Abstract
The HIV type 1 (HIV-1) Tat protein stimulates transcription elongation by recruiting P-TEFb (CDK9/cyclin T1) to the transactivation response (TAR) RNA structure. Tat-induced CDK9 kinase has been shown to phosphorylate Ser-5 of RNA polymerase II (RNAP II) C-terminal domain (CTD). Results presented here demonstrate that Tat-induced Ser-5 phosphorylation of CTD by P-TEFb stimulates the guanylyltransferase activity of human capping enzyme and RNA cap formation. Sequential phosphorylation of CTD by Tat-induced P-TEFb enhances the stimulation of human capping enzyme guanylyltransferase activity and RNA cap formation by transcription factor IIH-mediated CTD phosphorylation. Using an immobilized template assay that permits isolation of transcription complexes, we show that Tat/TAR-dependent phosphorylation of RNAP II CTD stimulates cotranscriptional capping of HIV-1 mRNA. Upon transcriptional induction of latently infected cells, accumulation of capped transcripts occurs along with Ser-5-phosphorylated RNAP II in the promoter proximal region of the HIV-1 genome. Therefore, these observations suggest that Tat/TAR-dependent phosphorylation of RNAP II CTD is crucial not only in promoting transcription elongation but also in stimulating nascent viral RNA capping.
The capping of mRNA occurs cotranscriptionally by a series of three enzymatic reactions in which the 5′ triphosphate terminus of the nascent transcript is cleaved to a diphosphate by RNA 5′ triphosphatase, capped with GMP by RNA guanylyltransferase (GT) and methylated at the N7 position of guanine by RNA (guanine-7) methyltransferase. The three activities are present in all eukaryotes examined. Yeast species encode three proteins corresponding to each enzyme activity, whereas in metazoans the first two activities are part of one protein consisting of N-terminal triphosphatase and C-terminal GT domains (1, 2).
Targeting of cap formation to RNA polymerase II (RNAP II) transcripts is achieved through physical interaction of components of the capping apparatus with the phosphorylated C-terminal domain (CTD) of the largest subunit of RNAP II (3–6). The mammalian RNAP II CTD is composed of 52 tandemly repeated heptads with a consensus, Tyr-1-Ser-2-Pro-3-Thr-4-Ser-5-Pro-6-Ser-7, conserved in most eukaryotes. Ser-2 and Ser-5 are targets of phosphorylation and dephosphorylation during transcription (7). Principal kinases responsible for RNAP II CTD phosphorylation during transcription include transcription factor IIH (TFIIH) (CDK7/cyclin H) and positive transcription elongation factor P-TEFb (CDK9/cyclin T1). Phosphorylation of the CTD Ser-2 and Ser-5 residues has differential effects on recruitment and activation of capping enzyme (CE) (8, 9). Although Ser-2 phosphorylation of CTD heptads is sufficient for mammalian GT binding, its activation requires Ser-5-phosphorylated CTD heptads (10). RNAP II CTD is phosphorylated at Ser-5 by TFIIH during transcription initiation through the promoter clearance stage and changes to Ser-2 phosphorylation when the polymerase is associated with the coding region (11). However, RNAP II CTD Ser-5 phosphorylation is sustained by Tat/transactivation response (TAR)-induced P-TEFb following release of TFIIH from HIV-1 transcription elongation complexes (TECs) at the promoter clearance stage (12, 13). Chromatin immunoprecipitation (ChIP) assays show an accumulation of RNAP II phosphorylated at Ser-5 in the promoter proximal regions of a number of genes (14, 15), suggesting that the role of RNAP II CTD phosphorylation in RNA processing is more than mere anchoring of transcription factors.
HIV-1 Tat transactivation provides a particularly useful model to study regulation of processive transcription elongation and mRNA capping. Tat stimulates HIV-1 transcription elongation by recruitment of P-TEFb to the TAR RNA structure, and Tat/TAR-associated CDK9 then phosphorylates RNAP II CTD and other RNAP II-associated proteins, leading to a transition from nonprocessive to processive transcription (16). P-TEFb phosphorylates both Ser-2 and Ser-5 of RNAP II CTD in the presence of Tat, whereas P-TEFb alone phosphorylates only Ser-2 (12). In this study, we used in vivo ChIP and in vitro staged transcription elongation assays to determine the function of Tat in regulating cotranscriptional capping of viral mRNA. Our results indicate that Tat promotes viral mRNA cap formation by inducing TAR-dependent phosphorylation of RNAP II CTD during transcription elongation, a crucial checkpoint in gene expression.
Materials and Methods
Antibody. Antibodies used were α-CTD H5 and H14 (Covance, Richmond, CA), α-RNAP II (N-20), α-p62 subunit of TFIIH (Santa Cruz Biotechnology), α-CE (3), and α-2,2,7-trimethylguanosine (Calbiochem).
CTD Phosphorylation in Vitro. In vitro kinase assays were performed by incubating purified RNAP II or GST-CTD, P-TEFb or purified TFIIH, and ATP for 60 min at 30°C in the absence or presence of Tat in buffer (50 mM Tris-HCl, pH 7.5/5 mM DTT/5 mM MnCl2/4 mM MgCl2). Phosphorylated RNAP II and GST-CTD were then fractionated by electrophoresis through SDS-polyacrylamide gels and analyzed by Western blot with α-CTD H5 (α-phosphoserine 2) or H14 (α-phosphoserine 5).
Purification of Phosphorylated GST-CTD or RNAP II. Phosphorylated GST-CTD was adsorbed to glutathione-Sepharose beads during a 2-h incubation at 4°C in buffer (50 mM Tris-HCl, pH 8.0/50 mM NaCl/5% glycerol/1mMDTT/0.03% Triton X-100). Bead-bound GST-CTD was then washed with 1 M NaCl to remove residual non-CTD proteins. Phosphorylated RNAP II was bound to protein A/G-Sepharose beads by α-RNAP II antibody N-20 (the epitope is in the N terminus of the large subunit of RNAP II).
In Vitro Binding Assay. Bead-bound GST-CTD was incubated with human CE (HCE) in binding buffer (20 mM Tris-HCl, pH 7.9/0.1 M NaCl/0.2 mM EDTA/1mMDTT/1 mM PMSF/0.2% Nonidet P-40) overnight at 4°C and then washed with buffer (20 mM Tris-HCl, pH 7.9/0.5 M NaCl/0.2 mM EDTA/1 mM DTT/1 mM PMSF/0.4% Nonidet P-40). Bead-bound proteins were fractionated by electrophoresis through SDS-polyacrylamide gels and analyzed by Western blot with α-CE antibody.
RNA GT Assay. The GT activity of HCE was assayed by formation of the covalent enzyme-GMP intermediate. HCE was incubated with [α-32P]GTP in the absence or presence of bead-bound RNAP II or GST-CTD in buffer [25 mM Tri-HCl, pH 7.5/5 mM MgCl2/0.5 mM DTT/0.1 μg of inorganic pyrophosphatase, (Roche Molecular Biochemicals)] for 15 min at 37°C. Guanylylation of HCE was assessed by electrophoresis through SDS-polyacrylamide gels followed by autoradiography.
RNA Cap Formation. T7 polymerase run-off RNA (72-mer) transcribed from BamHI-linearized pBluescript II+ (Stratagene) was incubated with HCE and [α-32P]GTP in the absence or presence of bead-bound RNAP II or GST-CTD for 15 min at 37°C. Samples were extracted with phenol/chloroform, precipitated with ethanol, and analyzed by electrophoresis through 10% polyacrylamide gels containing 7 M urea in TBE buffer (88 mM Tris/88 mM boric acid/2 mM EDTA, pH 8.2) followed by autoradiography.
Stepwise Transcription. Purification of HIV-1 preinitiation complexes (PICs) was carried out as described previously by using biotinylated HIV-1 LTR templates (12). The purified PICs were walked to position U14 by incubation with 50 μM dATP, CTP, GTP, and UTP for 10 min at 30°C followed by extensive washing with buffer. PICs and TECs stalled at U14 were analyzed by Western blot with α-RNAP II antibody (N-20). TECs stalled at U14 were walked stepwise along the DNA template by repeated incubations with different sets of two or three NTPs and washed extensively to remove the unincorporated NTPs. RNAP II was labeled with [γ-32P]ATP during stepwise transcription elongation and immunoprecipitated with α-RNAP II antibody (N-20). Incorporation of 32P into RNAP II was analyzed by electrophoresis through SDS-polyacrylamide gels followed by autoradiography. Transcripts were labeled with [α-32P]UTP and analyzed by electrophoresis through 10% polyacrylamide gels containing 7 M urea in TBE buffer followed by autoradiography.
Cotranscriptional Capping. HIV-1 PICs were analyzed by Western blot with α-CE antibody. TECs stalled at G36 and C81 were incubated with HCE and then analyzed by Western blot with α-CE antibody. TECs, which were supplemented with HCE, were incubated with [α-32P]GTP, and the GMP-labeled transcripts were analyzed by electrophoresis through 10% polyacrylamide gels containing 7 M urea in TBE buffer followed by autoradiography. GMP-labeled transcripts isolated from the indicated TECs were dissolved in S1 buffer (30 mM sodium acetate, pH 4.6/50 mM NaCl/1 mM zinc acetate/5% glycerol) and incubated with 20 units of nuclease S1 for 2 h at 37°C. Digests were spotted on polyethyleneimine-cellulose TLC plates that were developed with 0.2 M (NH4)2SO4. The S1-resistant caps were visualized by autoradiography.
ChIP of HIV-1 Latently Infected Cells. Five to 10 million OM10.1 cells, a promyelocytic line containing a single copy of integrated WT HIV-1, in log phase were incubated for 2 h with or without tumor necrosis factor α (TNFα, 5 μg/ml), which induces transcription of latent proviral DNA (17). After a 24-h induction, cells were cross-linked (1% formaldehyde, 10 min at 37°C), and samples were sonicated to reduce DNA fragments to ≈200–800 nt for ChIP assays (14), in which transcription complexes were immunoprecipitated with antibody α-CTD H5, H14, or α-2,2,7-trimethylguanosine. Specific DNA sequences in the immunoprecipitates were detected by PCR by using primers specific for the HIV-1 LTR between –92 to +180 nt (forward primer, 5′-ACTTTTCCGGGGAGGCGCGATC-3′; reverse primer, 5′-GCCACTGCTAGAGATTTCCACACTG-3′). The primers for GAPDH are located around the transcription start site (–85 and +81) (forward primer, 5′-TACTAGCGGTTTTACGGGCG-3′; reverse primer, 5′-TCGAACAGGAGGAGCAGAGA-3′).
Results
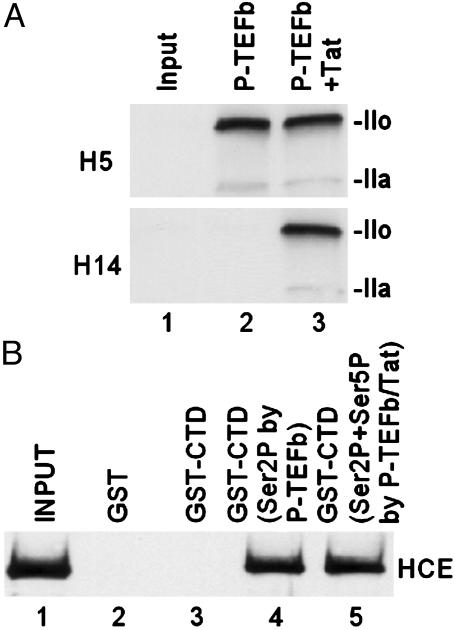
Interaction Between HCE and CTD Phosphorylated by P-TEFb. Consistent with our previous report (12), P-TEFb phosphorylated RNAP II CTD at both Ser-2 and Ser-5 in the presence of Tat, whereas P-TEFb alone phosphorylated RNAP II CTD only at Ser-2 (Fig. 1A). Similar results were observed when GST-CTD was used as substrate (data not shown). To determine whether CE interacts with CTD phosphorylated by P-TEFb, HCE was incubated with bead-bound GST-CTD. Results shown in Fig. 1B demonstrated that HCE bound to CTD phosphorylated by P-TEFb. There was no difference in binding to CTD phosphorylated either at Ser-2 by P-TEFb alone (lane 4) or at both Ser-2 and Ser-5 by P-TEFb plus Tat (lane 5).
Fig. 1.
Interaction between HCE and CTD phosphorylated by P-TEFb. (A) Tat modifies the substrate specificity of CDK9. In vitro kinase assays were performed by incubating purified RNAP II with P-TEFb and ATP (100 μM) in the absence or presence of Tat. Phosphorylated CTD was analyzed by Western blot with specific antibody H5 (α-phosphoserine 2) or H14 (α-phosphoserine 5). The hypophosphorylated (IIa) and hyperphosphorylated (IIo) forms of the largest subunit of RNAP II are indicated. (B) Interaction of HCE and CTD phosphorylated by P-TEFb. HCE was incubated with bead-bound GST-CTD. Bead-bound proteins were eluted by boiling in SDS loading buffer and then analyzed by Western blot with α-CE antibody.
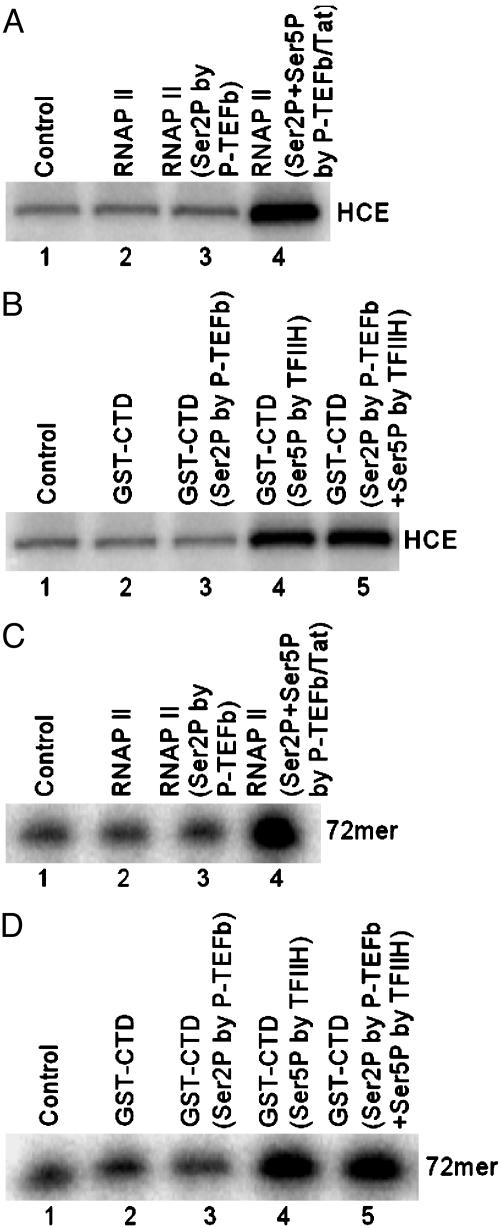
Tat-Induced Ser-5 Phosphorylation of CTD Stimulates the GT Activity of HCE and RNA Cap Formation. To test whether CTD phosphorylation by P-TEFb stimulates CE activity, HCE was incubated with [α-32P]GTP in the absence or presence of phosphorylated RNAP II. Results shown in Fig. 2A demonstrated that phosphorylation of RNAP II CTD at both Ser-2 and Ser-5 by P-TEFb plus Tat resulted in an ≈4-fold increase in enzyme–GMP complex formation (lane 4). When GST-CTD was used as substrate for P-TEFb, CTD phosphorylated at both Ser-2 and Ser-5 by P-TEFb plus Tat resulted in a similar ≈4-fold increase in formation of enzyme–GMP complex (data not shown). To determine whether prior Ser-2 phosphorylation influences the stimulatory effect of Ser-5 phosphorylation on HCE, TFIIH, which specifically phosphorylates Ser-5 of CTD (18, 19), was used. Results shown in Fig. 2B indicate that Ser-2 phosphorylation by P-TEFb had little effect on stimulation of enzyme–GMP complex formation by TFIIH-mediated Ser-5 phosphorylation (lanes 4 and 5).
Fig. 2.
Tat-induced Ser-5 phosphorylation of CTD by P-TEFb stimulates the GT activity of HCE and RNA cap formation. (A) CTD phosphorylation by P-TEFb/Tat stimulates the GT activity of HCE. HCE was incubated with [α-32P]GTP in the absence or presence of bead-bound RNAP II. Guanylylation of HCE was assessed by SDS/PAGE followed by autoradiography. (B) Ser-2 phosphorylation does not influence the effect of CTD Ser-5 phosphorylation on HCE activation. Guanylylation of HCE was performed by incubating HCE with [α-32P]GTP in the absence or presence of bead-bound GST-CTD and then resolved by SDS/PAGE followed by autoradiography. (C) CTD phosphorylation by P-TEFb/Tat stimulates RNA cap formation. A T7 polymerase run-off RNA (72-mer) was incubated with HCE and [α-32P]GTP in the absence or presence of bead-bound RNAP II. GMP-labeled RNA was then analyzed by electrophoresis through 10% polyacrylamide gels containing 7 M urea in TBE buffer followed by autoradiography. (D) CTD Ser-2 phosphorylation does not influence the effect of CTD Ser-5 phosphorylation on RNA cap formation. Transfer of GMP was performed by incubating RNA with HCE and [α-32P]GTP in the absence or presence of bead-bound GST-CTD. GMP-labeled RNA was then analyzed by electrophoresis through 10% polyacrylamide gels containing 7 M urea in TBE buffer followed by autoradiography.
Tat-induced Ser-5 phosphorylation of RNAP II CTD by P-TEFb specifically induced RNA capping (Fig. 2C). Phosphorylation of RNAP II CTD at both Ser-2 and Ser-5 by P-TEFb plus Tat resulted in an ≈4-fold increase in transfer of GMP to RNA 5′ ends (lane 4). CTD phosphorylation at both Ser-2 and Ser-5 by P-TEFb plus Tat also resulted in a similar increase in RNA cap formation when GST-CTD was used as substrate for P-TEFb (data not shown). As in the case of enzyme–GMP complex formation (Fig. 2B), Ser-2 phosphorylation had little effect on stimulation of RNA cap formation in response to Ser-5 phosphorylation (Fig. 2D). The results indicate that Tat-induced Ser-5 phosphorylation by P-TEFb stimulates the GT activity of HCE and RNA cap formation.
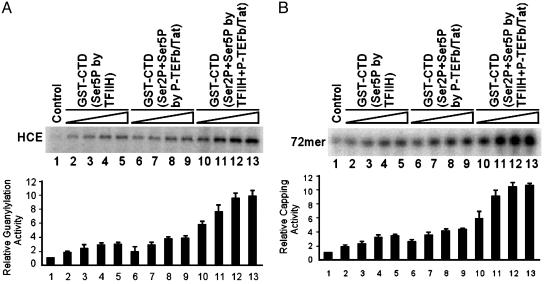
Additive Effects of Ser-5 Phosphorylation of CTD by TFIIH and Tat-Induced P-TEFb on the GT Activity of HCE and RNA Cap Formation. TFIIH and P-TEFb are two principal kinases responsible for RNAP II CTD phosphorylation during HIV-1 transcription. CTD phosphorylation by either TFIIH or Tat-induced P-TEFb stimulated HCE GT activity (Fig. 3A, lanes 1–9). Interestingly, a >3-fold further increase in enzyme–GMP complex formation was observed when GST-CTD was phosphorylated sequentially by TFIIH followed by P-TEFb plus Tat (lanes 10–13). Similarly, the cooperative effects of CTD phosphorylation sequentially by TFIIH and Tat-induced P-TEFb resulted in a >3-fold additional increase in RNA cap formation (Fig. 3B, lanes 10–13).
Fig. 3.
Additive effects of CTD phosphorylation sequentially by TFIIH and Tat-induced P-TEFb on the stimulation of the GT activity of HCE and RNA cap formation. (A) Stimulation of the GT activity of HCE. Guanylylation of HCE was performed by incubating HCE with [α-32P]GTP in the absence (lane 1) or presence of bead-bound GST-CTD phosphorylated by TFIIH (lanes 2–5), P-TEFb/Tat (lanes 6–9), or sequentially by TFIIH and P-TEFb/Tat (lanes 10–13) and then assessed by SDS/PAGE followed by autoradiography. Quantitation of three independent assays performed under similar conditions is shown at the bottom of the panel. (B) Stimulation of RNA cap formation. Transfer of GMP to RNA was performed by incubating RNA with HCE and [α-32P]GTP in the absence (lane 1) or presence of bead-bound GST-CTD phosphorylated by TFIIH (lanes 2–5), P-TEFb/Tat (lanes 6–9), or sequentially by TFIIH and P-TEFb/Tat (lanes 10–13) and then analyzed by electrophoresis through 10% polyacrylamide gels containing 7 M urea in TBE buffer followed by autoradiography. Quantitation of three independent assays performed under similar conditions is shown below.
TFIIH is a part of PICs and functions between stages of transcription initiation and promoter clearance when the nascent transcript is <30–50 bases long (20). Although TFIIH is dispensable for Tat function (21), our previous results demonstrated that TFIIH regulates Tat-dependent CDK9 activation during HIV-1 transcription (13). It was important therefore to determine whether TFIIH associates with HIV-1 TECs in vivo. Results of ChIP assays demonstrated that TFIIH is released from the HIV-1 TECs as the polymerase enters the ORF (data not shown). These results are consistent with the model in which the coordinated kinase activities of TFIIH and Tat-dependent P-TEFb promote RNAP II CTD hyperphosphorylation and polymerase processivity (22). Importantly, the results argue that Tat-induced CTD Ser-5 phosphorylation by P-TEFb augments the basal level of RNA capping by TFIIH-mediated CTD phosphorylation.
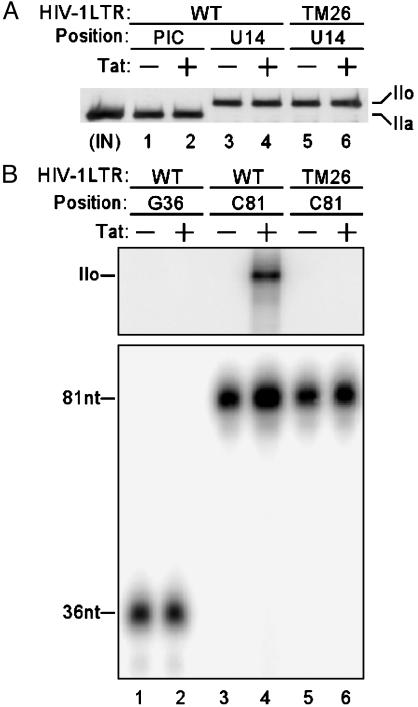
Tat/TAR-Dependent Phosphorylation of the RNAP II CTD by P-TEFb Stimulates HIV-1 Transcription Elongation. Several lines of evidence suggest that RNAP II CTD is phosphorylated during HIV-1 transcription in a biphasic pattern, initially during the transcription initiation stage and later during the elongation phase (12, 23). To determine the effects of Tat/TAR-dependent secondary phase of RNAP II CTD phosphorylation on HIV-1 transcription elongation, a stepwise transcription elongation assay was designed. HIV-1 PICs were purified by using a biotinylated HIV-1 LTR promoter and walked to position U14 by incubation with dATP, CTP, GTP, and UTP (12). RNAP II was phosphorylated during the initiation stage and converted from the hypophosphorylated form IIa in PICs to the hyperphosphorylated form IIo in TECs stalled at U14 with either the WT or the TAR mutant (TM26) HIV-1 LTR template (Fig. 4A). The TAR mutant TM26 has base substitutions in the TAR RNA pyrimidine bulge structure required for Tat binding as well as the TAR RNA loop required for cyclin T1 binding (24). TECs stalled at U14 were walked stepwise along the DNA template by repeated incubations with different sets of two or three NTPs. RNAP II was labeled with [γ-32P]ATP during stepwise transcription elongation and immunoprecipitated with α-RNAP II antibody. Consistent with our previous results (12), a Tat/TAR-dependent secondary phase of RNAP II CTD phosphorylation was observed during transcription elongation (Fig. 4B Upper, lane 4). The secondary RNAP II CTD phosphorylation occurred only in TECs with the WT template in the presence of Tat, when TECs were stalled at C81, a stage at which an intact TAR RNA structure is formed (lanes 3 and 4). The secondary phase of RNAP II CTD phosphorylation was not detected in TECs stalled at G36, a stage of transcript elongation at which the TAR RNA structure is not yet formed (lanes 1 and 2). Importantly, the secondary phase of RNAP II CTD phosphorylation was not observed in TECs stalled at C81 with the TAR mutant TM26 template (lanes 5 and 6). Western blot analysis showed an equivalent level of RNAP II in each lane (data not shown).
Fig. 4.
Tat/TAR-dependent phosphorylation of the RNAP II CTD stimulates HIV-1 transcription elongation. (A) RNAP II CTD is phosphorylated during the initiation stage or early stage of elongation. RNAP II associated with PICs or TECs stalled at U14 was analyzed by Western blot with α-RNAP II antibody N-20. The WT and TAR mutant (TM26) HIV-1 LTR templates and the hypophosphorylated (IIa) and hyperphosphorylated (IIo) forms of the large subunit of RNAP II are indicated. IN, input. (B) Tat/TAR-dependent phosphorylation of RNAP II CTD by P-TEFb stimulates transcription elongation. RNAP II CTD was labeled with [γ-32P]ATP during stepwise transcription elongation and immunoprecipitated with α-RNAP II antibody N-20. Incorporation of 32P into RNAP II was assessed by SDS/PAGE followed by autoradiography (Upper). Nascent transcripts were labeled with [α-32P]UTP and analyzed by electrophoresis through 10% polyacrylamide gels containing 7 M urea in TBE buffer followed by autoradiography (Lower).
To assess the effect of Tat/TAR-dependent phosphorylation of RNAP II CTD on transcription elongation, nascent transcripts were internally labeled with [α-32P]UTP during stepwise transcription elongation (Fig. 4B Lower). A 2-fold increase in transcript at C81 was observed when the TAR RNA structure was formed in response to Tat with the WT LTR template (lanes 3 and 4). However, no Tat-mediated increase was observed in transcript at G36 when the TAR RNA structure was not formed (lanes 1 and 2) or the longer transcript at C81with the mutant TAR, which blocks P-TEFb and Tat binding (lanes 5 and 6). Therefore, the results suggest that stimulation of transcription elongation depends on Tat/TAR-induced phosphorylation of RNAP II CTD.
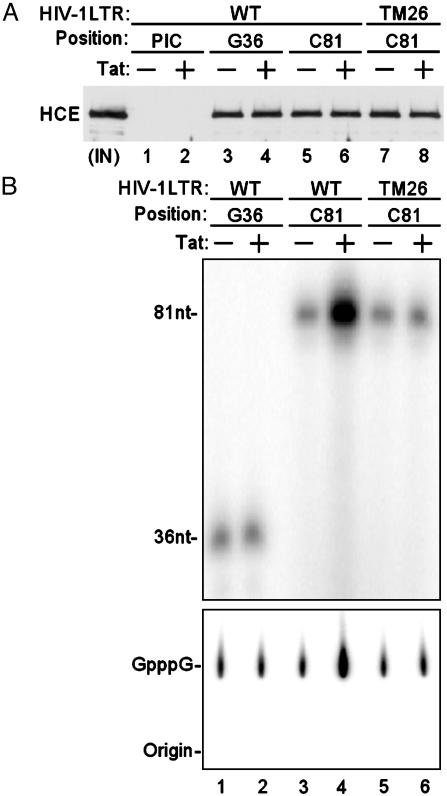
Tat/TAR-Dependent Phosphorylation of the RNAP II CTD by P-TEFb Stimulates Cotranscriptional Capping of HIV-1 Transcripts. To investigate association of HCE with HIV-1 transcription complexes during various stages of the transcription cycle, HIV-1 PICs were analyzed by Western blot. Consistent with a recent report (25), HCE is not a component of HIV-1 PICs (Fig. 5A, lanes 1 and 2). TECs stalled at G36 and C81 were incubated with HCE and extensively washed to remove unbound proteins. Results demonstrated that Tat did not influence recruitment of HCE into TECs stalled at either G36 or C81 (lanes 3–8). Because RNAP II CTD is hyperphosphorylated during the initiation stage or early stage of elongation (Fig. 4A), the results suggest that association of HCE with the transcription complexes depends on RNAP II CTD hyperphosphorylation.
Fig. 5.
Tat/TAR-dependent phosphorylation of the RNAP II CTD stimulates cotranscriptional capping of viral mRNA. (A) HCE is recruited into HIV-1 TECs through phosphorylated RNAP II CTD. PICs were analyzed by Western blotting with α-CE antibody. TECs stalled at G36 and C81 were incubated with HCE and extensively washed to remove unbound proteins before Western blot analysis with α-CE antibody. The WT and TAR mutant (TM26) HIV-1 LTR templates are indicated. IN, input. (B) Tat/TAR-dependent phosphorylation of the RNAP II CTD stimulates capping of viral mRNA. The indicated TECs were supplemented with HCE and then incubated with [α-32P]GTP. GMP-labeled transcripts were analyzed by electrophoresis through 10% polyacrylamide gels containing 7 M urea in TBE buffer followed by autoradiography (Upper). GMP-labeled RNAs isolated from the indicated TECs were analyzed by S1 nuclease digestion followed by polyethyleneimine-cellulose TLC (Lower).
To assess whether Tat/TAR-dependent phosphorylation of RNAP II CTD stimulates cap formation of nascent viral transcripts, TECs stalled at G36 and C81 were supplemented with HCE and then incubated with [α-32P]GTP. Results shown in Fig. 5B demonstrated that Tat/TAR-dependent phosphorylation of RNAP II CTD resulted in an 8-fold increase in GMP labeling of nascent viral transcript at C81 with WT template (Fig. 5B Upper, lanes 3 and 4). No increase was observed either in transcript at C81 with the mutant TAR template (lanes 5 and 6) or transcript at G36 with WT template when the TAR structure is not yet formed (lanes 1 and 2). To document that the GMP labeling is in the 5′ cap structure of the nascent viral transcripts, RNAs were isolated from the indicated TECs and analyzed by S1 nuclease digestion followed by polyethyleneimine-cellulose TLC. The increase in S1 nuclease-resistant GpppG cap structure was similar to Tat/TAR-dependent GMP labeling of transcript at C81 with WT template (Fig. 5B Lower). The results suggest that stimulation of viral RNA capping is directly in response to Tat/TAR-dependent phosphorylation of RNAP II CTD. If the 2-fold increase in viral transcripts in response to Tat (Fig. 4B Lower, lanes 3 and 4) is taken into account, Tat/TAR-dependent phosphorylation of RNAP II CTD increased cotranscriptional capping of viral mRNA by 4-fold.
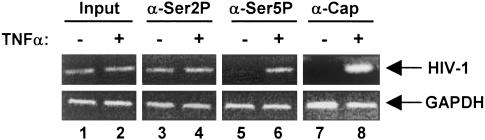
Increase of Capped HIV-1 RNA Is Coupled with Ser-5 Phosphorylation in Infected Cells. To explore the role of Tat/TAR-dependent phosphorylation of RNAP II CTD in HIV-1 mRNA capping in vivo, ChIP assays were performed with OM10.1 cells, HIV-1 latently infected cells in which <1 in 50 uninduced cells expresses HIV-1 RNAs (17). OM10.1 cells were treated with TNFα to induce production of full-length viral transcripts and infectious virus particles. Transcription complexes including capped viral mRNA were immunoprecipitated with α-CTD Ser-2 (H5), α-CTD Ser-5 (H14), or α-2,2,7-trimethylguanosine antibody (26). Results shown in Fig. 6 demonstrated that TNFα activation of OM10.1 cells resulted in an increase of Ser-5 phosphorylation of RNAP II CTD and capped transcripts at the HIV-1 promoter. However, no change was observed in Ser-2 phosphorylation upon TNFα activation. Importantly, no increase of Ser-5 phosphorylation or capped transcripts was observed at the promoter proximal region of a cellular gene, GAPDH, following TNFα treatment. The results show that an increase in capped transcripts at the HIV-1 promoter in vivo is associated with Ser-5 phosphorylation of the RNAP II CTD.
Fig. 6.
Increase of capped HIV-1 transcripts coupled with accumulation of Ser-5 phosphorylation in infected cells. OM10.1 cells were treated with TNFα to induce virus production and then subjected to ChIP assays. Transcription complexes were immunoprecipitated with antibody α-CTD H5 or H14 or α-2,2,7-trimethylguanosine, and recovered DNA was used for PCR amplification of the HIV-1 and GAPDH promoter regions.
Discussion
The mRNA capping reactions occur cotranscriptionally, and the phosphorylated CTD of RNAP II provides key molecular contacts with these capping reactions throughout transcriptional elongation. TFIIH and P-TEFb are two principal CTD kinases during HIV-1 transcription (12, 13). In the HIV-1 PICs, TFIIH phosphorylates Ser-5 of RNAP II CTD whereas P-TEFb phosphorylates Ser-2 of RNAP II CTD (12). After TFIIH is released from TECs between +14 and +36, CDK9 is activated and induces Tat/TAR-dependent phosphorylation of the RNAP II CTD (13). In vivo crosslinking experiments in budding yeast showed that recruitment of CEs to the 5′ ends of nascent transcripts requires Kin 28, the kinase subunit of TFIIH that phosphorylates Ser-5 (27, 28). Moreover, the mammalian GT is activated allosterically by binding to Ser-5-phosphorylated heptads (10, 29). The results presented here suggest that Tat-independent TFIIH-mediated Ser-5 phosphorylation of RNAP II CTD induces a basal level of mRNA cap formation. Following release of TFIIH from TECs, stimulation of mRNA cap formation is enhanced by the sequential Tat/TAR-dependent phosphorylation of RNAP II CTD by P-TEFb. Synergistic effects of RNAP II CTD phosphorylation sequentially by TFIIH and Tat-induced P-TEFb during Tat-activated transcription promotes enhanced capping of nascent viral mRNA. It will be of interest to determine whether different heptad repeats are also sequentially phosphorylated.
It is possible that transcription elongation factor SPT5 plays an important role in the cotranscriptional process of mRNA cap formation during Tat-activated HIV-1 transcription, as SPT5 has been shown to stimulate mRNA capping (29). Recently, it has been reported that Tat directly interacts with mammalian CE and stimulates HIV-1 mRNA capping (25, 30). The results presented here suggest that the association of P-TEFb with Tat/TAR during transcription elongation induces CDK9 to phosphorylate RNAP II CTD and other RNAP II-associated proteins, such as SPT5 and Tat-SF1, not only leading to the transition from nonprocessive to processive transcription but also stimulating cotranscriptional mRNA capping.
The coupling of mRNA capping and Tat/TAR-dependent Ser-5 phosphorylation of the RNAP II CTD is further demonstrated by the localized concentration of the cap structure and RNAP II phosphorylated at CTD Ser-5 in the promoter proximal region of the integrated HIV-1 gene (Fig. 6). The concentration of promoter proximal transcription factors and a relative paucity of RNAP II in the downstream regions of the gene are consistent with the immobilized gene expression factory model (4). It is intriguing that the processive transcription induced by HIV-1 Tat is centered on stimulation of the essential steps in transcript stabilization and elongation, based on its ability to activate P-TEFb-mediated Ser-5 phosphorylation of RNAP II CTD. Therefore, selective inhibition of Tat/TAR-dependent P-TEFb kinase could be an effective target for developing antiviral drugs.
Acknowledgments
We thank Dr. Danny Reinberg for purified TFIIH and RNAP II, Chun Chu for affinity-purified α-CE, and Dr. Priya Srinivasan for HCE. M.Z. was supported by the King Fahd endowment to George Washington University.
Abbreviations: TAR, transactivation response; RNAP II, RNA polymerase II; CTD, C-terminal domain; TFIIH, transcription factor IIH; CE, capping enzyme; HCE, human CE; PIC, preinitiation complex; TEC, transcription elongation complex; GT, guanylyltransferase; ChIP, chromatin immunoprecipitation; TNFα, tumor necrosis factor α.
References
- 1.Furuichi, Y. & Shatkin, A. J. (2000) Adv. Virus Res. 55 135–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shuman, S. (2001) Prog. Nucleic Acid Res. Mol. Biol. 66 1–40. [DOI] [PubMed] [Google Scholar]
- 3.Yue, Z., Maldonado, E., Pillutla, R., Cho, H., Reinberg, D. & Shatkin, A. J. (1997) Proc. Natl. Acad. Sci. USA 94 12898–12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCracken, S., Fong, N., Rosonina, E., Yankulov, K., Brothers, G., Siderovski, D., Hessel, A., Foster, S., Shuman, S. & Bentley, D. L. (1997) Genes Dev. 11 3306–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho, E. J., Takagi, T., Moore, C. R. & Buratowski, S. (1997) Genes Dev. 11 3319–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho, C. K., Sriskanda, V., McCracken, S., Bentley, D., Schwer, B. & Shuman, S. (1998) J. Biol. Chem. 273 9577–9585. [DOI] [PubMed] [Google Scholar]
- 7.Kobor, M. S. & Greenblatt, J. (2002) Biochim. Biophys. Acta 1577 261–275. [DOI] [PubMed] [Google Scholar]
- 8.Bentley, D. (2002) Curr. Opin. Cell Biol. 14 336–342. [DOI] [PubMed] [Google Scholar]
- 9.Proudfoot, N. J., Furger, A. & Dye, M. J. (2002) Cell 108 501–512. [DOI] [PubMed] [Google Scholar]
- 10.Ho, C. K. & Shuman, S. (1999) Mol. Cell 3 405–411. [DOI] [PubMed] [Google Scholar]
- 11.Komarnitsky, P., Cho, E. J. & Buratowski, S. (2000) Genes Dev. 14 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou, M., Halanski, M. A., Radonovich, M. F., Kashanchi, F., Peng, J., Price, D. H. & Brady, J. N. (2000) Mol. Cell. Biol. 20 5077–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou, M., Nekhai, S., Bharucha, D. C., Kumar, A., Ge, H., Price, D. H., Egly, J. M. & Brady, J. N. (2001) J. Biol. Chem. 276 44633–44640. [DOI] [PubMed] [Google Scholar]
- 14.Cheng, C. & Sharp, P. A. (2003) Mol. Cell. Biol. 23 1961–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morillon, A., O'Sullivan, J., Azad, A., Proudfoot, N. & Mellor, J. (2003) Science 300 492–495. [DOI] [PubMed] [Google Scholar]
- 16.Price, D. H. (2000) Mol. Cell. Biol. 20 2629–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butera, S. T., Roberts, B. D., Lam, L., Hodge, T. & Folks, T. M. (1994) J. Virol. 68 2726–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy, R., Adamczewski, J. P., Seroz, T., Vermeulen, W., Tassan, J. P., Schaeffer, L., Nigg, E. A., Hoeijmakers, J. H. & Egly, J. M. (1994) Cell 79 1093–1101. [DOI] [PubMed] [Google Scholar]
- 19.Trigon, S., Serizawa, H., Conaway, J. W., Conaway, R. C., Jackson, S. P. & Morange, M. (1998) J. Biol. Chem. 273 6769–6775. [DOI] [PubMed] [Google Scholar]
- 20.Zawel, L., Kumar, K. P. & Reinberg, D. (1995) Genes Dev. 9 1479–1490. [DOI] [PubMed] [Google Scholar]
- 21.Chen, D. & Zhou, Q. (1999) Mol. Cell. Biol. 19 2863–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yankulov, K. & Bentley, D. (1998) Curr. Biol. 8 R447–R449. [DOI] [PubMed] [Google Scholar]
- 23.Isel, C. & Karn, J. (1999) J. Mol. Biol. 290 929–941. [DOI] [PubMed] [Google Scholar]
- 24.Rounseville, M. P. & Kumar, A. (1992) J. Virol. 66 1688–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiu, Y. L., Ho, C. K., Saha, N., Schwer, B., Shuman, S. & Rana, T. M. (2002) Mol. Cell 10 585–597. [DOI] [PubMed] [Google Scholar]
- 26.Moteki, S. & Price, D. (2002) Mol. Cell 10 599–609. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez, C. R., Cho, E. J., Keogh, M. C., Moore, C. L., Greenleaf, A. L. & Buratowski, S. (2000) Mol. Cell. Biol. 20 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroeder, S. C., Schwer, B., Shuman, S. & Bentley, D. (2000) Genes Dev. 14 2435–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen, Y. & Shatkin, A. J. (1999) Genes Dev. 13 1774–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiu, Y. L., Coronel, E., Ho, C. K., Shuman, S. & Rana, T. M. (2001) J. Biol. Chem. 276 12959–12966. [DOI] [PubMed] [Google Scholar]