-directed mutagenesis (11). Here we examine ASADH from Haemophilus influenzae (hiASADH), a small Gram-negative bacterium whose only natural host is human and that is further distinguished as the first free-living organism to have its complete genome sequenced (12). The focus of these studies is to examine enzyme-complex structures in the catalytic cycle of ASADH, to gain detailed insight into the catalytic mechanism. This knowledge will aid in the design of more potent and selective inhibitors of this key metabolic enzyme. To provide molecular details of both the active and phosphate-binding sites, we have solved three structures of hiASADH: the apoenzyme, the enzyme treated with ASA, and the enzyme with ASA plus phosphate. Structure refinement of the complexes reveals not the expected stable ground-state complexes but the capture of a covalently bound tetrahedral reaction intermediate derived from ASA during the catalytic cycle of hiASADH.
Methods
Crystallization of the Apoenzyme. Recombinant H. influenzae (TIGR locus HI0646, Swiss-Pro P44801) ASADH was purified as described (13), concentrated by ultrafiltration (Millipore) to 15 mg/ml, and dialyzed against 10 mM Hepes/1 mM EDTA/1 mM DTT at pH 7.0. Initial crystallization conditions were obtained by hanging-drop vapor diffusion by using the polyethylene glycol (PEG)/Ion Screen (Hampton Research, Riverside, CA). After further optimization, high-quality crystals of the apoenzyme were grown at 20°C in 1:1 mixtures of enzyme and precipitant solution (24–28% PEG 3350/0.2 M ammonium acetate/0.1 M Tris, pH 8.5). Crystals were produced overnight and subsequently frozen for data collection. Harvesting solutions for freezing the crystals were 24–28% PEG 3350, 0.2 M ammonium acetate, and 20% glycerol in 0.1 M Tris, pH 8.5.
Formation of Enzyme–Substrate Complexes. Crystals of native apoenzyme were complexed by soaking with ASA. A suitable crystal was introduced into artificial mother liquor (26% PEG 3350/0.2 M ammonium acetate/0.1 M Tris, pH 8.5). The substrate ASA was then added to the mother liquor from a stock concentration of 100 mM to a final concentration of 2 mM. In the case where phosphate was introduced, potassium phosphate was added to a final concentration of 50 mM in addition to the ASA. The crystals were allowed to soak in this solution for 1 h before harvesting. A harvesting cryosolution was prepared (26% PEG 3350/0.2 M ammonium acetate/2 mM ASA/0.1 M Tris, pH 8.5, with 20% glycerol and either with or without 50 mM phosphate as appropriate), and this solution was introduced stepwise over ≈1 h to prevent damage to the crystals.
Data Collection and Processing. A data set was collected from a single frozen crystal of apoenzyme on a Quantum IV imaging plate (detector distance 175 mm; 1° oscillation per image) at the BioCARS beamline (14-D) at Argonne National Laboratory (APS). For the ASADH crystals soaked with ASA, a complete data set was collected up to 2.0 Å at the same beamline (detector distance 150 mm; 1° oscillation per image). Data on the ASADH crystals soaked with either ASA or ASA and phosphate were collected on a home source with a Rigaku (Tokyo) generator and an R-AXIS IV imaging plate (detector distance 160 mm; 1° oscillation per image). The images were processed and scaled by using the program hkl2000 (14). Data collection statistics for each of these data sets are summarized in Table 1.
Table 1. X-ray data collection and structure refinement statistics.
| Parameter | ASADH | ASADH-ASA | ASADH-ASA-Pi |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P212121 | P21212 |
| Cell dimensions, Å | 53.8 × 109.9 × 113.2 | 54.6 × 111.0 × 113.7 | 54.9 × 57.7 × 113.8 |
| Resolution, Å | 2.04 | 2.00 | 2.15 |
| I/σ(I) | 29.2 (8.3) | 19.8 (5.1) | 27.8 (5.6) |
| Total reflections | 279,972 | 150,757 | 143,627 |
| Unique reflections | 42,970 | 39,040 | 19,724 |
| Completeness, % | 99.4 (98.0) | 83.9 (76.7) | 99.9 (100) |
| Rsym, % | 5.5 (19.2) | 6.2 (29.4) | 6.8 (35.3) |
| Refinement | |||
| Number of atoms | |||
| Protein | 5,497 | 5,497 | 5,497 |
| Waters | 371 | 335 | 143 |
| Ligands | 0 | 14 | 24 |
| rms deviation | |||
| Bonds, Å | 0.006 | 0.012 | 0.012 |
| Angles, ° | 1.4 | 1.7 | 1.6 |
| Dihedral angles, ° | 22.7 | 24.1 | 23.8 |
| Rcryst, %* | 19.6 | 20.2 | 22.8 |
| Rfree, %† | 25.7 | 26.2 | 28.4 |
| Average B | |||
| Protein, Å2 | 19.8 | 20.3 | 34.4 |
| Ligands, Å2 | — | 21.7 | 35.9 |
Numbers in parentheses are related to the highest-resolution shell.
Rcryst = 100(ΣhklΣi|Ihkl,i — 〈Ihkl〉|/Σhkl,i/ Ihkl,i), where Ihkl,i is the intensity of an individual measurement of the reflection with Miller indices h, k, and I; and 〈Ihkl〉 is the mean intensity of that reflection.
Rfree = R for a randomly selected 5% of the data excluded from the refinement.
Structure Solution and Refinement. Data sets of the apoenzyme and the ASADH–hemithioacetal intermediate belong to a P212121 space group and give a dimer in the asymmetric unit. The ASADH–hemithioacetal intermediate with phosphate was indexed and processed in a P21212 space group, giving a monomer per asymmetric unit (Table 1). The H. influenzae ASADH–hemithioacetal structure was solved by molecular replacement by using a monomer of E. coli apo-ASADH (PDB 1BRM, chain A) as the search model. The H. influenzae apoenzyme and the ternary ASADH–hemithioacetal plus phosphate structures were subsequently solved by molecular replacement by using the H. influenzae ASADH–hemithioacetal structure as the search model. The cns package was used for the rotation and translation searches and structural refinement (15). Optimal solutions for the rotation matrix corresponding to two subunits of ASADH per asymmetric unit were obtained for the apoenzyme and the ASADH–hemithioacetal structure. The first translation search clearly identified a solution well above the next highest peak. The second translation search, performed after crystallographic symmetry had been applied to the coordinates of the first subunit, yielded a solution corresponding to the position of the second subunit. The model containing both subunits was subjected to rigid body refinement considering the two subunits as independent rigid structures. Rigid body refinement of the data to moderate resolution, followed by simulated annealing of the data extended to 2.0 Å, improved the R factor from 49% to 25%. The nonconserved amino acids in the search model were then replaced on the basis of the sequence of H. influenzae ASADH. Model building was carried out by using xtalview (16), with several iterations of refinement and model building leading to improvement in the final structure. The same method was used to determine the final structure of the ASADH–hemithioacetal structure with phosphate, with the minor modification of having only one subunit in the asymmetric unit. The final refinement statistics for the structures are listed in Table 1.
Results and Discussion
Strategy of Complex Formation. Although ASA and phosphate are the products of the ASADH-catalyzed reaction, they can also function as substrates in the nonphysiological direction due to the freely reversible nature of this reaction. Because the substrate in the biological direction, β-aspartyl phosphate, is unstable in aqueous solution, the reaction is monitored by the rate of NADPH formation after combining enzyme, ASA, and inorganic phosphate with NADP. Kinetics based on this assay have shown that the reaction has a preferred but not obligatory binding order in which ASA binds more efficiently when phosphate is already present (17). Thus ASADH with ASA and phosphate bound in the active site pocket represents an active form of the enzyme that is poised for catalytic turnover on addition of NADP.
There is no evidence for significant conformation changes in response to the binding of ASA or phosphate. In contrast, major conformational changes to the overall structure occur when the enzyme cofactor NADP binds (10). Consequently, apoenzyme hiASADH crystals are grown overnight and then soaked with either ASA or ASA and phosphate in the cryogenic mother liquor to isolate reaction intermediates that are unable to produce β-aspartyl phosphate until NADP is introduced. In most cases, the crystals remain intact during the soaks and retain their low degree of mosaicity, with typical values from 0.4° to 0.6°.
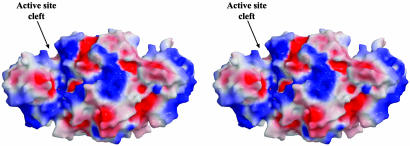
Overall Enzyme Conformation. These structures of the H. influenzae enzyme confirm that ASADH is a functional homodimer with one active site per monomer. The overall fold and tertiary structure of hiASADH is essentially the same as the ASADHs from each microorganism examined to date (8–10). The C-terminal domain is involved primarily in hydrophobic intersubunit contacts, whereas the more hydrophilic N-terminal domain forms the active site and the NADP-binding pocket. These sites are located in a large cleft in the surface of the enzyme, with the active site clefts in each subunit found on opposite sides of the ASADH dimer (Fig. 1). In the absence of NADP, these hiASADH structures are found in an open conformation, with a disordered loop (residues 39–54) that is normally involved in cofactor binding. The loop was not modeled in these structures due to lack of convincing electron density, and the failure of this loop to adopt a distinct well ordered conformation in the absence of NADP is indicative of the flexibility required for cofactor binding. Binding of NADP causes the cavity to rearrange from an open to a closed conformation, as evidenced by the movement in a hinge region of the enzyme and the repositioning of two surface loops on the cleft of the cofactor binding cavity (9, 10).
Fig. 1.
Stereoview of a surface representation colored according to electrostatic potential, such that regions with potential <–10 kBT are red, and those >+10 kBT are blue (kB, Boltzmann constant; T, absolute temperature). The active sites are located in broad clefts, with the cleft on the front of the dimer annotated.
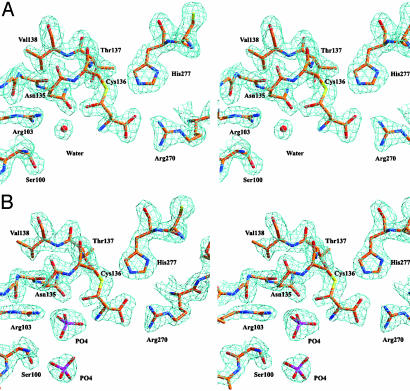
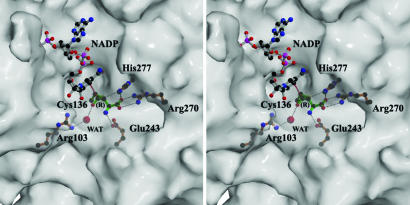
Comparison of Substrate and Inhibitor Binding. The hiASADH structures display well defined electron density for ASA (Fig. 2A) and for ASA and phosphate (Fig. 2B), thereby allowing comparison between these structures and that of a previously solved ASADH structure with a bound covalent active site inhibitor. That V. cholerae ASADH structure was cocrystallized as an ASADH/NADP/SMCS ternary complex, with a subsequent enzyme-mediated modification of the SMCS inhibitor, resulting in a cysteine moiety attached to the active site thiol in a disulfide linkage (Fig. 3A) (10). The amino and carboxyl group interactions of ASA with key active site residues in the H. influenzae structure are virtually identical to those observed in the V. cholerae ASADH–inhibitor complex. The ASA carboxyl group interacts in a bidentate manner with Arg-270, and the ASA amino group is similarly stabilized by hydrogen bonding to Glu-243. However, the ASA has now reacted to form a hemithioacetal on binding to the active site (Fig. 3B), rather than the disulfide linkage that was formed in the V. cholerae ASADH–inhibitor complex. The major structural differences between the covalently bound inhibitor and the binding of the natural substrate ASA are the hydrogen bonding with the backbone amide hydrogen of Asn-135 and the additional interactions that are observed with bound phosphate (Fig. 4). These interactions cause a shift in the position of the carbonyl carbon of bound ASA by >1.5 Å relative to the corresponding sulfur atom of bound SMCS. This shift occurs despite the nearly identical positions of their α-amino and α-carboxyl groups and may represent an essential difference between a catalytically active and catalytically inactive structure.
Fig. 2.
(A) Stereoview of the (2Fo–Fc) electron density map, contoured at 1.3σ, of the active site of hiASADH with the hemithioacetal derived from ASA covalently bound to active site Cys-136. (B) Stereoview of the (2Fo–Fc) electron density map, contoured at 1.2σ, of the active site of the hiASADH with the covalent hemithioacetal and two molecules of phosphate.
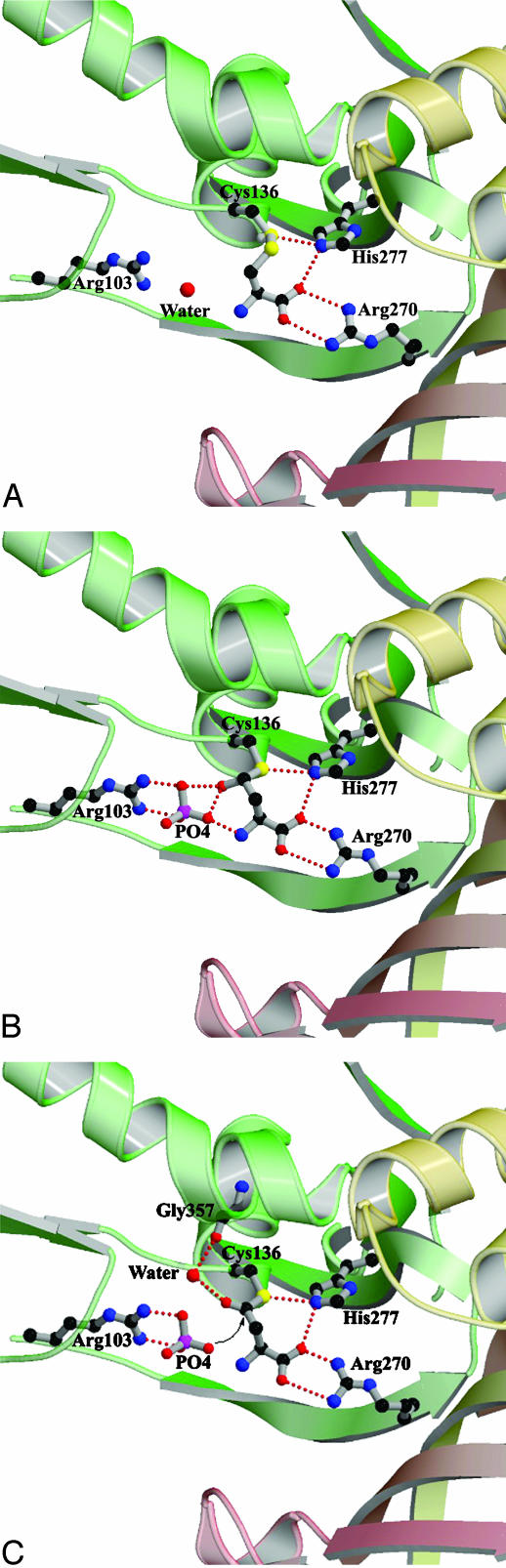
Fig. 3.
A comparison of the interactions and orientations between key active site residues and the intermediate complexes. (A) Structure of the ASADH–SMCS complex. (B) Structure of the ASADH–hemithioacetal complex. (C) Model of the acyl-enzyme intermediate, showing the likely repositioning of the oxygen after oxidation by NADP and the proposed attack by phosphate.
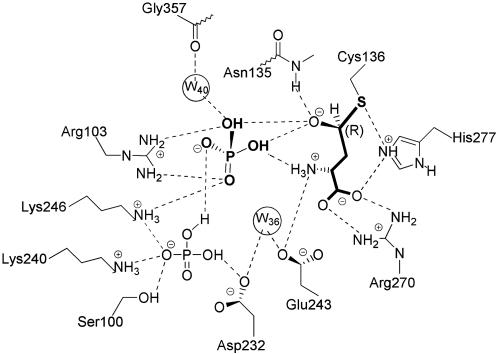
Fig. 4.
Schematic representation of the ASADH active site with a bound hemithioacetal reaction intermediate. Interactions between ASA and phosphate and key active site residues are shown.
The Phosphate-Binding Site. The bound phosphate engages in a bidentate interaction with Arg-103, and its position is further defined by an electrostatic interaction with Lys-246. Interestingly, the phosphate oxyanion is positioned through these interactions to engage with both the hemithioacetal oxygen and the amino group of ASA, thereby placing it optimally for subsequent nucleophilic attack. The bound inorganic phosphate substrate thus associates more intimately with ASA than previously suspected, and the interactions between these substrates provide structural support for the synergistic binding previously observed kinetically (17).
In the absence of phosphate, a water molecule occupies this binding pocket, and the ASA carbonyl oxygen can be stabilized by interaction with this water. A bound water molecule can also function catalytically as a phosphate surrogate, as evidenced by the slow conversion of NADP to NADPH in the presence of ASA (J.B. and R.E.V., unpublished observations). This conversion is presumably due to the ability of water to attack the activated carbonyl group of the thioester intermediate that forms after NADP reduction.
Unexpectedly, a second phosphate oxyanion was observed bound in the structure of the ternary complex (Fig. 2B). Although this second phosphate has no direct contact with ASA, it is within hydrogen-bonding distance to the first phosphate and to Lys-246, while engaging in additional interactions with Lys-242 and Ser-100 (Fig. 4). The distance between Lys-242 and Ser-100 is 8 Å in both the apoenzyme and the ASADH/ASA structures. However, this distance decreases to 4 Å when phosphate is bound to the enzyme, demonstrating that these residues move in response to the second phosphate. It seems likely that this second bound phosphate molecule is an artifact of the harvesting conditions, in which native hiASADH crystals were soaked with 50 mM phosphate, a concentration that is ≈30 times the Km value for this substrate. Even under these conditions, this second phosphate-binding site displays a lower occupancy, indicated by a weaker electron density (Fig. 2B). This phosphate also has a higher temperature factor (B = 66 Å2) than the substrate phosphate (B = 33 Å2). There are no kinetic data to support either an inhibitory or an activator role for a second phosphate, because the reaction velocity is unchanged even at phosphate concentrations up to 50 mM (13).
The Tetrahedral Intermediate Structure. The observed electron density at the active site of hiASADH is clearly that of a covalently bound intermediate (Fig. 2 A) and not that of an enzyme–ASA complex. The best fit to the electron density is obtained by assuming a tetrahedral geometry with R configuration at the original aldehydic carbon, thus raising the question of how this reactive intermediate is stabilized. We propose that hydride transfer from this intermediate structure to NADP will lead to the formation of a relatively stable acyl-enzyme intermediate (18). However, conversion of this tetrahedral intermediate to a trigonal planar species by this oxidative route is not possible without the presence of the physiological electron acceptor, NADP. This tetrahedral intermediate is stabilized by hydrogen bonding with the side chain functional groups of Glu-243, His-277, and Arg-270 (Fig. 5). The developing negative charge on the hemithioacetal oxygen is stabilized by the backbone amide group of Asn-135 in an interaction reminiscent of the stabilization of the tetrahedral intermediates in the mechanism of serine proteases (19). However, instead of using only backbone amide groups for stabilization, ASADH incorporates direct hydrogen bonding with the bound substrate phosphate to complete the oxyanion hole. The additional stabilization of the hemithioacetal oxygen afforded by these interactions will lower the energy barrier to formation of the covalent acyl-enzyme intermediate.
Fig. 5.
Superimposition of NADP from the V. cholerae ASADH/NADP complex onto the hiASADH/ASA active site, showing the orientation of NADP relative to the tetrahedral intermediate. The substrate oxygen is pointed away from NADP in a position that would facilitate removal of the hydrogen, with the proposed trajectory for hydride transfer shown (· · ·).
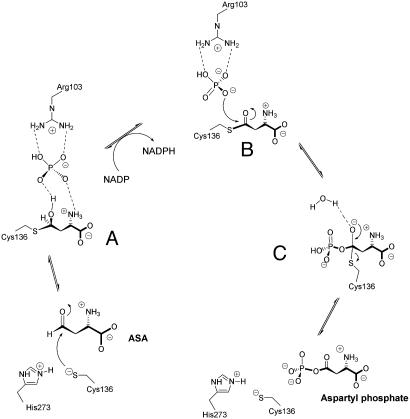
Catalytic Mechanism. The ASADH structures examined to date (8–10) support the roles for those active site amino acids previously implicated in catalysis (11, 20). The active site cysteine, histidine, and glutamine in ASADH are proposed to serve the same function as the catalytic triad found in cysteine proteases (21). The proposed role for His-277 as the acid/base catalyst that facilitates deprotonation of active site Cys-136 is validated by the current structures. Nucleophilic attack by Cys-136 to the re face of ASA results in the formation of a covalently bound hemithioacetal (Fig. 6A). The Nε atom of His-277 remains within hydrogen-bonding distance of both the active site sulfur atom and the substrate carboxyl group. The aldehydic carbon of ASA must form a tetrahedral stereocenter with R configuration in this reaction intermediate to be positioned correctly relative to NADP for hydride transfer. A similar stereochemical arrangement is observed in the intermediate that is formed after attack of the active site thiol of papain (22). This positioning of the ASADH reaction intermediate is demonstrated graphically by an overlay of the bound NADP from the V. cholerae ASADH/NADP structure onto the active site of the hiASADH structure (Fig. 5). This model suggests that the hemithioacetal oxygen points away from NADP, thereby providing the correct orientation of the hydrogen for subsequent hydride transfer. We postulate that hydride transfer triggers the formation of a trigonal planar thioester intermediate (Fig. 6B). The phosphate anion, previously engaged in hydrogen bonding to the hemithioacetal oxygen, now encounters electrostatic repulsion against the newly formed carbonyl oxygen. A model of the acyl-enzyme intermediate in which the thioester is a trigonal planar species shows that the carbonyl oxygen would tilt away from the phosphate oxygens, thus making the carbonyl carbon more susceptible to nucleophilic attack by phosphate (Fig. 3C). The enhanced electrophilicity of the thioester carbonyl carbon relative to its tetrahedral precursor facilitates nucleophilic attack by phosphate at this position. This thioester model now places the carbonyl oxygen within hydrogen-bonding distance to water 40, which is stabilized by the backbone carbonyl of Gly-357. This interaction is expected to further activate the carbonyl group.
Fig. 6.
Proposed mechanism for the catalytic cycle of ASADH shown in reverse biological direction. (A) Tetrahedral intermediate derived from reaction with ASA. (B) Proposed acyl intermediate with trigonal planar geometry. (C) Second tetrahedral intermediate with covalently bound aspartyl phosphate.
It is unlikely that His-277 is involved in proton transfers to the hemithioacetal oxygen as was previously postulated (9). The imidazole ring of His-277 is 5.7 Å away from this oxygen, and these residues are pointed directly away from each other. Such a proton transfer from His-277 would also require the disruption of the well established interaction between the Nε and the substrate carboxyl group. Similar interactions are seen between the active site histidine in papain and the carboxyl group of a covalently bound inactivator (22). Formation of NADPH presumably triggers an opening of the binding cleft, allowing the reduced cofactor to diffuse out of the binding cavity. Nucleophilic attack by phosphate at the carbonyl carbon leads to the second tetrahedral intermediate (Fig. 6C), which collapses to release β-aspartyl phosphate.
Conclusion
Three structures of the potential antimicrobial target hiASADH have been solved to high resolution. The structure with ASA and phosphate bound represents a true reaction intermediate in the catalytic cycle of ASADH and yields deeper insight into the stabilization of this intermediate and the catalytic mechanism of this enzyme. The precise position that phosphate occupies had been a longstanding uncertainty that is now resolved. The improved clarity of the active site presented here provides a structural framework on which to develop more effective ASADH inhibitors that could lead to a new generation of antimicrobial compounds.
Acknowledgments
We thank Drs. Tim Mueser and Venkataraman Kabaleeswaran for helpful discussions on the analysis and refinement of the enzyme structures and the staff members at BioCARS for their assistance with data collection. This work was supported by a grant from the National Science Foundation. The use of the Argonne National Laboratory at the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Energy Research, under Contract No. W-31-109-ENG-38.
Abbreviations: ASA, l-aspartate-β-semialdehyde; ASADH, ASA dehydrogenase; SMCS, S-methyl cysteine sulfoxide; hiASADH, Haemophilus influenzae ASADH; PEG, polyethylene glycol.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org [PDB ID codes 1NWC (apoenzyme), 1NWH (ASADH-hemithioacetal structure), and 1NX6 (ASADH-hemithioacetal structure with phosphate)].
References
- 1.Cohen, G. N. (1983) in Amino Acids: Biosynthesis and Genetic Regulation, eds. Herrmann, K. M. & Somerville, R. L. (Addison–Wesley, Reading, MA), pp. 147–171.
- 2.Viola, R. E. (2001) Acc. Chem. Res. 34 339–349. [DOI] [PubMed] [Google Scholar]
- 3.Patte, J. C. (1983) in Amino Acids: Biosynthesis and Genetic Regulation, eds. Herrmann, K. M. & Somerville, R. L. (Addison–Wesley, Reading, MA), pp. 213–228.
- 4.Harb, O. S. & Kwaik, Y. A. (1998) Infect. Immun. 66 1898–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox, R. J., Hadfield, A. T. & Mayo-Martin, M. B. (2001) Chem. Commun. 18 1710–1711. [DOI] [PubMed] [Google Scholar]
- 6.Cox, R. J., Sutherland, A. & Vederas, J. C. (2000) Bioorg. Med. Chem. 8 843–871. [DOI] [PubMed] [Google Scholar]
- 7.Han, S., Moore, R. A. & Viola, R. E. (2003) SYNLETT 845–846.
- 8.Hadfield, A. T., Kryger, G., Ouyang, J., Petsko, G. A., Ringe, D. & Viola, R. E. (1999) J. Mol. Biol. 289 991–1002. [DOI] [PubMed] [Google Scholar]
- 9.Hadfield, A. T., Shammas, C., Kryger, G., Ringe, D., Petsko, G. A., Ouyang, J. & Viola, R. E. (2001) Biochemistry 40 14475–14483. [DOI] [PubMed] [Google Scholar]
- 10.Blanco, J., Moore, R. A., Kalabeeswaran, V. & Viola, R. E. (2003) Protein Sci. 12 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouyang, J. & Viola, R. E. (1995) Biochemistry 34 6394–6399. [DOI] [PubMed] [Google Scholar]
- 12.Fleischmann, R. D., Adams, M. D., White, O., Clayton, R. A., Kirkness, E. F., Kerlavage, A. R., Bult, C. J., Tomb, J. F., Dougherty, B. A., Merrick, J. M., et al. (1995) Science 269 496–512. [DOI] [PubMed] [Google Scholar]
- 13.Moore, R. A., Bocik, W. E. & Viola, R. E. (2002) Protein Expression Purif. 25 189–194. [DOI] [PubMed] [Google Scholar]
- 14.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- 15.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54 905–921. [DOI] [PubMed] [Google Scholar]
- 16.McRee, D. E. (1999) J. Struct. Biol. 125 156–165. [DOI] [PubMed] [Google Scholar]
- 17.Karsten, W. E. & Viola, R. E. (1991) Biochim. Biophys. Acta 1077 209–219. [DOI] [PubMed] [Google Scholar]
- 18.Biellmann, J. F., Eid, P., Hirth, C. & Jornvall, H. (1980) Eur. J. Biochem. 104 53–58. [DOI] [PubMed] [Google Scholar]
- 19.Wilmouth, R. C., Edman, K., Neutze, R., Wright, P. A., Clifton, I. J., Schneider, T. R., Schofield, C. J. & Hajdu, J. (2001) Nat. Struct. Biol. 8 689–694. [DOI] [PubMed] [Google Scholar]
- 20.Karsten, W. E. & Viola, R. E. (1992) Biochim. Biophys. Acta 1121 234–238. [DOI] [PubMed] [Google Scholar]
- 21.Storer, A. C. & Ménard, R. (1994) Methods Enzymol. 244 486–500. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto, D., Matsumoto, K., Ohishi, H., Ishida, T., Inoue, M., Kitamura, K. & Mizuno, H. (1991) J. Biol. Chem. 266 14771–14777. [DOI] [PubMed] [Google Scholar]