Abstract
Cell-free genetic circuit elements were constructed in a transcription–translation extract. We engineered transcriptional activation and repression cascades, in which the protein product of each stage is the input required to drive or block the following stage. Although we can find regions of linear response for single stages, cascading to subsequent stages requires working in nonlinear regimes. Substantial time delays and dramatic decreases in output production are incurred with each additional stage because of a bottleneck at the translation machinery. Faster turnover of RNA message can relieve competition between genes and stabilize output against variations in input and parameters.
Cell-free expression systems offer an alternative to in vivo protein synthesis and present numerous advantages: gene and polymerase concentrations can be controlled, reporter measurements are quantitative, and a large parameter space can be studied. Cell-free systems based on cell extracts are continuously being optimized for long-lived synthesis and high yield (1–5). They are being adapted to high-throughput methodologies, large-scale protein production, and in vitro evolution of proteins (6–8). Recently, a cell-free transcription–translation system was reconstituted from almost one hundred purified components (9).
Since the discovery of the lac operon (10), transcriptional regulatory elements have been uncovered and intensively studied (11–16). In addition, synthetic genetic circuits, such as a flip-flop switch and an oscillator, were introduced into cells in a forward engineering approach (17–20).
We were motivated to construct functional in vitro circuit elements that would be amenable to a detailed investigation difficult to achieve in vivo. More generally, we posed the question: is it possible to assemble simple regulatory elements within a cell-free expression system, as a prelude to more complex synthetic circuits? We chose a system of in vitro transcription–translation based on wheat germ extracts. Transcription is performed by bacteriophage T7, SP6, and Escherichia coli RNA polymerase (RNAP) enzymes. The following genes were used and subcloned into vectors: firefly luciferase (luc), enhanced GFP (egfp), T7RNAP, SP6RNAP, sigma factor F (rpoF gene), E. coli lacI repressor. Circuit elements were realized by embedding interaction schemes between the genes such that the protein expressed from one vector affects transcription from another vector.
We use the formalism of electrical network: input, output, gain, and cascading stages. Input/output response functions are measured with RNAP input and reporter gene output. Plasmid concentrations are set as “parameters.” Genes with the same promoter define a stage. We first measured the response functions of single genes. We then built two- and three-stage cascades and mapped the input/parameter space in which they function. Finally, a three-gene, inducible system was constructed in which both the RNAP that transcribes the reporter gene and a repressor that prevents transcription were produced in situ.
Materials and Methods
General. Phosphocreatine, creatine phosphokinase, and polyguanylic acid 5′ [poly(G)] were purchased from Sigma. DNA primers were provided by Operon Technologies (Alameda, CA). E. coli RNA polymerase core enzyme was purchased from Epicentre Technologies (Madison, WI).
Coupled Transcription–Translation. Reactions were carried out in a commercial cell-free wheat germ extract and T7/SP6 RNA polymerase system (TNT-T7/SP6, Promega). To prolong expression from 3 to 6 h, the reaction was supplemented with 30–40 mM phosphocreatine, 1–2 mM magnesium acetate, and 0.15–0.45 mg/ml creatine phosphokinase (short-lifetime messenger extract, SLM). Long-lifetime messenger extract (LLM) was obtained by adding to SLM extract 0.075 mg/ml (final concentration) poly(G) (21). T7 and SP6 RNA polymerases were provided with the kit, and their concentrations were estimated at ≈1 μM by Bradford assay.
Plasmids. Cloning was performed by routine procedures (22). The T7-luc plasmid was provided with the wheat germ extract kit. T7-egfp plasmid was obtained as follows. The sequence of egfp was amplified by PCR from the plasmid pEGFP-N1 (Clontech), and the PCR product was inserted into the BamHI and SacI restrictions site of plasmid T7-luc. For the plasmids SP6-egfp and SP6-luc, the egfp and luc sequences from the plasmid T7-egfp and T7-luc were inserted into the BamHI and SacI restriction sites of the pSP64-poly(A) plasmid (Promega). The plasmid Ptar-luc was then obtained in one step: the E. coli tar promoter (23) was inserted into the sites NheI and BamHI of plasmid SP6-luc. Two vectors were used to construct two different plasmids T7/lacO-luc with equal expression: pET21(+) (Novagen) and pIVEX2.3d (Roche Applied Science). The luc gene was amplified by PCR from the plasmid T7-luc, the PCR product was inserted into the BamHI and XhoI restriction sites of the vector pET21(+). Plasmid pIVEX2.3d-lacO-luc was constructed in two steps. The T7/lacO sequence was inserted into the BglII and XbaI restriction sites of the vector pIVEX2.3d. The luc sequence was amplified by PCR from the plasmid T7-luc and inserted into the NcoI and SacI restriction sites of the vector pIVEX2.3d-lacO. For plasmid SP6-T7rnap, the T7 RNA polymerase coding sequence was amplified by PCR from E. coli strain BL21(DE3), the PCR product was inserted into HindIII and BamHI restriction sites of plasmid pSP64-poly(A). For plasmid T7-SP6rnap, the SP6 RNA polymerase coding sequence was amplified by PCR from the SP6 bacteriophage and inserted into the BamHI and SacI restriction sites of the plasmid T7-luc. The gene rpoF was amplified by PCR from E. coli and inserted into the BamHI and SacI restriction sites of plasmid pSP64-poly(A) to give the plasmid SP6-rpoF. For the constructions of T7-lacI and SP6-lacI, lacI gene was amplified from plasmid pET21(+) by PCR. The GTG codon was changed to ATG. The PCR product was inserted into the BamHI and SacI restriction sites of plasmids T7-luc and pSP64-poly(A). DNA quantifications were done by using the Picogreen kit reagent (Molecular Probes).
Luciferase Luminescence and EGFP Kinetics. Luminescence was measured with a photon multiplier tube (Electron Tube limited, type P10PC) calibrated with purified Firefly Luciferase (Sigma) and Luciferase Assay Reagent (LAR, Roche Applied Science). Data acquisition was done by a PC counter board with labview interface (National Instruments, Austin, TX). Luminescence was measured at the end of reactions, after 6 h of incubation. Two microliters of sample were mixed with 18 μl of LAR and measured immediately thereafter. For real-time kinetics of luciferase, 2 μl of LAR were added to 18 μl of translation reaction. Time courses of EGFP expression were done on an Olympus IX70 inverted microscope equipped with a 75 W Xenon lamp and the proper set of filters. Fluorescence was collected through a ×40 objective and amplified with a photon multiplier tube (Hamamatsu, GaAsP H7421–40). Signals were acquired with a PC as previously described for bioluminescence. A silicone chamber was made as follows: a hole of 3-mm diameter was punched in a 5-mm-thick PDMS layer (Poly-DiMethylSiloxane, Sylgard 184, Corning) then stuck on glass coverslip. Ten microliters of the reaction mixture were added to the bottom of the well and closed with a glass coverslip.
mRNA Saturation Level. Transcript saturation level in the cell extract was measured by incubating varying amounts of Luc mRNA (Promega) in LLM. Luc production measured after 30 min indicated no increase with mRNA levels >20 nM, which defines the concentration of mRNA above which translation machinery saturates.
ATP Concentration Measurement. ATP concentration in the extract during expression was measured with an ATP bioluminescence assay kit (Sigma). Expression ends concomitantly with ATP decrease from 1.5 to 1.2 mM.
Results and Discussion
Cell-Free Extract Expression Boosting. Protein expression reactions were carried out in batch mode, without continuous exchange of nutrients and byproducts, in a volume of 10 μl at room temperature (25°C). Firefly luciferase (Luc) and EGFP were the reporter proteins. We found that expression in the commercial wheat germ extract is low: protein synthesis stops after 3 h at a maximum reporter production of 100 nM (Fig. 1A). Two modifications enhance the protein production and prolong the time course of synthesis. First, polyguanylic acid 5′ [poly(G)] boosts the protein production 5-fold by increasing the messenger RNA lifetime (21). The average lifetime of transcripts increases from 20–30 min in the nonmodified extract to ≈2 h by addition of poly(G). Hereafter, long and short lifetime messenger extracts are denoted LLM and SLM, respectively. In agreement with published results, we found that the end of expression was concomitant with the decrease of the ATP level in the extract (Fig. 1B, refs. 24–26). In agreement with the manufacturer, the initial 1.5 mM ATP concentration was found to be optimal, and addition of ATP or other nucleotides does not improve protein production. Adjustment of creatine phosphate concentration for ATP regeneration and adjustment of magnesium concentration extend the expression for up to 6 h, doubling both the time of synthesis and the total production (Fig. 1 A). With such modifications, up to 1 μM Luc can be synthesized in 6 h. We have measured that 20–100 nM transcript concentration can be processed simultaneously in the extract; below this value, the expression is linearly proportional to the concentration of template; above this value, the translation machinery is saturated; at 300 nM transcripts, the expression decreases by a factor of 100; at 500 nM transcripts, expression decreases by a factor of 1,000. We do not know whether an excess messenger inhibits translation initiation. Variation of amino acids and transfer RNA concentrations did not bring any improvement of expression. Plasmids are stable in the wheat germ system, only 20% of their activity is lost after 4 h in the extract. By decreasing 10 times the concentration of the input RNA polymerase, the expression lasts 3 h instead of 6 h, suggesting that the enzyme degrades slowly. Finally, with continuous translation systems, it has been shown that wheat germ extract can maintain expression for tens of hours (5); it is thus unlikely that degradation of the translation machinery limits the production time of our system.
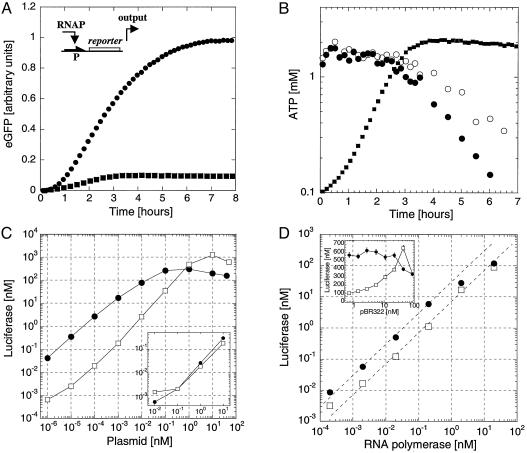
Fig. 1.
One-stage network. (A) A reporter gene (luc or egfp, promoter P) is expressed in the coupled transcription/translation T7/SP6 RNA polymerase wheat germ extract. Kinetics of expression of EGFP in LLM extract (0.1 nM SP6-egfp, filled circles) and SLM extract (0.5 nM T7-egfp, filled squares) with 20 nM T7 RNA polymerase. Three phases are observed: a lag phase of 10 min, an accumulation of synthesized EGFP, and a slowing down to a plateau. The initial lag phase is for transcription and translation. In the SLM extract, expression stops after 3 h, whereas, in the LLM extract, EGFP keeps accumulating beyond 6 h. (B) ATP concentration measurement in the SLM extract. With 20 nM T7 RNA polymerase and 0.5 nM T7-eGFP plasmid (filled circles) and without expression (open circles). The kinetics of expression of EGFP in SLM extract from A has been superimposed (gray filled squares). (C) Luc production in LLM extract measured after 6 h as a function of T7-luc plasmid concentration (filled circles) and SP6-Luc (open squares) with 20 nM RNA polymerase. (Inset) endogenous expression from the extract without addition of T7 or SP6 RNA polymerase as a function of the T7-luc (filled circles) and SP6-luc (open squares) plasmids. (D) Luc production in LLM extract as a function of T7 or SP6 RNA polymerase at 0.1 nM plasmid concentration, T7-luc (filled circles), and SP6-luc (open squares). (Inset) Luc production in LLM extract as a function of neutral plasmid pBR322 concentration, 20 nM RNA polymerase, 0.1 nM T7-luc plasmid (filled circles), and 0.1 nM SP6-luc (open squares).
Single-Stage Response Characteristics. The T7-luc and SP6-luc plasmids were used to determine the expression properties in the LLM extract. These constructs are the simplest circuit elements, with RNA polymerase as input, protein production as output, and plasmid concentration as parameter. In the first experiment, the maximum yield, the limits of detection, and the response functions were determined by varying plasmid concentration at a fixed polymerase concentration of 20 nM for both T7 and SP6, the concentration recommended by the extract manufacturer (Fig. 1C). Expression saturates at high plasmid concentrations to give a maximum yield of 1 μM Luc. We are able to detect Luc production from plasmid concentrations as low as femtomolar. Endogenous RNA polymerase activity in the wheat germ extract is similar for both T7-luc and SP6-luc plasmids as measured in the absence of input RNA polymerase (Fig. 1C Inset). For the SP6 system, production is less efficient at low plasmid concentrations than in the T7 system, but the maximum yield at higher plasmid concentrations is greater (Table 1). We measured how production is affected by the presence of a “neutral” plasmid (Fig. 1D Inset). SP6 RNA polymerase activity, but not T7 RNA polymerase activity, is enhanced in the presence of nonspecific DNA, which may also explain why SP6 is less efficient than T7 at low plasmid concentration (Fig. 1C). SP6 may be more sensitive to the presence of proteins bound nonspecifically to the plasmid that hinder transcription, and the addition of plasmid (neutral or template) may disperse these proteins and clear up transcription.
Table 1. Characteristics of the cascading networks used in this study.
| No. of stages | Cascade | Time delay, min | Maximum Luc production, nM |
|---|---|---|---|
| 1 | T7-luc | 15 | 500 |
| SP6-luc | 2,000 | ||
| 2 | T7-SP6rnap → SP6-luc | 60-90 | 100 |
| SP6-T7rnap → T7-luc | 100 | ||
| 3 | T7-SP6rnap → SP6-σ28 → Ptar-luc | 180 | 1 |
To further characterize expression from single genes, we studied the input/output relations at fixed parameter (plasmid) concentrations (Fig. 1D). We searched for a regime in which the gain, defined as the output per input per unit plasmid concentration, is constant. The T7 system is colinear below 0.1 nM RNA polymerase input and below 0.1 nM T7-luc with a maximum gain of 300 Luc proteins per RNA polymerase molecule per plasmid molecule. Although at 20 nM RNAP, the SP6 system is linear with plasmid concentration in the range of 10–4 to 1 nM (Fig. 1C), there is no colinear regime with respect to the two variables. We found a maximum gain of 400 at 5 nM plasmid below 0.2 nM SP6 RNAP. The maximum output/input ratio was 50 for the T7 system (10 nM Luc at 0.2 nM T7 RNAP and 1 nM T7-luc) and 2,000 for the SP6 system (400 nM Luc at 0.2 nM SP6 RNAP and 5 nM SP6-luc). Finally, we tested expression from the T7-luc plasmid in the presence of SP6 RNAP and vice versa. We found that T7 and SP6 are orthogonal systems, in that the mutual leaks are at the level of endogenous expression from the extract alone. The expression from T7-luc and SP6-luc with both polymerases equals the sum of expression from the separate systems (data not shown).
Cascaded Circuit Elements. For the construction of in vitro genetic circuits, one would like to concatenate single-gene elements in analogy to electrical circuits. Ideally, we would like to design genetic circuits such that the total gain for a series of concatenated elements is the product of the individual gains for each element, as achievable in linear electrical amplifiers. An obvious difference between genetic and electrical circuits, however, is that input and output of electrical circuits are both voltages and are established instantaneously, whereas genetic circuit elements convert input into a molecularly distinct output with an inevitable time delay. Furthermore, the battery maintaining the electrical circuit is not depleted, whereas in vitro and in batch mode, addition of stages in genetic circuits drains the limited protein synthesis resources. For these reasons, multiplicity of gain cannot be achieved in our system but could be in a continuous one. Nevertheless, we demonstrate successful concatenation of genetic circuit elements in vitro. As described below, we were forced to work in the nonlinear regime to obtain sufficient output to cascade through two or three stages.
Implementation of two- and three-stage cascaded elements is presented in Fig. 2. A two-stage cascade was constructed with the plasmids T7-SP6RNAP and SP6-luc. The T7 RNAP input drives production of SP6 RNAP, which in turn induces expression of Luc from the SP6-luc plasmid. Because SP6 RNAP accumulates slowly (Fig. 1 A), Luc production is detectable only after a long delay of 1.5 h (Fig. 2 A). The system cascades only above 0.1 nM input T7 RNAP. For various T7-SP6RNAP and SP6-luc plasmid concentrations tested, Luc output is reduced by at least one order of magnitude as compared with the single-gene elements (Table 1). For example, 130 nM Luc was produced from 20 nM input T7 RNAP, 0.5 nM T7-SP6RNAP, and 10 nM SP6-luc plasmids. The transcription leak of T7 RNAP on the SP6 promoter is significantly lower than the total output of this two-stage cascade (Fig. 2 A). In contrast, the leaks become significant for a three-stage cascade.
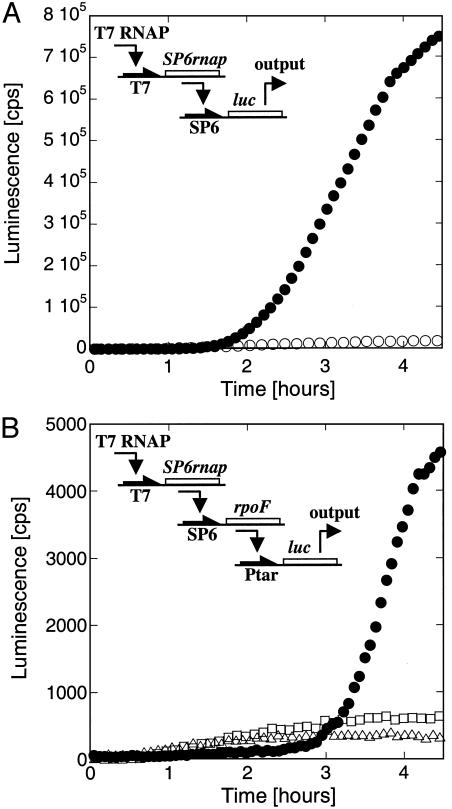
Fig. 2.
Two and three-stage networks in LLM extract. (A) The SP6 RNA polymerase gene is transcribed by the T7 RNA polymerase. The synthesized SP6 RNA polymerase transcribes the luc gene. Kinetics of expression of the cascade with 20 nM input, 0.5 plasmid T7-SP6rnap, and 2 nM plasmid SP6-luc (filled circles), or 2 nM SP6-luc plasmid only (open circles). (B) The SP6 RNA polymerase gene is transcribed by the T7 RNA polymerase. The SP6 RNA polymerase transcribes the rpoF gene (E. coli sigma factor F), and the synthesized sigma F interacts with the E. coli core enzyme (added to the extract) to induce the transcription of luc gene. To test the kinetics of Luc synthesis: the three-step transcriptional cascade introduces a time delay of 3 h in Luc synthesis (filled circles: 5 nM T7-SP6rnap, 5 nM SP6-rpoF, and 0.5 nM Ptar-luc plasmids); as a control, in the absence of the first or the first two stages, the luc gene is transcribed from the leak of both the T7 RNA polymerase and the E. coli RNA polymerase core enzyme (open triangles, 0.5 nM Ptar-luc plasmid; open squares, 5 nM SP6-rpoF and 0.5 nM Ptar-luc plasmids).
We realized a three-stage transcriptional cascade with the plasmids T7-SP6RNAP, SP6-rpoF, and Ptar-luc. In addition, T7 RNAP and the E. coli RNAP core enzyme were added as input (Fig. 2B). SP6 RNAP is synthesized first and induces the expression of the rpoF gene encoding the E. coli sigma factor F (σF). The σF interacts with the E. coli core enzyme to induce the expression of Luc via the promoter of the E. coli tar gene (23). The LLM extract was used to ensure sufficient protein production. Because of the low output level and the presence of three different RNA polymerases and promoters, transcription leaks become important in this construction. Parameters were scanned, and a working point was found only at a low concentration of input T7 RNAP (2 nM) and a high first-stage plasmid (T7-SP6RNAP) concentration (5 nM). Under these conditions, we found that leaks are reduced and endogenous expression, although increased, is still significantly lower than the output. Elsewhere in the parameter space, the leaks were dominant. As shown in Fig. 2B, with the three plasmids, Luc production appeared sharply only after ≈3 h. A low yield of 1 nM Luc was obtained, which was nevertheless 15 times higher than production from leaks. When the first-stage gene was removed and T7 RNAP was added, expression due to leaks appeared after only 30 min because the T7 RNAP, in the absence of its own promoter, was available for interaction with other promoters. We predict that with one or two more stages, no working points would be found. In our system, each new stage reduces the yield by at least an order of magnitude (Table 1). We hypothesized that the severe decrease in Luc output with each additional stage is caused by a bottleneck at the translation machinery.
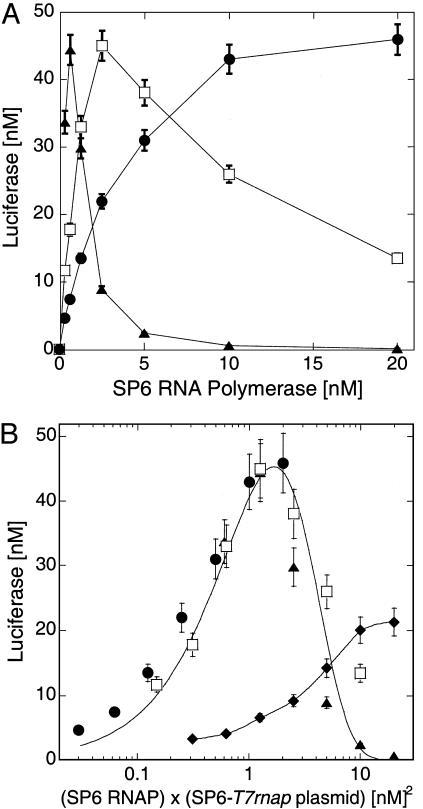
Sharing of Translation Resources. To understand better the properties of the translation machinery and its limitations, we compared synthesis in SLM and LLM for a two-gene transcriptional cascade. We inverted the cascade presented in Fig. 2 A: the T7 RNAP is synthesized first and then activates expression of Luc via the SP6-luc plasmid. Here too, there is a time delay of ≈1 h for Luc synthesis (not shown). In the LLM extract, we varied the SP6 RNAP input concentration and measured Luc output. Increasing the SP6-T7RNAP plasmid concentration induced a dramatic narrowing of the SP6 RNAP concentration band over which Luc was produced (Fig. 3A). This observation demonstrates that, in the LLM extract, information transfer is sensitive to parameter variation. A maximum output was obtained for the same value of the product of SP6 RNAP and the SP6-T7RNAP plasmid concentrations (Fig. 3B), indicating that there is a value of first stage transcription rate at which protein production can optimally cascade. Indeed, all of the data collapse onto a single curve as a function of this product (Fig. 3B). Above the first-stage transcription rate that maximizes Luc production, LLM extract operates in sharing mode such that overproduction of T7RNAP mRNA occupies the translation machinery and inhibits luc mRNA translation. In contrast, Luc production in the SLM extract, with mRNA turnover ≈5-fold faster than in the LLM extract, showed no fall-off in the range of parameters investigated (Fig. 3B). Without rapid turnover, messages are “stored” and saturate the translation machinery. Presumably, SLM extract could enter sharing mode as well, if more genes were added to compete for translation resources.
Fig. 3.
Two-stage network, sharing of resources. The T7 RNA polymerase gene is transcribed by SP6 RNA polymerase. The synthesized T7 RNA polymerase transcribes the luc gene in LLM extract. Expression of the cascade is measured after 6 h. (A) Luc output as a function of SP6 RNA polymerase input in LLM extract with 1 nM T7-luc plasmid and different concentrations of SP6-T7rnap plasmid: 0.1 nM (filled circles), 0.5 nM (open squares), 2 nM (filled triangles). (B) Input–output curves of A (in LLM extract) collapse onto one curve when plotting data as a function of (SP6-T7rnap plasmid) × (SP6 polymerase) (symbols as in A). A monotonous response is observed in SLM extract (filled diamonds), 1 nM SP6-T7rnap and T7-luc plasmid. Solid lines are smoothing fits.
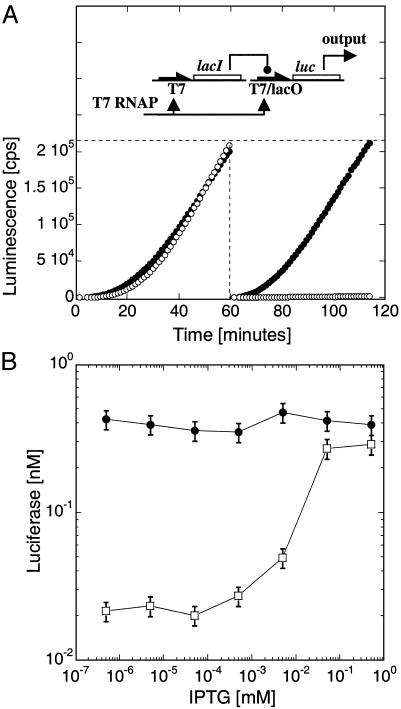
Negative Regulatory Elements. To develop synthetic circuits with diverse and useful features, we must include elements of negative as well as positive regulation. To introduce a negative regulatory element, the repressor gene lacI was cloned under the T7 promoter, whereas expression of the reporter protein Luc was controlled by the T7/lacO promoter/operator system (ref. 27 and Fig. 4A Upper). The LacI repressor bound to its target lacO acts as a “roadblock” for transcription of the downstream luc gene. LacI binding to DNA in vivo is cooperative, requiring dimers and tetramers. Tight repression occurs with a 40 nM protomer concentration, necessitating a basal level of repressor in our cell-free system. This basal repressor concentration was obtained by incubating the T7-lacI plasmid for 1 h in the extract before adding the T7/lacO-luc plasmid (Fig. 4A Right). We measured 40-fold repression, which could be entirely lifted with 100 μM isopropyl β-d-thiogalactoside (IPTG), a lactose analog that inhibits repressor binding to DNA (Fig. 4B). No repression was observed without the preincubation (Fig. 4A Left), because Luc was synthesized before LacI could reach its critical concentration for effective DNA binding.
Fig. 4.
T7/lacO LacI repression. (A) E. coli LacI repressor under the T7 promoter represses the plasmid T7/lacO-luc [pET21(+)-luc] by binding its recognition site, lacO, located downstream of the T7 promoter. Kinetics of coexpression of T7-lacI with T7-luc (filled circles) or T7/lacO-luc (open circles), without (Left) or with (Right) 1 h of preincubation of the plasmid T7-lacI, expression in LLM extract with 0.1 nM of each plasmid. (B) Induction of Luc synthesis as a function of IPTG concentration after 1 h of preincubation of T7-lacI plasmid, followed by 1 h of coexpression with T7/lacO-luc (open squares) or T7-luc (filled circles) (0.5 nM T7-lacI plasmid and 0.1 nM of either T7-luc or T7/lacO-luc plasmids).
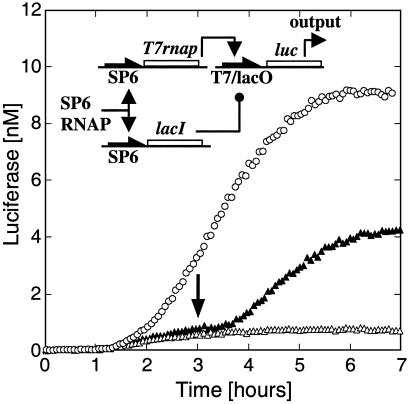
A Three-Gene Circuit. Our next goal was to engineer a functional circuit by using positive and negative regulatory elements. The circuit was constructed with the plasmids SP6-T7RNAP, SP6-lacI and T7/lacO-luc (Fig. 5). Both T7 RNAP and LacI repressor are synthesized in parallel. Expression of Luc at the second stage is then determined by T7 activation versus LacI repression. The function of the circuit, which we aimed to optimize, was the amplitude of induction. To have sufficient protein production for all genes, the experiment was carried out in the LLM extract. To maintain information flow and repress efficiently at the same time, the most critical parameters were the ratio and the total concentration of plasmids participating in the first stage. The competition between transcriptional activation and repression could be used as a control parameter to bias one tendency or the other by tuning the ratio between SP6-T7RNAP and SP6-lacI plasmids in the extract. We found a 20-fold maximum repression with 10-fold more plasmid coding for the repressor. With IPTG in the extract, Luc was synthesized continuously for 6 h to yield 10 nM, whereas without IPTG, synthesis was repressed (Fig. 5). Induction after 3 h of incubation lifted the repression, demonstrating the ability to switch on Luc synthesis at will. Late induction resulted in a 2-fold lower expression level than induction at the outset, because expression stops after 6 h.
Fig. 5.
Three-gene circuit. A three-gene circuit with plasmids SP6-lacI, SP6-T7rnap, and T7/lacO-luc (pIVEX2.3d-lacO-luc). Coexpression of the three genes with (open circles) and without (open triangles) IPTG at t = 0 (expression in LLM extract, 0.005 nM SP6-T7rnap, 0.1 nM SP6-lacI and 0.5 nM T7/lacO-luc plasmids). IPTG (0.5 mM) was added after3hto induce Luc production (arrow, filled triangles).
Summary and Conclusions
We constructed one-, two-, and three-stage cascades of in vitro gene expression to illustrate principles of cell-free genetic circuit assembly. Linear regimes were found for single-gene systems, but sequential expression from genes in series required working in nonlinear regimes. The final protein output dropped by at least one order of magnitude and a delay of ≈1.5 h was introduced for each additional gene in the series. An optimal transcription rate for the first stage of a two-stage cascade was found. This optimum, which generates sufficient output from the first stage to induce transcription at the second stage, while reserving translation machinery to process message from the second stage, appears to be a function of messenger lifetime in the cell-free extract. Finally, we assembled a three-gene circuit that uses the lac system to externally control the induction of gene expression.
Most applications of cell-free protein expression have so far been directed toward maximizing protein synthesis and therefore focused on stability of mRNA and reduction of nuclease activity (28). We have shown that engineering in vitro genetic circuits using cell-free expression systems requires a different optimization approach, in particular for the bookkeeping of cell extract resources. Rapid turnover of mRNA is required to avoid saturation of the translation machinery, which is the bottleneck for coordinated synthesis in an extract with finite resources. Moreover, the constraints imposed by batch-mode cell-free expression may be relaxed in continuous mode or partly avoided by circuit designs that implement gene autoregulation. Protein lifetime is an important issue that we did not touch, especially for reconstitution of oscillatory networks (17).
Acknowledgments
We thank D. Fass, S. Liu, G. Bonnet, M. Levine, R. Golsteyn, D. Thaler, T. Tlusty, and D. Braun for useful suggestions. The SP6 bacteriophage was provided by I. J. Molineux (University of Texas, Austin). V.N. and R.B.-Z. were supported by the Burroughs Wellcome Foundation.
Abbreviations: RNAP, RNA polymerase; EGFP, enhanced GFP; IPTG, isopropyl β-d-thiogalactoside; LLM, long-lifetime messenger extract; SLM, short-lifetime messenger extract.
References
- 1.Roberts, B. E. & Paterson, B. M. (1973) Proc. Natl. Acad. Sci. USA 70 2330–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, C. W., Straus, J. W. & Dudock, B. S. (1983) Methods Enzymol. 101 635–644. [DOI] [PubMed] [Google Scholar]
- 3.Erickson, A. H. & Blobel, G. (1983) Methods Enzymol. 96 38–50. [DOI] [PubMed] [Google Scholar]
- 4.Spirin, A. S., Baranov, V. I., Ryabova, L. A., Ovodov, S. Y. & Alakhov, Y. B. (1988) Science 242 1162–1164. [DOI] [PubMed] [Google Scholar]
- 5.Madin, K., Sawasaki, T., Ogasawara, T. & Endo, Y. (2000) Proc. Natl. Acad. Sci. USA 97 559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanes, J. & Pluckthun, A. (1997) Proc. Natl. Acad. Sci. USA 94 4937–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amstutz, P., Forrer, P., Zahnd, C. & Pluckthun, A. (2001) Curr. Opin. Biotechnol. 12 400–405. [DOI] [PubMed] [Google Scholar]
- 8.Sawasaki, T., Ogasawara, T., Morishita, R. & Endo, Y. (2002) Proc. Natl. Acad. Sci. USA 99 14652–14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimizu, Y., Inoue, A., Tomari, Y., Suzuki, T., Yokogawa, T., Nishikawa, K. & Ueda, T. (2001) Nat. Biotechnol. 19 751–755. [DOI] [PubMed] [Google Scholar]
- 10.Jacob, F. & Monod, J. (1961) J. Mol. Biol. 3 318–356. [DOI] [PubMed] [Google Scholar]
- 11.Davidson, E. H. (2001) Genomic Regulatory Systems: Development and Evolution (Academic, San Diego).
- 12.Ptashne, M. & Gann, A. (2001) Genes and Signals (Cold Spring Harbor Lab. Press, Plainview, NY).
- 13.Shen-Orr, S. S., Milo, R., Mangan, S. & Alon, U. (2002) Nat. Genet. 31 64–68. [DOI] [PubMed] [Google Scholar]
- 14.Milo, R., Shen-Orr, S., Itzkovitz, S., Kashtan, N., Chklovskii, D. & Alon, U. (2002) Science 298 824–827. [DOI] [PubMed] [Google Scholar]
- 15.Lee, T. I., Rinaldi, N. J., Robert, F., Odom, D. T., Bar-Joseph, Z., Gerber, G. K., Hannett, N. M., Harbison, C. T., Thompson, C. M., Simon, I., et al. (2002) Science 298 799–804. [DOI] [PubMed] [Google Scholar]
- 16.Barkai, N. & Leibler, S. (1997) Nature 387 913–917. [DOI] [PubMed] [Google Scholar]
- 17.Elowitz, M. B. & Leibler, S. (2000) Nature 403 335–338. [DOI] [PubMed] [Google Scholar]
- 18.Gardner, T. S., Cantor, C. R. & Collins, J. J. (2000) Nature 403 339–342. [DOI] [PubMed] [Google Scholar]
- 19.Becskei, A. & Serrano, L. (2000) Nature 405 590–593. [DOI] [PubMed] [Google Scholar]
- 20.Becskei, A., Seraphin B. & Serrano, L. (2001) EMBO J. 20 2528–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen, X. C., Yao, S. L., Fukano, H., Terada, S., Kitayama, A., Nagamune, T. & Suzuki, E. (1999) J. Biotechnol. 75 221–228. [Google Scholar]
- 22.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 23.Arnosti, D. N. & Chamberlin, M. J. (1989) Proc. Natl. Acad. Sci. USA 86 830–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawarasaki, Y., Nakano, H. & Yamane, T. (1994) Biosci. Biotechnol. Biochem. 58 1911–1913. [DOI] [PubMed] [Google Scholar]
- 25.Kawarasaki, Y., Kawai, T., Nakano, H. & Yamane, T. (1995) Anal. Biochem. 226 320–324. [DOI] [PubMed] [Google Scholar]
- 26.Matveev, S. V., Vinokurov, L. M., Shaloiko, L. A., Davies, C., Matveeva, E. A. & Alakhov, Y. B. (1996) Biochim. Biophys. Acta 1293 207–212. [DOI] [PubMed] [Google Scholar]
- 27.Giordano, T. J., Deuschle, U., Bujard, H. & McAllister, W. T. (1989) Gene 84 209–219. [DOI] [PubMed] [Google Scholar]
- 28.Jermutus, L., Ryabova, L. A. & Pluckthun, A. (1998) Curr. Opin. Biotechnol. 9 534–548. [DOI] [PubMed] [Google Scholar]