Abstract
Covalent posttranslational protein modifications by eukaryotic transglutaminases proceed by a kinetic pathway of acylation and deacylation. Ammonia is released as the acylenzyme is formed, whereas the cross-linked product is released later in the deacylation step. Superposition of the active sites of transglutaminase type 2 (TG2) and the structurally related cysteine protease, papain, indicates that in the formation of tetrahedral intermediates, the backbone nitrogen of the catalytic Cys-277 and the Nε1 nitrogen of Trp-241 of TG2 could contribute to transition-state stabilization. The importance of this Trp-241 side chain was demonstrated by examining the kinetics of dansylcadaverine incorporation into a model peptide. Although substitution of the Trp-241 side chain with Ala or Gly had only a small effect on the Michaelis constant Km (1.5-fold increase), it caused a >300-fold lowering of the catalytic rate constant kcat. The wild-type and mutant TG2-catalyzed release of ammonia showed kinetics similar to the kinetics for the formation of cross-linked product, indicating that transition-state stabilization in the acylation step was rate-limiting. In papain, a Gln residue is at the position of TG2-Trp-241. The conservation of Trp-241 in all eukaryotic transglutaminases and the finding that W241Q-TG2 had a much lower kcat than wild-type enzyme suggest evolutionary specialization in the use of the indole group. This notion is further supported by the observation that transition-state-stabilizing side chains of Tyr and His that operate in some serine and metalloproteases only partially substituted for Trp.
Transglutaminase type 2 (TG2) is a member of a superfamily of calcium-dependent enzymes that catalyze a variety of posttranslational protein-modifying reactions, including transamidation (1). This process may result in cross-linking of proteins by Nε-(γ-glutamyl)lysyl bridges between a Gln acceptor and a donor Lys, in incorporation of an amine into a Gln or in acylation of a Lys side chain in a protein. Transglutaminases (TGs) display exquisite specificities for selecting acceptor and donor residues in their substrates. Formation of the isopeptide bond confers extra rigidity to the resulting supramolecular structure and resistance to proteolysis, and it is an essential component of important biological processes such as blood coagulation, hardening of the fertilization envelope, and extracellular matrix assembly (1). In addition to transamidation reactions, TGs can esterify and deamidate proteins; these reactions are critical to the barrier function of skin and to the pathophysiology of gluten-induced enteropathy (celiac disease). Specialized noncatalytic actions of TGs have also been identified. For example, TG2 functions as a G protein in intracellular signaling (2, 3) and is involved in extracellular protein–protein interactions with integrins (4) that are mediated by the tight binding of TG2 to fibronectin (5); a catalytically inactive TG, band 4.2, is involved in maintenance of membrane integrity in erythrocytes (1).
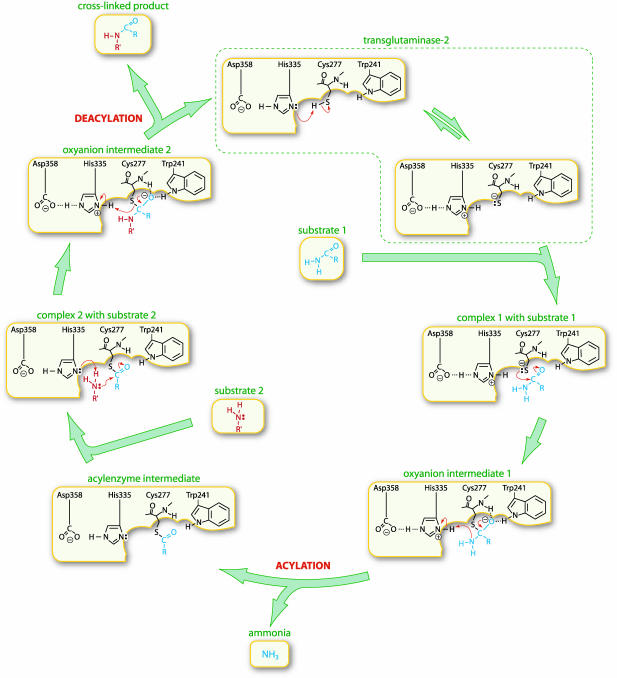
Crystal structures are available for TG2 (6) and three other TGs: factor XIIIA (7), sea bream TG (8), and TG3 (9). All eukaryotic TGs share common catalytic centers with the papain-like family of cysteine proteases, composed of a triad of Cys-His-Asp or Cys-His-Asn. Reactions of TGs, similar to those of papain, proceed by a kinetic pathway of acylation and deacylation; hence, by analogy with papain, the TG active-site Cys and His are likely to form a thiolate–imidazolium ion pair (10). The Gln-containing substrate binds to calcium-activated TG to form a Michaelis complex, which enables the active-site thiolate to attack the γ-carboxamide group of the substrate. This attack would lead to formation of the first tetrahedral adduct, the breakdown of which produces an acylenzyme intermediate with release of ammonia. In transamidation reactions, acylation of the enzyme would be rate-limiting (11, 12). In contrast to papain proteases, in which no preequilibration with the second substrate occurs, in TGs, the acylenzyme intermediate forms a saturable Michaelis complex with the second substrate, whether it is a small molecular weight amine or the ε-amino group of some peptide-bound Lys residue. Nucleophilic attack by these substrates would generate a second tetrahedral intermediate, the breakdown of which completes a forward cycle of catalysis with release of cross-linked product and enzyme.
We have recently shown that Trp-241 in TG2 was essential for catalysis (13), and here we provide evidence that the side chain of Trp-241 makes an important contribution to stabilizing the transition-state intermediates. This finding is in contrast to the catalytic apparatus used by the papain family, which uses a Gln residue at the equivalent position for transition-state stabilization. The conservation of a Trp in all enzymatically active TGs at the position corresponding to Trp-241 in TG2 indicates that transition-state stabilization by this residue is a general feature of catalysis in all eukaryotic TGs, from slime mold to mammals.
Materials and Methods
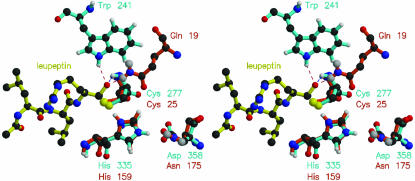
Superposition of Active-Site Side Chains of the Papain–Leupeptin Complex and TG2. insightii (Accelrys, San Diego) was used to superimpose GDP-bound human TG2 (ref. 6; Protein Data Bank ID code 1KV3) on the papain–leupeptin complex (ref. 14; ID code 1POP), by using atoms of catalytic triad side chains: CB, SG of Cys (A25, 1POP; A277, 1KV3), CG, ND1, CD2, CE1, NE2 of His (A159, 1POP; A335, 1KV3), CG, OD1 and ND2/OD2 of Asn/Asp (A175-Asn, 1POP; A358-Asp, 1KV3). rms deviation between the two structures for these 20 atoms was 0.66 Å.
Purification of Recombinant Proteins. Equivalent yields of WT and site-directed mutant rat TG2 (13) expressed as thrombin-cleavable GST fusion proteins (3, 13, 15) were obtained and stored at –80°C. Protein concentration was determined by using WT TG2 (quantitated by amino acid analysis) as the standard.
Photoaffinity Labeling. Proteins were labeled with 2.86 μM 8-azido-[α-32P]GTP (ICN) (3) in the absence of DTT and MgCl2. Coomassie-stained gels and autoradiographs were analyzed by densitometry. Mutant protein concentration was adjusted relative to a linear standard curve of WT protein, intensity of GTP labeling was normalized for the amount of protein, and mutant labeling was plotted as a ratio of WT labeling.
Peptide–Amine Conjugation Assay for TG Activity and Enzyme Kinetics. To determine Km and Vmax values, initial velocities (v) were determined. Eleven concentrations (between 0.08 and 4 mM) of the synthetic β-casein161–175 pentadecapeptide, Ser-Val-Leu-Ser-Leu-Ser-Gln-Ser-Lys-Val-Leu-Pro-Val-Pro-Glu (Aus-pep, Parkville, Australia), were incubated (37°C; final volume of 50 μl) with 100 mM Tris·HCl (pH 8)/4 mM CaCl2/1.5 mM dansylcadaverine (DC; ref. 16) and WT (10 pmol for 4 min) or mutant TG2 (50 pmol of W241Y for 4 min; 100 pmol of W241F for 8 min; 100 pmol of W241H for 20 min; 500 pmol of W241A for 20 min; 200 pmol of W241G for 20 min). β-Casein161–175–amine conjugate formation was linear over time. Reactions were terminated (50 μl of 1% orthophosphoric acid), 80 μl was applied to a C18 RP-HPLC column [XTerra RP18 3.5 μm, 4.6 × 50 mm column, Waters; 5-min linear gradient of acetonitrile/0.1% trifluoroacetic acid (5–80%; flow rate of 1.5 ml/min)], and column effluent was monitored with a diode array HPLC detector (Agilent 1100 Series, Agilent Technologies, Melbourne). Linear product standard curves were generated from reactions in which β-casein161–175 substrate (0–150 μM) had been completely converted to product by WT TG2 (100 pmol) over 60 min. Peak areas corresponding to product were integrated and quantitated against standard curves. Unmodified and modified β-casein161–175 peaks (generated after 60-min incubation with 100 pmol of TG2, 1.5 mM DC, and 0.25 mM β-casein161–175) were analyzed by matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) MS with a PE Biosystems Voyager System 4038 (Foster City, CA), in both linear and reflectron mode, by using calibration masses that bracketed the mass range of interest. Km and Vmax values were calculated (nonlinear regression with prism, GraphPad, San Diego) by using v = (Vmax × [S])/(Km + [S]), where [S] is the concentration of peptide substrate. Correlation coefficients were 0.998 for WT, 0.987 for W241Y, 0.993 for W241F, 0.985 for W241H, 0.995 for W241A, and 0.991 for W241G. The overall rate constant kcat was calculated by using kcat = Vmax/[E0], where [E0] is total enzyme concentration. Values represent means ± SEM of triplicate determinations. ΔΔGbinding, ΔΔGcatalysis, and ΔΔG‡T were calculated (17) and represent, respectively, the difference between WT and mutant enzymes in free energy required to form the enzyme–substrate complex (E·S) from E + S, to convert the E·S complex to the transition-state complex (E·S‡), and to stabilize the transition-state complex (i.e., difference in free energy required to reach E·S‡ from E + S).
Ammonia Release Assays. Proteins were dialyzed (4 h at 4°C) in buffer [50 mM Hepes, pH 8/300 mM NaCl/1 mM EDTA/10% (vol/vol) glycerol/0.15 mM PMSF; three changes] to remove residual DTT. NH3 release (18) by WT (40 pmol) or mutant TG2 (200 pmol of W241Y; 400 pmol of W241F, W241H; 2,000 pmol of W241A, W241G) was quantitated over time (37°C, 0.1 mM β-casein161–175 substrate; final volume of 200 μl). kcat/Km was calculated by using v = [E0][S]kcat/Km (19); kcat was derived from kcat/Km by using Km values for β-casein161–175 obtained from the DC–β-casein161–175 conjugate assay.
Results and Discussion
Superposition of Cysteine Protease and TG Active Sites Reveals Candidates for Transition-State Stabilization. In the x-ray structure of the cysteine protease papain (active site shown in red in Fig. 1), complexed to its transition-state inhibitor leupeptin (yellow), the oxyanion of leupeptin is hydrogen-bonded to the backbone amide of the active-site Cys-25 (3.0 Å) and the Nε2 nitrogen of Gln-19 (2.9 Å) of papain (14). Superposition (blue in Fig. 1) of the structure of the TG2 active site (6) reveals that the backbone nitrogen atom of the active-site Cys-277 would be 2.9 Å and the Nε1 nitrogen atom of Trp-241 is 3.41 Å from the papain inhibitor. Additional superpositions of the structures of the active-site residues of factor XIIIA (13, 20) and TG3 (9) show similar positioning for both the active-site Cys and the Trp equivalent of Trp-241 in TG2 (data not shown), suggesting that this may be a common feature for all eukaryotic TGs. In sea bream TG the side-chain χ2 torsion angle of this Trp (Trp-236) is rotated by 180° (8, 13), suggesting possibly significant repositioning in the transition state.
Fig. 1.
Stereo image showing superposition of active sites of TG2 (Cys-277, His-335, Asp-358; blue) and the papain (Cys-25, His-159, Asn-175; red)–leupeptin (yellow) complex. Distances (dotted lines) between the leupeptin oxyanion and the side-chain amide nitrogen of Gln-19 (red) of papain, or of Trp-241 (blue) of TG2, are 2.9 and 3.4 Å, respectively. Distances between the leupeptin oxyanion and the backbone amide nitrogen of Cys-25 of papain, or of Cys-277 of TG2, are 3.0 and 2.9 Å, respectively.
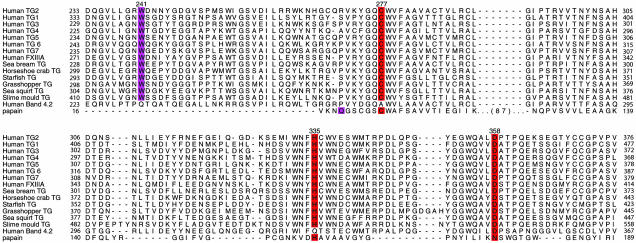
Trp-241 Is Conserved Among Enzymatically Active Eukaryotic TGs. The core regions of TGs are highly conserved; a 52–64% identity exists between human TG2 and all the other human TGs. Similar degrees of identity are seen between the core regions of human TG2 and fish (65%), invertebrate (≈55%), chordate (59%), and slime-mold (49%) TGs (data not shown). Trp-241, or its equivalent, is conserved in all enzymatically active forms of eukaryotic TGs, representatives of which are shown in Fig. 2. Significantly, this Trp residue is missing from the catalytically inactive member of the TG family, band 4.2, providing further support for the notion that Trp-241 might play an important role for stabilizing the transition states in TG catalysis.
Fig. 2.
Partial alignment (29) of human TGs, TGs representative of major branches of the TG phylogenetic tree (1), and papain. Active site Cys, His, and Asp (Asn in papain) are highlighted in red (active-site numbering is for TG2). Trp-241 equivalent of TG2 and transition-state-stabilizing Gln-19 of papain are highlighted in purple.
A Peptide–Amine Conjugation Assay for TG Activity. To evaluate transition-state stabilization of TG2 by Trp-241, a sensitive assay was required to allow kinetic parameters (Km, kcat, kcat/Km, ΔΔG values) to be determined, even when catalytic activity in Trp-241-substituted mutants was significantly impaired. The peptide, Ser-Val-Leu-Ser-Leu-Ser-Gln-Ser-Lys-Val-Leu-Pro-Val-Pro-Glu, incorporating residues 161–175 of β-casein (β-casein161–175), contains the primary site (Gln-167) for amine incorporation into β-casein (21). Incubation of β-casein161–175 with WT TG2 in the presence of saturating levels (1.5 mM) of the fluorescent amine DC results in release of NH3 and formation of DC-conjugated peptide:
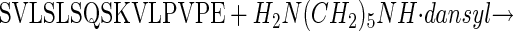 |
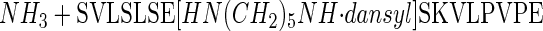 |
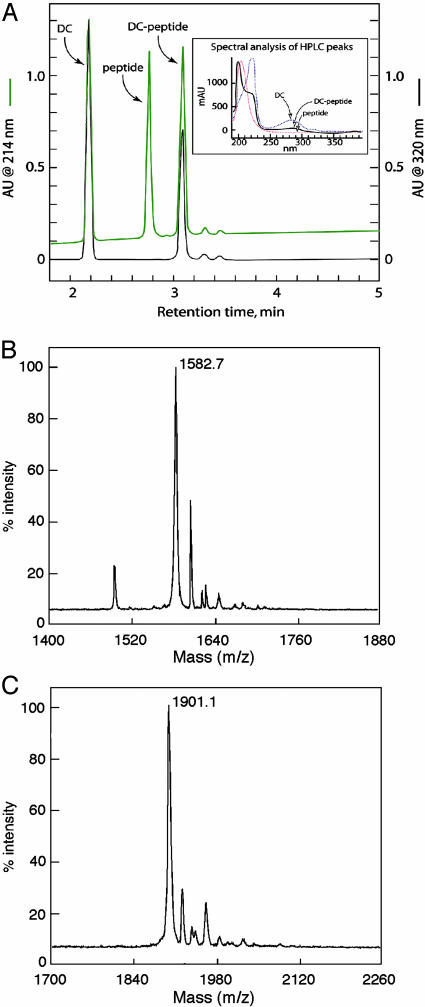
Kinetic constants for WT-catalyzed formation of DC–β-casein161–175 conjugate are shown in Table 1. This product only appears on addition of TG2 (data not shown) and can be separated from substrates by RP-HPLC. Monitoring column effluent at 320 nm (Fig. 3A) demonstrated that β-casein161–175 does not absorb at this wavelength, whereas DC does. Incorporation of DC into β-casein161–175 was evidenced by a peak of absorbance at 320 nm, at the position of the DC–β-casein161–175 conjugate. Spectral profiles from 180 to 400 nm of substrate and product peaks verified formation of DC–β-casein161–175 conjugate (Fig. 3A Inset). Thus, DC is characterized by absorbance peaks at 220 and 280 nm. β-Casein161–175 absorbs only at 214 nm (because the peptide contains no Trp or Tyr residues, no absorbance occurs at 280 nm). The conjugated DC–β-casein161–175 peak shows spectral features of both DC and β-casein161–175, with absorbance peaks at 214, 220, and 280 nm.
Table 1. Formation of DC—β-casein161–175 peptide conjugate and release of NH3 by WT and Trp-241 mutant TG2.
| Formation of DC—β-casein161-175 peptide conjugate |
NH3 release |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme | Km, mM | ΔΔGbinding, kcal·mol-1 | kcat, s-1 | ΔΔGcatalysis, kcal·mol-1 | kcat/Km, s-1·mM-1 | ΔΔGT‡, kcal·mol-1 | kcat, s-1 | ΔΔGcatalysis, kcal·mol-1 | kcat/Km, s-1·mM-1 | ΔΔGT‡, kcal·mol-1 |
| WT | 1.45 ± 0.06 | 2.450 ± 0.034 | 1.69 | 3.74 | 2.58 | |||||
| W241Y | 2.12 ± 0.24 | 0.23 | 0.149 ± 0.008 | 1.74 | 0.069 (25) | 1.97 | 0.138 | 2.03 | 0.065 (40) | 2.27 |
| W241F | 3.49 ± 0.34 | 0.54 | 0.082 ± 0.008 | 2.10 | 0.023 (70) | 2.65 | 0.199 | 1.81 | 0.057 (45) | 2.35 |
| W241H | 3.26 ± 0.40 | 0.50 | 0.031 ± 0.005 | 2.64 | 0.010 (165) | 3.14 | 0.088 | 2.31 | 0.027 (95) | 2.81 |
| W241A | 2.31 ± 0.16 | 0.29 | 0.008 ± 0.0003 | 3.50 | 0.0036 (470) | 3.79 | 0.018 | 3.29 | 0.008 (320) | 3.56 |
| W241G | 2.12 ± 0.19 | 0.23 | 0.006 ± 0.0004 | 3.72 | 0.0028 (615) | 3.95 | <0.017 | >3.32 | <0.008 (>320) | >3.56 |
Kinetic constants and ΔΔG values were determined as described (see Materials and Methods). Values represent means ± SEM of triplicate determinations. Values in parentheses represent the fold decrease relative to WT.
Fig. 3.
Verification of components of the DC–β-casein161–175 conjugate assay for TG activity. (A) HPLC elution and spectral analysis (Inset) of TG assay substrates (DC and β-casein161–175 peptide) and the DC–β-casein161–175-conjugated product. WT TG2 (10 pmol) was incubated for 60 min with 0.5 mM DC and 0.75 mM β-casein161–175. HPLC elution was monitored at 214 nm (green trace) and 320 nm (black trace). (Inset) DC (blue dotted line), β-casein161–175 (red dotted line), and DC–β-casein161–175 conjugate (solid black line). (B and C) MALDI-TOF MS analysis (linear mode) of β-casein161–175 and DC–β-casein161–175, respectively.
MALDI-TOF MS verified the mass of β-casein161–175 and DC-conjugated β-casein161–175 (Fig. 3B). Unmodified β-casein161–175 gave a major species with a mass (m/z) of 1,582.7 Da (Fig. 3B), in agreement with the monoisotopic molecular weight predicted on the basis of amino acid sequence. The mass of the predominant peak for the modified β-casein161–175 peptide (1,901.1 Da; Fig. 3C) increased by 318.4 Da. This finding is consistent with removal of NH2 (17 Da) from the Gln side chain in β-casein161–175 and addition of DC (335 Da).
The Acylation Step Is Rate-Limiting. WT TG2 catalyzed NH3 release from β-casein161–175 with a kcat of 3.7 s–1 (Table 1), which is similar to the kcat value of 2.5 s–1 obtained for transamidation of β-casein161–175 by DC (Table 1). This finding indicates that formation of the acylenzyme intermediate is rate-limiting in the reaction.
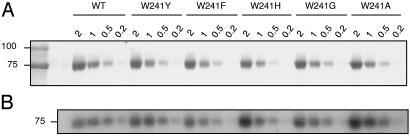
Trp-241 Is a Transition-State-Stabilizing Residue. To evaluate transition-state stabilization by Trp-241 in TG2, the Trp side chain was replaced with that of Ala or Gly. GTP binding, which allosterically regulates TG activity by binding close to the TG active site of TG2 (6), was not adversely affected in the mutants (Fig. 4). This finding suggested that substitutions for the indole side chain did not cause the protein to misfold. Replacement of Trp-241 with Ala or Gly had minimal influence on the Km or ΔΔGbinding values for the rate-limiting acylation step (Table 1), indicating that the Trp-241 side chain is not involved in formation of the Michaelis complex in the ground-state enzyme. These side-chain substitutions, however, caused marked decreases in catalytic rates (kcat) and specificity constants (kcat/Km, 470- to 605-fold reduction; Table 1). The large increases in Gibbs free energy for transition-state stabilization (ΔΔG‡T of 3.5–3.7 kcal/mol) for the mutants relative to WT TG2 indicate that the main role of the Trp-241 side chain is to stabilize the transition state.
Fig. 4.
GTP photolabeling of WT and Trp-241 mutant proteins (0.2–2.0 μg). Gels were visualized by Coomassie blue staining (A) and autoradiographed for 7 days at –80°C (B). After densitometry, GTP-labeling intensity was normalized for amount of protein. Labeling efficiencies for mutant proteins, relative to WT TG2, were 1.4 ± 0.06 for W241Y, 1.1 ± 0.06 for W241F, 2.1 ± 0.5 for W241H, 1.4 ± 0.3 for W241G, and 1.4 ± 0.5 for W241A.
Tyr Partially Compensates for Trp in Transition-State Stabilization. To investigate whether residues other than Trp can stabilize the transition state in TG2, Trp-241 was replaced with Gln, His, or Tyr. Gln stabilizes the transition-state oxyanion in papain (22). His has a five-membered ring similar to the indole ring of Trp, with a nitrogen atom in a similar position, and has been shown to stabilize the transition-state oxyanion in metalloproteases (23). Tyr, like Trp, has a ring structure, which may participate in hydrophobic and aromatic interactions, and a hydroxyl group that potentially stabilizes the oxyanion in zinc metallopeptidases (24) by hydrogen bonding. Purified mutants resolved as single bands on SDS/PAGE and retained WT GTP binding (Fig. 4; ref. 13). The activity of W241Q was lower than that of W241G and was not quantitated. Both His and Tyr replacement mutants showed decreases in kcat/Km (attributable to decreases in kcat and not Km), which were associated with substantial increases in ΔΔG‡T (3 kcal/mol for W241H and 2 kcal/mol for W241Y; Table 1). The smaller ΔΔG‡T associated with Trp-241 replacement by Tyr indicates some stabilization of the transition state in this mutant, caused either by hydrogen-bonding or by hydrophobic/aromatic interactions with the substrate. To determine whether this stabilization might be due to hydrogen bonding between the hydroxyl group of Tyr and the oxyanion, Trp-241 was replaced by Phe (Fig. 4). The small additional increase in ΔΔG‡T observed for W241F, compared with W241Y (0.7 kcal/mol; Table 1), suggests that hydrogen-bonding interactions play little role in stabilization of the transition state by Tyr at position 241. Clearly, none of the substituted residues are suitable replacements for Trp in the active-site pocket, an observation that is supported by the evolutionary conservation of this residue (Fig. 2).
Trp-241 Stabilizes the First Transition-State Intermediate in the Acylation Step. Mutant TG2-catalyzed release of NH3 from β-casein161–175 yielded kcat/Km and ΔΔG‡T values similar to those obtained for DC conjugation of β-casein161–175 (Table 1), showing that, as with WT TG2, the acylation step was rate-limiting.
Conclusions
TG-catalyzed reactions proceed by consecutive steps of acylation and deacylation, presumably involving tetrahedral adducts in the transition states for both. In this study, we identified Trp-241 of TG2, a residue essential for catalysis (13), to be critical for stabilizing the transition-state intermediates (Fig. 1). Evaluation of the reaction kinetics for various Trp-241 substitutions indicated that the rate-limiting step in the transamidation reaction examined was the formation of the acylenzyme intermediate that is concomitant with NH3 release (Table 1). Because GTP binding by the mutants did not differ significantly from WT, it could be assumed that the substitutions did not cause misfolding of the protein (Fig. 4). Replacement of Trp-241 with Ala or Gly did not affect Km (Table 1), indicating that the Trp side chain does not play a role in formation of the ground-state Michaelis complex (Fig. 5). The mutations, however, resulted in marked reductions in kcat and increases of nearly 4 kcal/mol in free energy required for transition-state stabilization (Table 1). Similar increases in ΔΔG‡T have been reported for substitution of the oxyanion-stabilizing Gln-19 side chain of papain (G19A, ΔΔG‡T = 2.4 kcal/mol; G19S, ΔΔG‡T = 3.8 kcal/mol; ref. 22). Strikingly, neither Gln nor His, which act as transition-state-stabilizing residues in cysteine proteases and metalloproteases, were able to stabilize the transition state in TG2, when substituted for the Trp residue in position 241 (Table 1). Phe or Tyr partially compensated for Trp-241, and only a small additional increase occurred in ΔΔG‡T for W241F, compared with W241Y (Table 1). ΔΔG‡T values for ammonia release by the TG2 mutants were similar to those obtained for the incorporation of DC into peptide substrate, confirming a role for Trp-241 in stabilizing the transition-state intermediate in the rate-limiting acylation step (Table 1).
Fig. 5.
Reaction pathway and a proposed mechanism for TG2-catalyzed transamidations, based on the papain-reaction mechanism. Active-site Cys-277 and His-335 form a thiolate–imidazolium ion pair. Binding of the Gln-containing substrate 1 enables the thiolate to attack the γ-carboxamide group in the substrate. The reaction proceeds through oxyanion intermediate 1 that is stabilized by H-bonding both to the backbone nitrogen of Cys-277 and to the Nε1 nitrogen of Trp-241. In the ensuing acylation step, NH3 is released as acylenzyme intermediate is formed. A distinguishing feature of TGs, which sets these enzymes apart from papain proteases, is a high specificity for the second substrate manifested in a saturable complex between substrate 2 and the acylenzyme intermediate. Nucleophilic attack by the amino group of substrate 2 leads to formation of oxyanion intermediate 2 that is again stabilized by H-bonding interactions to both Cys-277 and Trp-241. In the final deacylation step, cross-linked product is released and TG2 is regenerated.
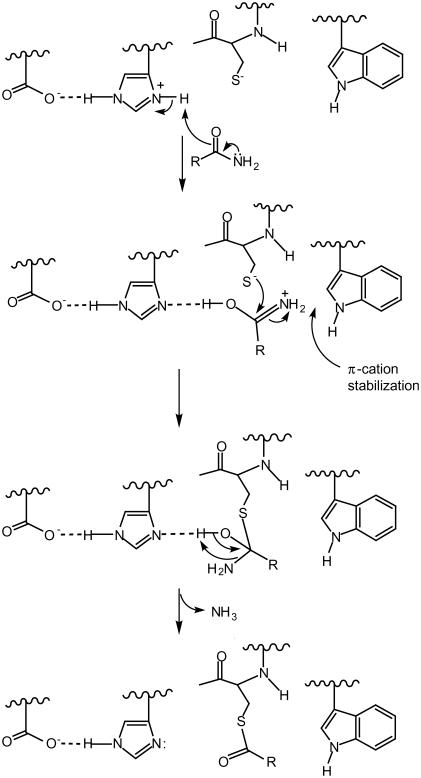
Based on our findings, it is now possible to offer mechanistic schemes for the acylation step in TG2 catalysis that are consistent with the thermodynamic values presented for the Trp-241 mutants. One of these postulates stabilization of the transition-state intermediate by a π-cation type of interaction (25) between the amide group of the Gln-containing substrate and the indole ring of Trp-241 as outlined (Scheme 1). The data in Table 1 show that substitution of Trp-241 by the aromatic ring-containing amino acids of Tyr and Phe, which can form such amino–aromatic interactions, support a substantial degree of transition-state stabilization.
Scheme 1.
Another mechanistic scheme for formation of the thiolester acylenzyme intermediate (Fig. 5) is based on the reaction mechanism proposed for papain (22). By analogy with papain and other enzymes (e.g., tyrosyl-tRNA synthetase; ref. 26), a ΔΔG‡T of ≈4 kcal/mol might be attributed to loss of a hydrogen-bonding interaction (27, 28) between the Trp-241 side chain and the substrate oxyanion. Trp-241 might similarly stabilize oxyanion intermediate 2 in the deacylation step (Fig. 5).
The conservation of Trp-241 or its equivalent in all enzymatically active eukaryotic TGs (Fig. 2) indicates evolutionary specialization in the TG catalytic mechanism for transition-state stabilization by the indole group of this residue.
Acknowledgments
We thank Professor Rick Silverman (Northwestern University) for suggesting π-cation interactions as a possible mechanism for acylation in TG catalysis, Drs. Ke Liu and Nawazish Navqi for technical help with HPLC, and Drs. Kieran Scott and Gillian Begg for helpful discussions. This work was supported by Australian National Health and Medical Research Council Grants 142000 and 256305, National Institutes of Health Grant HL-16346, and a Freedman Foundation Postdoctoral Fellowship (to M.A.W.).
Abbreviations: TG, transglutaminase; TG2, TG type 2; dansyl, 5-(dimethylamino)naphthalene-1-sulfonyl; cadaverine, 1,5-diaminopentane; DC, dansylcadaverine; MALDI-TOF, matrix-assisted laser desorption ionization–time-of-flight.
References
- 1.Lorand, L. & Graham, R. M. (2003) Nat. Rev. Mol. Cell Biol. 4 140–157. [DOI] [PubMed] [Google Scholar]
- 2.Nakaoka, H., Perez, D. M., Baek, K. J., Das, T., Husain, A., Misono, K., Im, M.-J. & Graham, R. M. (1994) Science 264 1593–1596. [DOI] [PubMed] [Google Scholar]
- 3.Iismaa, S. E., Wu, M.-J., Nanda, N., Church, W. B. & Graham, R. M. (2000) J. Biol. Chem. 275 18259–18265. [DOI] [PubMed] [Google Scholar]
- 4.Akimov, S. S., Krylov, D., Fleischmann, L. F. & Belkin, A. M. (2000) J. Cell Biol. 148 825–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lorand, L., Dailey, J. E. & Turner, P. M. (1988) Proc. Natl. Acad. Sci. USA 85 1057–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu, S., Cerione, R. & Clardy, J. (2002) Proc. Natl. Acad. Sci. USA 99 2743–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yee, V. C., Pedersen, L. C., Le Trong, I., Bishop, P. D., Stenkamp, R. E. & Teller, D. C. (1994) Proc. Natl. Acad. Sci. USA 91 7296–7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noguchi, K., Ishikawa, K., Yokoyama, K., Ohtsuka, T., Nio, N. & Suzuki, E. (2001) J. Biol. Chem. 276 12055–12059. [DOI] [PubMed] [Google Scholar]
- 9.Ahvazi, B., Kim, H. C., Kee, S.-H., Nemes, Z. & Steinert, P. M. (2002) EMBO J. 21 2055–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Micanovic, R., Procyk, R., Lin, W. & Matsueda, G. (1994) J. Biol. Chem. 269 9190–9194. [PubMed] [Google Scholar]
- 11.Curtis, C. G., Stenberg, P., Brown, K. L., Baron, A., Chen, K., Gray, A., Simspon, I. & Lorand, L. (1974) Biochemistry 13 3257–3262. [DOI] [PubMed] [Google Scholar]
- 12.Stenberg, P., Curtis, C. G., Wing, D., Tong, Y. S., Credo, R. B., Gray, A. & Lorand, L. (1975) Biochem. J. 147 155–163. [PMC free article] [PubMed] [Google Scholar]
- 13.Murthy, S. N., Iismaa, S. E., Begg, G., Freymann, D. M., Graham, R. M. & Lorand, L. (2002) Proc. Natl. Acad. Sci. USA 99 2738–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schröder, E., Phillips, C., Garman, E., Harlos, K. & Crawford, C. (1993) FEBS Lett. 315 38–42. [DOI] [PubMed] [Google Scholar]
- 15.Iismaa, S. E., Chung, L., Wu, M.-J., Teller, D. C., Yee, V. C. & Graham, R. M. (1997) Biochemistry 36 11655–11664. [DOI] [PubMed] [Google Scholar]
- 16.Lorand, L., Rule, N. G., Ong, H. H., Furlanetto, R., Jacobsen, A. J. D., Öner, N. & Bruner-Lorand, J. (1968) Biochemistry 7 1214–1223. [DOI] [PubMed] [Google Scholar]
- 17.Wilkinson, A. J., Fersht, A. R., Blow, D. M. & Winter, G. (1983) Biochemistry 22 3581–3586. [DOI] [PubMed] [Google Scholar]
- 18.Fickenscher, K., Aab, A. & Stüber, W. (1991) Thromb. Haemostasis 65 535–540. [PubMed] [Google Scholar]
- 19.Fersht, A. (1999) Structure and Mechanism in Protein Science (Freeman, New York).
- 20.Pedersen, L. C., Yee, V. C., Bishop, P. D., Le Trong, I., Teller, D. C. & Stenkamp, R. E. (1994) Protein Sci. 3 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorman, J. J. & Folk, J. E. (1980) J. Biol. Chem. 255 419–427. [PubMed] [Google Scholar]
- 22.Ménard, R., Carrière, J., Laflamme, P., Plouffe, C., Khouri, H. E., Vernet, T., Tessier, D. C., Thomas, D. Y. & Storer, A. C. (1991) Biochemistry 30 8924–8928. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez, M., Liu, X., Wouters, M. A., Heyberger, S. & Husain, A. (2001) J. Biol. Chem. 276 4998–5004. [DOI] [PubMed] [Google Scholar]
- 24.Brown, C. K., Madauss, K., Lian, W., Beck, M. R., Tolbert, W. D. & Rodgers, D. W. (2001) Proc. Natl. Acad. Sci. USA 98 3127–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma, J. C. & Dougherty, D. A. (1997) Chem. Rev. 97 1303–1324. [DOI] [PubMed] [Google Scholar]
- 26.Fersht, A. R., Shi, J. P., Knill-Jones, J., Lowe, D. M., Wilkinson, A. J., Blow, D. M., Brick, P., Carter, P., Waye, M. M. & Winter, G. (1985) Nature 314 235–238. [DOI] [PubMed] [Google Scholar]
- 27.Ippolito, J. A., Alexander, R. S. & Christianson, D. W. (1990) J. Mol. Biol. 215 457–471. [DOI] [PubMed] [Google Scholar]
- 28.Yun, Y. S., Lee, T.-H., Nam, G. H., Jang, D. S., Shin, S., Oh, B.-H. & Choi, K. Y. (2003) J. Biol. Chem. 278 28229–28236. [DOI] [PubMed] [Google Scholar]
- 29.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]