Abstract
The Rad23 family of proteins, including the human homologs hHR23a and hHR23b, stimulates nucleotide excision repair and has been shown to provide a novel link between proteasome-mediated protein degradation and DNA repair. In this work, we illustrate how the proteasomal subunit S5a regulates hHR23a protein structure. By using NMR spectroscopy, we have elucidated the structure and dynamic properties of the 40-kDa hHR23a protein and show it to contain four structured domains connected by flexible linker regions. In addition, we reveal that these domains interact in an intramolecular fashion, and by using residual dipolar coupling data in combination with chemical shift perturbation analysis, we present the hHR23a structure. By itself, hHR23a adopts a closed conformation defined by the interaction of an N-terminal ubiquitin-like domain with two ubiquitin-associated domains. Interestingly, binding of the proteasomal subunit S5a disrupts the hHR23a interdomain interactions and thereby causes it to adopt an opened conformation.
Nucleotide excision repair (NER) is the major process for the removal of bulky DNA adducts caused by genotoxic agents such as UV radiation. The first step in this process is the recognition of damaged DNA, which is accomplished by a complex of the xeroderma pigmentosum complementation group C (XPC) protein and either of the hHR23 proteins (hHR23a or hHR23b). This protein complex preferentially binds UV-damaged DNA and recruits other components of the NER pathway to the lesion (1). This pathway is crucial for the maintenance of genome integrity. Defects in NER result in the disease xeroderma pigmentosum, which is characterized by severe sensitivity to UV radiation and a 1,000-fold increase in the risk of developing skin cancer (2).
In addition to their well characterized function in NER, the hHR23 proteins play a role in ubiquitin-mediated protein degradation and provide a novel link between these two biologically important pathways. They can bind several components of the ubiquitin–proteasome pathway, including ubiquitin (3) and two different proteasomal subunits (4, 5). One of these subunits, S5a, also recognizes polyubiquitin chains (6, 7) via either of two sequence motifs that are comprised of alternating long and short hydrophobic side chains, and it is the second motif that binds the hHR23 proteins (4). Studies suggest that the hHR23 proteins inhibit the degradation of other proteins by regulating polyubiquitin chain formation (8, 9), and in yeast, Rad23 (which is the hHR23a/b homolog) is reported to inhibit the ubiquitination of Rad4 (which is the XPC homolog) (10, 11). Additionally, hHR23 itself is ubiquitinated and degraded by this pathway (12). Hence these two prominent biological pathways are intricately linked via hHR23. Indeed hHR23 protein function is essential as indicated by the embryonic lethal phenotype of mice that lack both hHR23 genes (13).
To better understand the regulation and function of this biologically important protein family, we have used NMR to elucidate the structure and dynamic properties of the 40-kDa hHR23a protein. We have found that hHR23a contains four structural domains that are connected by flexible linker regions and that the domains interact to form an interesting protein structure. Additionally, we reveal that the proteasomal subunit S5a disrupts the hHR23a protein structure by blocking its interdomain interactions and discuss the functional significance of this induced structural change.
Materials and Methods
Sample Preparation. HHR23a and the UBA1 (C&P Biotech, Thornhill, ON, Canada) and XPC-binding domains (C&P Biotech) were each cloned into the pGEX-6p expression vector for the expression of GST fusion proteins. After expression in BL21(DE3) cells the proteins of interest were purified on glutathione-Sepharose resin and separated from the GST tag by cleavage with PreScission protease. Further purification was achieved by FPLC (Pharmacia), using either Superdex 200 or 75 preparative columns.
Patrick Young (Stockholm University, Stockholm) generously provided us with plasmid PUb2, for expression of S5a (196–307) (14). The ubiquitin-like (UBL) and ubiquitin-associated (UBA) 2 domains were cloned into a pET expression vector in-frame with a polyhistidine tag and expressed in BL21(DE3) cells. These proteins were purified by using affinity chromatography followed by FPLC.
In vitro protein ligation was achieved by using the IMPACT-TWIN protein fusion system (New England Biolabs), which exploits the inducible self-cleavage activity of protein splicing elements (inteins) (15). We expressed separately two hHR23a protein fragments, including 1–117 and 118–363, as fusion proteins with a chitin binding domain and an intein. Residue 118 was mutated from an alanine to a cysteine to enable the in vitro ligation reaction, and the ligated product was purified by FPLC.
We produced labeled samples for NMR spectroscopy, including perdeuterated samples, by growth and expression in M9 minimal media with 15NNH4Cl, 13C glucose, or 2H2O as the only sources of nitrogen, carbon, or water.
NMR Spectroscopy Used to Determine the hHR23a Structure. All spectra were processed by using nmrpipe (16) and visualized in xeasy (17). To assign chemical shift values to atoms within hHR23a, we acquired three pairs of triple resonance experiments and found that >95% of the spin systems identified in the [1H,15N] heteronuclear single quantum coherence (HSQC) spectrum of hHR23a appear in these spectra. The triple resonance experiments included 3D [1H,15N,13C] HNCA (18), [1H,15N,13C] HN(CO)CA (19), [1H,15N,13C] HNCO, [1H,15N,13C] HN(CA)CO (20, 21), [1H,15N,13C] HNCACB (19), and [1H,15N,13C] HN(CO)CACB (19). Spectra were acquired on a Varian INOVA 600-MHz spectrometer.
Distance constraints were obtained by using 15N- and 13C-dispersed NOESY experiments. An 15N-dispersed NOESY experiment was acquired at 800 MHz on a 50% deuterated 15N-labeled sample with a 200-ms mixing time (22). 13C-dispersed NOESY spectra were acquired with an 80-ms mixing time for the aliphatic and aromatic regions on a 13C-labeled sample dissolved in 2H2O. Additional distance constraints involving the aromatic side chains were provided by a 2D NOESY spectrum with an 80-ms mixing time on a sample dissolved in 2H2O. By using the various NOESY spectra, we obtained 4,355 distance constraints that were used directly in our structure calculations. Additionally, we used a program (talos) to obtain backbone torsion angles Φ (C′(i–1)-Ni-Cαi-C′i) and ϕ (Ni-Cαi-C′-Ni+1) for 74 residues (23).
To test for interdomain nuclear Overhauser effect (NOE) interactions involving the UBL domain, we used asymmetric isotope labeling, in combination with isotope editing techniques. We used the in vitro protein ligation protocol described above to produce a protein sample in which only hHR23a (C118–363) is 13C-labeled. A 13C half-filtered NOESY experiment, which contains a 13C purge filter followed by a 13C selection period, was acquired on this sample to test for aliphatic-aliphatic NOEs between the UBL domain and the other domains (24). We also tested for interdomain NOEs involving the UBL domain by producing an hHR23a sample in which only hHR23a (C118–363) is 2H- and 15N-labeled. An 15N-dispersed NOESY experiment on such a sample yields amide to aliphatic NOE crosspeaks that are exclusively interdomain (25). Both experiments were acquired at 800 MHz.
Residual Dipolar Coupling Measurements. Based on the reported success of aligning ubiquitin (26), we produced an hHR23a sample with a liquid crystalline phase at 7.5 mg/ml Pf1 phage in 100 mM NaCl, 20 mM sodium phosphate, pH 6.5. Changes in splitting relative to the isotropic 1JNH values were measured at 600 MHz on 0.55 mM 15N-labeled hHR23a at 25°C by using in-phase antiphase [1H,15N] HSQC experiments (27). These values were then used to determine the relative orientation of each domain by using the program module 1.0 (28). An alignment tensor was calculated for each of the structural domains and a model was generated by applying only translational motions in the common frame.
NMR Dynamics Studies. Rates for 15N longitudinal [RN(NZ)] and transverse [RN(NX)] relaxation and magnitudes of the heteronuclear NOE enhancements were recorded on hHR23a, hHR23a (C118–363), and hHR23a in complex with S5a (196–307). These data were collected at 800 MHz, 25°C, pH 6.5 on 0.5 mM hHR23a, on 0.5 mM hHR23a (C118–363), and on 0.5 mM hHR23a-S5a (196–307) protein complex. In the hHR23a-S5a (196–307) complex, hHR23a was 15N-labeled and fully deuterated, whereas S5a (196–307) was unlabeled. RN(NZ) and RN(NX) were derived by fitting data acquired with different relaxation delays to a single exponential decay function, and error values were determined by repeating one data point. Two sets of spectra were recorded for steady-state NOE intensities, one with a 4-s proton saturation to achieve steady-state intensity and the other as a control spectrum with no saturation to obtain the Zeeman intensity. NOE enhancements were then calculated from the ratio described below in Eq. 1.
 |
[1] |
The data of each domain were analyzed according to model-free formulism to determine a rotational correlation time (τc) (29, 30), using the program modelfree 4.15 (29, 31).
NMR Structure Calculations. Structure calculations were performed based on the NOE distance and dihedral angle restraints described in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org, with the use of simulated annealing (32) in x-plor 3.851 (33) on R12000 Octane Silicon Graphics workstations. A total of 45 random structures were subjected to 120,000 simulated annealing and cooling steps of 0.005 ps. Of these structures, 12 converged with no NOE violation >0.3 Å and no dihedral angle violation >5°.
Results
HHR23a Contains Four Structural Domains. We have solved the structure of the 40-kDa hHR23a protein by using modern solution NMR techniques. The hHR23a protein has four structural domains (Fig. 1A), which are connected by unstructured linker regions. Each of the four structural domains and their secondary structural elements were readily identified by comparing assigned backbone chemical shift values of Cα and carbonyl atoms with those of randomly coiled peptides (34). The two UBA domains and the XPC-binding domain are entirely α-helical, whereas the UBL domain contains β-strands and an α-helix. These results are confirmed by the NOE patterns observed in NOESY spectra.
Fig. 1.
The DNA repair protein hHR23a has four well defined structural domains. (A) The hHR23a protein sequence is provided with the UBL, UBA, and XPC-binding domains shaded in blue, gray, and yellow, respectively. The structures of the UBL (B), UBA1 (C), XPC-binding (D), and UBA2 (E) domains of hHR23a are illustrated with α-helices and β-strands colored red and blue, respectively. The average backbone rms deviations to the mean structure for the UBL (3–78), UBA1 (161–200), XPC-binding (232–282), and UBA2 (317–359) domains are 0.410, 0.530, 0.461, and 0.402, respectively. The linker regions connecting each of these domains are not shown in B–E.
We were able to produce highly refined structures for each of the four domains (Fig. 1). Structures have been previously solved for each of the UBA domains of hHR23a when expressed as single domain constructs (35, 36). We found that these structures are preserved in the context of full-length hHR23a, as their backbone rms deviation is within 1.5 Å to their respective structures in the 40-kDa protein. The UBA domains each contain three α-helices, which span residues S161-S172, R177-S187, and H192-L199 in UBA1 and E320-L330, E334-Y341, and N349-N359 in UBA2. UBA2 is slightly larger in the full-length protein compared with the single domain construct as V316-E319 forms an ordered strand that extends toward α2. This additional strand serves to block hydrophobic residues, including I324 and Y341, from solvent exposure.
The UBL domain is structurally homologous to ubiquitin and belongs to a large family of structurally homologous proteins. We had previously built a model of the hHR23a UBL domain structure based on that solved for another UBL domain, hPLIC-2 (37). Within regions of secondary structural elements, the backbone atoms of the experimental and theoretical structures are within 1.9 Å of each other. The UBL domain contains five β-strands spanning residues V3-L10, Q13-M19, Q46-I49, I54-D57, and K69-M75. The first and last β-strands are oriented parallel to each other whereas all other neighboring β-strands are antiparallel. There is also an α-helix that spans residues V25-E35.
The XPC-Binding Domain Forms an α-Helical Bundle. We have found that the XPC-binding domain structure consists of a globular arrangement of four α-helices, which span residues E233-D237 (α1), Q240-I248 (α2), A253-G263 (α3), and L269-M283 (α4) (Fig. 1D). The last α-helix is terminated at both ends by proline residues and bends at the third turn of the helix to pack against α2 and α3. This tightly packed structural domain contains seven prolines, including P256, which is located in the middle of α3. It is largely hydrophobic and the absence of charged surface regions suggests that hHR23a binding to XPC is driven by hydrophobic interactions.
We used blast and the Dali Server (38) to test whether this structural domain has either sequence or structural homology with other known proteins. Although no sequence homology was identified, the XPC-binding domain was found to have slight structural homology with five other proteins. Potentially, the most interesting of these may be the mitochondrial protein import receptor Tom20 (Dali score of 2.1). Tom20 (39) has structural homology with α2-α4 of the XPC-binding domain but has an additional C-terminal α-helix positioned in the hydrophobic groove formed by the corresponding helices of α3 and α4. XPC may interact with the XPC-binding domain in an analogous manner to this helix. The XPC-binding domain also has slight structural homology with Nk-lysin (40) and As-48 (41) (Dali score for each of 2.3), which are both associated with antibacterial activity, as well as the C-terminal region of the vesicular transport protein Sec17 (42) (Dali score of 2.3). These proteins, however, all have charged surfaces, whereas that of the XPC-binding domain is largely hydrophobic. Finally, the XPC-binding domain is also structurally similar to the N-terminal region of the Tet Repressor of class D (43) (Dali score of 2.1), which forms the TetR DNA-binding domain. Indeed the region of the XPC protein that is required for binding DNA overlaps with that used for hHR23-binding (44). It is therefore possible that DNA binds at the interface of the XPC-hHR23 protein complex.
The Linker Regions Connecting the Domains Are Flexible. To assess the flexibility of the unstructured linker regions of hHR23a, we performed NMR relaxation experiments, which probe the internal dynamics of each residue. To quantify the mobility of the amide bond vectors of hHR23a, we recorded 15N longitudinal [RN(NZ)] and transverse [RN(NX)] relaxation measurements as well as heteronuclear dipole-dipole cross relaxation [RIN(IZ ↔ NZ)] measurements (Fig. 5 A–C, which is published as supporting information on the PNAS web site). These experiments revealed that residues located in the linker regions have much slower RN(NX) and much larger RIN(IZ ↔ NZ) values. In fact, the average value for RN(NX) and RIN(IZ ↔ NZ) of residues within these regions is 4.9 s–1 and 0.9, respectively, which indicates that they are indeed highly flexible.
The Structured Domains in hHR23a Interact in an Intramolecular Manner. In contrast with the linker regions, residues within the structured domains have fast RN(NX) and small RIN(IZ ↔ NZ) values. The average RN(NX) and RIN(IZ ↔ NZ) value within the domains is 35.1 s–1 and 0.25, respectively. Comparison of these values with those published for other proteins indicates that the structured domains have the dynamic properties of a large protein. We confirmed this observation by subjecting our data to the model-free analysis from which we derived τc for each domain (29, 30). Such values reveal the rate at which a molecule tumbles in solution and therefore large, bulky molecules have larger τc than do small ones. The values that we obtained are provided in Table 1 and do indeed reflect those of a large protein (45). Because each domain is <8 kDa and connected by long flexible linker regions, this indicates that they are not behaving individually as “beads-on-a-string” but rather are interacting.
Table 1. Rotational correlation times of hHR23a alone and with S5a (196–307).
| Times, ns |
||||
|---|---|---|---|---|
| Protein | UBL | UBA1 | XPC binding | UBA2 |
| hHR23a | 39 | 28 | 27 | 22 |
| hHR23a (C118-363) | N/A | 7 | 14 | 8 |
| hHR23a/S5a (196-307) | 25 | 18 | 24 | 16 |
N/A, not available.
Previous studies indicate that the yeast homolog Rad23 is a dimer (46). We therefore investigated the oligomeric state of hHR23a to determine whether the structured domains interact in an intermolecular manner. We mixed a sample containing fully deuterated 15N-labeled hHR23a with unlabeled hHR23a. When such a sample is used in an 15N-dispersed NOESY experiment, the presence of resonance crosspeaks between amide and aliphatic protons must arise from intermolecular interactions (25). In our NOESY experiment on this sample, recorded at 800 MHz, we observed no such crosspeaks, suggesting that hHR23a is not a dimer. Additionally, hHR23a behaved as a monomer in GST pull-down experiments using recombinant proteins and/or cellular lysates, in native gel electrophoresis, and in transfection experiments followed by coaffinity purification (data not shown). Interaction of the structured domains must therefore be in an intramolecular manner.
The UBL Domain of hHR23a Is Essential to Its Interdomain Interactions. Evidence for the importance of the UBL domain in hHR23a interdomain structure comes from several different sources. First, we found that deleting the UBL domain results in changes in the amide chemical shift positions of certain residues within the other three domains. As described above the structure of the UBL and UBA domains are preserved in the full-length protein compared with their single domain constructs. Therefore, the observed chemical shift changes are not caused by structural changes within the domains and are more likely caused by a loss of interaction with the UBL domain. Second, the UBL domain has a large τc of 39 ns. Because this 80-residue domain is at the N terminus and separated from the rest of the protein by an 80-residue flexible linker region, this value must be caused by interaction with another domain of the protein. Therefore with the hypothesis that deletion of the UBL domain would decrease the τc of the other domains, we conducted the relaxation experiments described above to determine τc for each domain in an hHR23a protein construct in which the UBL domain is deleted [hHR23a (C118–363)].
RN(NZ) and RN(NX) relaxation in addition to RIN(IZ ↔ NZ) measurements were recorded on hHR23a (C118–363) under identical conditions as those used for the full protein. As expected the linker regions remained flexible with almost identical values as those of the full-length protein and residues in these regions have average RN(NX) and RIN(IZ ↔ NZ) values of 5.2 s–1 and 0.9, respectively. In contrast, the RN(NX) and τc values of the UBA and XPC-binding domains were much reduced, with an average RN(NX) value of 16.8 s–1. These reduced τc values (Table 1) reflect a much faster tumbling rate for hHR23a when the UBL domain is deleted. In fact, the new values indicate that in the absence of the UBL domain the other domains adopt τc that are characteristic for the domain size (45). Taken together, our data indicate that the UBL domain plays an essential role in the interdomain interactions of hHR23a.
Interdomain Interactions Do Not Rigidly Lock hHR23a into a Single Protein Structure. If the domains of hHR23a interact to form a single rigid protein structure, then we would expect to able to record interdomain NOE interactions, which could be converted to interdomain distance constraints. We have assigned >95% of all NOE crosspeaks present in 15N- and 13C-dispersed NOESY spectra and found that all of these originate from intradomain interactions. However, because interdomain NOEs may be small or may overlap with intradomain NOEs, we used in vitro protein ligation to generate hHR23a protein samples that were suitable for obtaining unambiguous interdomain NOE interactions between the UBL domain and the rest of the protein.
To perform in vitro protein ligation (15) we expressed and purified two protein constructs separately, one containing residues 1–117 and the other containing residues 118–363. A118 was mutated to a cysteine to enable in vitro ligation. An [1H,15N]-HSQC spectrum on the ligated protein closely resembled that of the WT protein, indicating that the in vitro protein ligation protocol did not cause any changes in hHR23a protein structure.
To test for interdomain NOE interactions between the UBL domain and the C-terminal domains, we conducted two separate isotope-edited NOESY experiments. First, we ligated 13C-labeled hHR23a (C118–363) to unlabeled hHR23a (1–117) and acquired a 13C half-filtered NOESY experiment in 2H2O on this sample (24). Second, we recorded an 15N-dispersed NOESY experiment on a sample containing 2H-,15N-labeled hHR23a (C118–363) and unlabeled hHR23a (1–117) (25). Neither of these experiments contained NOE interactions between hHR23a (1–117) and hHR23a (C118–363), which suggests that the hHR23a protein structure is not rigidly locked into one conformation.
The hHR23a UBL Domain Interacts with Each UBA Domain. To define which domains interact directly, we analyzed the spectral changes in [1H,15N]-HSQC experiments that result from mixing the single domain constructs. Each of the four domains was expressed and purified separately, and [1H,15N]-HSQC spectra were recorded on each single domain construct alone and in the presence of each of the others. When the XPC-binding domain was mixed with each of the other three domains spectral changes were not observed for any of the amide resonances in any of the domains. These experiments were conducted at both 25°C and 10°Con 15N-labeled XPC-binding domain and each of the other domains at 1:1 and 1:2 molar ratio. We conducted additional experiments whereby 15N-labeled XPC-binding domain was mixed with 15N-labeled samples of each of the other domains at equimolar concentration. In all cases no spectral changes were observed, indicating that the XPC-binding domain does not directly contact any other domain when expressed as a single domain construct. We performed similar experiments to test whether the UBA domains contact each other. Likewise these domains did not show any spectral changes when mixed and therefore do not directly bind each other when expressed as single domain constructs.
In contrast, when the UBL domain was mixed with either of the UBA domains, we observed spectral changes in each of the domains. In particular, we found that residues within the C-terminal α-helix of each of the UBA domains experience chemical shift perturbations upon addition of the UBL domain; specifically residues R193, E196, L199, N349, A351, N353, F354, L356, S357, Q358, and N359 are shifted by UBL domain addition. These residues are also shifted in the hHR23a (C118–363) protein construct.
On the UBL domain, there is significant overlap in the binding surfaces for each UBA domain, which is formed by residues located in β-strands. T9, L10, T14, F15, K47, Y50, A51, G52, V76, and K78 experience chemical shift changes upon addition of either UBA domain, whereas Q11, L48, V73, and T77 are only shifted by the UBA2 domain. The surface on the UBL domain formed by these residues is not large enough to simultaneously bind each of the UBA domains (Fig. 2). Furthermore the significantly larger rotational correlation time of the UBL domain compared with the other domains is likely to be a result of chemical exchange between multiple states. Taken together these data suggest that the UBL domain exists in an equilibrium of exchange with each of the UBA domains.
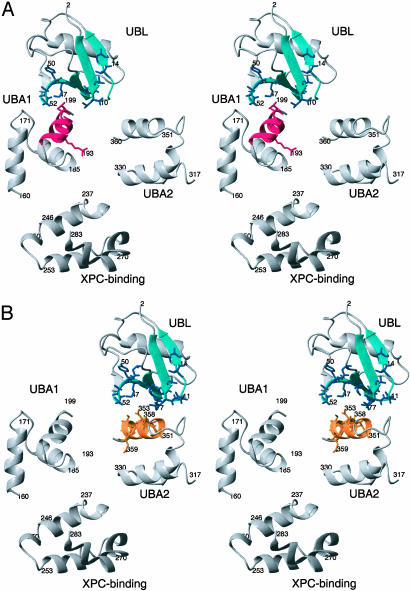
Fig. 2.
Stereoview of hHR23a protein structure. HHR23a undergoes chemical exchange between the conformation shown in A, whereby UBL contacts UBA1, and that in B, whereby UBL contacts UBA2. The relative angular orientation of each domain was determined by using residual dipolar coupling data and the program module 1.0 (28). The side-chain atoms of residues that experience chemical shift changes in [1H,15N]-HSQC experiments upon addition of the UBL domain are displayed in red and orange for the UBA1 and UBA2 domains, respectively. Residues of the UBL domain that undergo chemical shift changes upon addition of UBA1 (A) or UBA2 (B) are colored blue.
To test whether the XPC-binding domain influences the complex containing the other three hHR23a domains, we compared [1H,15N]-HSQC spectra acquired on a complex in which all three components were 15N-labeled, in the presence and absence of the XPC-binding domain. Addition of the XPC-binding domain did not cause spectra changes in resonances of the UBL or UBA domains and therefore does not bind any of the other domains when all four are present.
Model of the hHR23a Protein Structure. The relative orientation of each domain within a multidomain protein can be determined by using residual dipolar coupling constants in partially aligned samples (reviewed in ref. 47). Therefore to define the relative orientation of each of the hHR23a domains, we used in-phase antiphase [1H,15N]-HSQC experiments (27) to measure residual dipolar coupling constants on an 15N-labeled hHR23a protein sample that was partially aligned with 7.5 mg/ml Pf1 phage. The aligned sample has amide chemical shift values characteristic of the 40-kDa protein, suggesting that the interdomain interactions were preserved. The residual dipolar coupling data were used in the program module 1.0 (28) to produce the angular alignment illustrated in Fig. 2. As shown in Fig. 2, the interaction surfaces defined above can contact each other and maintain their relative orientations. Inspection of Fig. 2 reveals that although each of the UBA domains binds the β-sheet face of the UBL domain, the orientation of UBA1 is rotated by ≈90° relative to that of UBA2. In each case the UBL/UBA interaction surface is largely hydrophobic; however, the more acidic surface of UBA1 caused by E196 is likely why these two domains bind in different orientations. More specifically UBA1 is oriented such that E196 contacts K8 of the UBL domain. The larger binding surface of the UBL domain can accommodate each of the UBA domains, but not simultaneously. The competition between these different binding orientations is likely to be the origin of hHR23a conformational exchange.
The Binding Surfaces of S5a and the UBA Domains Overlap. We used an S5a construct spanning residues M196 to D307 to study the effect of S5a-binding on hHR23a protein structure. S5a (196–307) contains the two ubiquitin-binding domains and was previously shown to bind hHR23a with equal affinity to full-length S5a (4). To identify the residues of hHR23a that directly contact S5a (196–307), we acquired an 15N-dispersed NOESY experiment on a complex containing 15N-labeled and 100% 2H-labeled hHR23a and unlabeled S5a (196–307). In such an experiment all amide-to-aliphatic NOE interactions must be intermolecular (25). Indeed we observed numerous intermolecular NOEs between S5a (196–307) and residues within the UBL domain, but none involving the other domains of hHR23a. Therefore, S5a interacts directly with only the UBL domain of hHR23a.
Fig. 3A shows a surface diagram of a representative structure of the UBL domain of hHR23a with the residues that show intermolecular NOE interactions with S5a (196–307) displayed in blue. This interaction surface was confirmed by making point mutations in the binding surface such that interaction with S5a (196–307) is abrogated. An hHR23a protein construct in which T9 and I49 were substituted with alanine and threonine, respectively was generated, and the structural integrity of the UBL domain was confirmed by an [1H,15N]-HSQC spectrum. The hHR23a T9A/I49T mutant no longer binds S5a (196–307), possibly because of the substitution of the hydrophilic threonine for the hydrophobic isoleucine (Fig. 6, which is published as supporting information on the PNAS web site). The S5a-binding surface also overlaps significantly with that of the UBA domains (Fig. 3), which suggests that interaction with S5a and a UBA domain is mutually exclusive. Hence we hypothesized that hHR23a undergoes a conformational change upon binding S5a.
Fig. 3.
The S5a contact surface on the UBL domain overlaps significantly with that used to bind the UBA domains. (A) Residues whose amide protons showed NOE interaction with S5a (196–307) are colored blue. (B) Those that shift upon addition of either UBA domain are colored dark blue, and those that only shift upon addition of the UBA2 domain are light blue. (Left) The orientation is identical to that of Fig. 1B (Left), and these are rotated 180° (Right). The program grasp (51) was used.
S5a Binding Disrupts hHR23a Protein Structure. Upon binding S5a (196–307), those residues at the contact surface within the UBL domain experience large chemical shift changes because of the altered chemical environment. Interestingly, additional smaller chemical shift pertubations were observed in the other domains. Because S5a interacts directly with only the UBL domain, these smaller chemical shift changes in the other domains must be caused by changes in hHR23a protein structure.
To test whether S5a causes structural changes in hHR23a we performed NMR relaxation experiments on the protein complex, using 15N-labeled hHR23a complexed with fully deuterated S5a (196–307). We recorded RN(NZ) and RN(NX) relaxation in addition to RIN(IZ ↔ NZ) measurements and determined τc values for each domain by analyzing the data according to model-free formulism (29, 30). The relaxation experiments were acquired under conditions identical to those performed for the free protein. The values of the linker regions changed very little, indicating that they remain flexible when hHR23a is bound to S5a (196–307). In contrast, we found that the RN(NX) values for residues within the structured domains of the S5a (196–307)-bound hHR23a decreased substantially, to an average value of 27.6 s–1 (Fig. 5 D–F). The τc values also decreased (Table 1), indicating that addition of S5a (196–307) causes each domain to tumble faster in solution, which is suggestive of a less bulky structure. Because the hHR23a-S5a protein complex is larger than free hHR23a, the observed changes in the relaxation data indicate that the structural domains no longer interact. Therefore the proteasomal subunit S5a effectively competes with the UBA domains for the UBL domain of hHR23a and thereby traps it in an opened conformation (Fig. 4).
Fig. 4.
Model of S5a-induced conformational change in hHR23a. In the absence of S5a, hHR23a undergoes conformational exchange between two states as each of the UBA domains competes for the same binding surface on the UBL domain. Addition of S5a causes hHR23a to adopt an opened conformation as S5a blocks the UBA-binding surface of the UBL domain.
Discussion
The Rad23 family of proteins has recently attracted much attention for their ability to control the degradation of other proteins. Indeed this protein family is intricately connected to the ubiquitin-proteasome pathway as its UBA domains interact with ubiquitin (3) and its UBL domain interacts with S5a and Rpn1 (4, 5). To better understand how these interactions occur in the context of the full protein, we have solved the structure of hHR23a. Here we show that hHR23a contains four structured domains that are connected by long unstructured flexible linkers. Interestingly, we have found that the UBL domain interacts with the C-terminal helix of each of the UBA domains. However, the structure of hHR23a appears to maintain entropic freedom and does not rigidly adopt one domain arrangement in solution. We used residual dipolar coupling data in combination with chemical shift perturbation analysis to produce a model of the hHR23a protein structure.
Interestingly the proteasomal subunit S5a binds the hHR23a UBL domain at a surface that largely overlaps that used to interact with the UBA domains, thus causing hHR23a to adopt a more opened conformation in solution. We predict that ubiquitin binds each of the UBA domains in a manner similar to that of the UBL domain and also causes hHR23a to adopt an opened conformation. We conclude that interaction with members of the ubiquitin-proteasome pathway alters hHR23 structure and predict that such changes may serve as an important mechanism in regulating the degradation of certain proteins such as XPC (10, 11). This regulatory mechanism likely extends to other proteins that contain both UBL and UBA domains, such as members of the hPLIC family (48). These proteins also bind S5a through a conserved hydrophobic surface within the UBL domain (37, 49).
The UBA domains of hHR23a can inhibit the degradation of other proteins by binding to the K48-linked polyubiquitin chains that serve as signals for degradation (9, 50). This inhibitory effect is increased in hHR23a constructs that lack the UBL domain (9), and our structural data indicate that this is because of an increased accessibility of the UBA domains. Because S5a similarly increases accessibility to the UBA domains it likely functions with the hHR23 proteins to promote the inhibition of proteasome-mediated degradation.
Additionally, we reveal that the XPC-binding domain forms a globular α-helical fold. The surface of this domain is almost entirely hydrophobic, suggesting that hHR23 proteins bind XPC by using hydrophobic interactions. Comparison with a structural homolog, namely protein import receptor Tom20, suggests that XPC may bind in the hydrophobic groove formed by α3 and α4 of the XPC-binding domain.
We present here that the UBL domain can interact with each UBA domain in the absence of the XPC-binding domain. Based on this finding we hypothesize that binding to XPC does not result in a loss of UBL/UBA domain interaction. In fact, trapping the hHR23 proteins in a “closed” conformation may be important for its NER function.
Supplementary Material
Acknowledgments
We are grateful to Dr. Patrick Young for generously providing us with S5a (196–307) plasmid and Dr. Hiroshi Matsuo for useful discussions. NMR data were acquired at the NMR facility of the University of Minnesota, and we thank Dr. David Live and Dr. Beverly Ostrowsky for their technical assistance. NMR instrumentation was provided with funds from the National Science Foundation (Grant BIR-961477), the University of Minnesota Medical School, and the Minnesota Medical Foundation. Data processing and structure calculations were performed in the Basic Sciences Computing Lab of the University of Minnesota Supercomputing Institute. This work was in part funded by the Minnesota Medical Foundation (K.J.W.), an American Cancer Society Institutional Grant (to K.J.W.), National Institutes of Health Grants CA097004-01A1 (to K.J.W.) and CA37-CA64888 (to P.M.H.), and a scholarship from the Agency for Science, Technology, and Research of Singapore (to A.M.G.).
Abbreviations: NER, nucleotide excision repair; NOE, nuclear Overhauser effect; τc, rotational correlation time; UBA, ubiquitin-associated; UBL, ubiquitin-like; XPC, xeroderma pigmentosum complementation group C; HSQC, heteronuclear single quantum coherence.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1OQY).
References
- 1.Sugasawa, K., Ng, J. M., Masutani, C., Iwai, S., van der Spek, P. J., Eker, A. P., Hanaoka, F., Bootsma, D. & Hoeijmakers, J. H. (1998) Mol. Cell 2 223–232. [DOI] [PubMed] [Google Scholar]
- 2.Bootsma, D., Kraemer, K. H., Cleaver, J. E. & Hoeijmakers, J. H. (1998) in The Genetic Basis of Human Cancer, eds. Vogelstein, B. & Kinzler, K. W. (McGraw–Hill, New York), pp. 245–274.
- 3.Bertolaet, B. L., Clarke, D. J., Wolff, M., Watson, M. H., Henze, M., Divita, G. & Reed, S. I. (2001) Nat. Struct. Biol 8 417–422. [DOI] [PubMed] [Google Scholar]
- 4.Hiyama, H., Yokoi, M., Masutani, C., Sugasawa, K., Maekawa, T., Tanaka, K., Hoeijmakers, J. H. & Hanaoka, F. (1999) J. Biol. Chem. 274 28019–28025. [DOI] [PubMed] [Google Scholar]
- 5.Elsasser, S., Gali, R. R., Schwickart, M., Larsen, C. N., Leggett, D. S., Muller, B., Feng, M. T., Tubing, F., Dittmar, G. A. & Finley, D. (2002) Nat. Cell Biol. 4 725–730. [DOI] [PubMed] [Google Scholar]
- 6.Deveraux, Q., Ustrell, V., Pickart, C. & Rechsteiner, M. (1994) J. Biol. Chem. 269 7059–7061. [PubMed] [Google Scholar]
- 7.Ferrell, K., Deveraux, Q., van Nocker, S. & Rechsteiner, M. (1996) FEBS Lett. 381 143–148. [DOI] [PubMed] [Google Scholar]
- 8.Ortolan, T. G., Tongaonkar, P., Lambertson, D., Chen, L., Schauber, C. & Madura, K. (2000) Nat. Cell Biol. 2 601–608. [DOI] [PubMed] [Google Scholar]
- 9.Raasi, S. & Pickart, C. M. (2003) J. Biol. Chem. 278 8951–8959. [DOI] [PubMed] [Google Scholar]
- 10.Lommel, L., Ortolan, T., Chen, L., Madura, K. & Sweder, K. S. (2002) Curr. Genet. 42 9–20. [DOI] [PubMed] [Google Scholar]
- 11.Ng, J. M., Vermeulen, W., van der Horst, G. T., Bergink, S., Sugasawa, K., Vrieling, H. & Hoeijmakers, J. H. (2003) Genes Dev. 17 1630–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar, S., Talis, A. L. & Howley, P. M. (1999) J. Biol. Chem. 274 18785–18792. [DOI] [PubMed] [Google Scholar]
- 13.Friedberg, E. C. & Meira, L. B. (2000) Mutat. Res. 459 243–274. [DOI] [PubMed] [Google Scholar]
- 14.Young, P., Deveraux, Q., Beal, R. E., Pickart, C. M. & Rechsteiner, M. (1998) J. Biol. Chem. 273 5461–5467. [DOI] [PubMed] [Google Scholar]
- 15.Evans, T. C., Jr., Benner, J. & Xu, M. Q. (1999) J. Biol. Chem. 274 3923–3926. [DOI] [PubMed] [Google Scholar]
- 16.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J. & Bax, A. (1995) J. Biomol. NMR 6 277–293. [DOI] [PubMed] [Google Scholar]
- 17.Bartels, C., Xia, T.-H., Billeter, M., Güntert, P. & Wüthrich, K. (1995) J. Biomol. NMR 6 1–10. [DOI] [PubMed] [Google Scholar]
- 18.Yamazaki, T., Lee, W., Revington, M., Mattiello, D. L., Dahlquist, F. W., Arrowsmith, C. H. & Kay, L. E. (1994) J. Am. Chem. Soc. 116 6464–6465. [Google Scholar]
- 19.Yamakazi, T., Lee, W., Arrowsmith, C. H., Muhandiram, D. R. & Kay, L. E. (1994) J. Am. Chem. Soc. 116 11655–11666. [Google Scholar]
- 20.Matsuo, H., Li, H. & Wagner, G. (1996) J. Magn. Reson. B 110 112–115. [DOI] [PubMed] [Google Scholar]
- 21.Matsuo, H., Kupce, E., Li, H. & Wagner, G. (1996) J. Magn. Reson. B 111 194–198. [DOI] [PubMed] [Google Scholar]
- 22.Venters, R. A., Huang, C. C., Farmer, B. T., 2nd, Trolard, R., Spicer, L. D. & Fierke, C. A. (1995) J. Biomol. NMR 5 339–344. [DOI] [PubMed] [Google Scholar]
- 23.Cornilescu, G., Delaglio, F. & Bax, A. (1999) J. Biomol. NMR 13 289–302. [DOI] [PubMed] [Google Scholar]
- 24.Zwahlen, C., Legault, P., Vincent, S. J. F., Greenblatt, J., Konrat, R. & Kay, L. E. (1997) J. Am. Chem. Soc. 119 6711–6721. [Google Scholar]
- 25.Walters, K. J., Matsuo, H. & Wagner, G. (1997) J. Am. Chem. Soc. 119 5958–5959. [Google Scholar]
- 26.Zweckstetter, M. & Bax, A. (2001) J. Biomol. NMR 20 365–377. [DOI] [PubMed] [Google Scholar]
- 27.Ottiger, M., Delaglio, F. & Bax, A. (1998) J. Magn. Reson. 131 373–378. [DOI] [PubMed] [Google Scholar]
- 28.Dosset, P., Hus, J. C., Marion, D. & Blackledge, M. (2001) J. Biomol. NMR 20 223–231. [DOI] [PubMed] [Google Scholar]
- 29.Mandel, A. M., Akke, M. & Palmer, A. G., 3rd (1995) J. Mol. Biol. 246 144–163. [DOI] [PubMed] [Google Scholar]
- 30.Lipari, G. & Szabo, A. (1982) J. Am. Chem. Soc. 104 4546–4559. [Google Scholar]
- 31.Palmer, A. G., Rance, M. & Wright, P. E. (1991) J. Am. Chem. Soc. 113 4371–4380. [Google Scholar]
- 32.Nilges, M., Clore, G. M. & Gronenborn A. M. (1988) FEBS Lett. 239 129–136. [DOI] [PubMed] [Google Scholar]
- 33.Brünger, A. T. (1993) XPLOR Version 3.1: A System for X-Ray Crystallography and NMR (Yale Univ. Press, New Haven).
- 34.Wishart, D. S. & Case, D. A. (2001) Methods Enzymol. 338 3–34. [DOI] [PubMed] [Google Scholar]
- 35.Mueller, T. D. & Feigon, J. (2002) J. Mol. Biol. 319 1243–1255. [DOI] [PubMed] [Google Scholar]
- 36.Dieckmann, T., Withers-Ward, E. S., Jarosinski, M. A., Liu, C. F., Chen, I. S. & Feigon, J. (1998) Nat. Struct. Biol 5 1042–1047. [DOI] [PubMed] [Google Scholar]
- 37.Walters, K. J., Kleijnen, M. F., Goh, A. M., Wagner, G. & Howley, P. M. (2002) Biochemistry 41 1767–1777. [DOI] [PubMed] [Google Scholar]
- 38.Holm, L. & Sander, C. (1993) J. Mol. Biol. 233 123–138. [DOI] [PubMed] [Google Scholar]
- 39.Abe, Y., Shodai, T., Muto, T., Mihara, K., Torii, H., Nishikawa, S., Endo, T. & Kohda, D. (2000) Cell 100 551–560. [DOI] [PubMed] [Google Scholar]
- 40.Liepinsh, E., Andersson, M., Ruysschaert, J. M. & Otting, G. (1997) Nat. Struct. Biol 4 793–795. [DOI] [PubMed] [Google Scholar]
- 41.González, C., Langdon, G. M., Bruix, M., Gálvez, A., Valdivia, E., Maqueda, M. & Rico, M. (2000) Proc. Natl. Acad. Sci. USA 97 11221–11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice, L. M. & Brunger, A. T. (1999) Mol. Cell 4 85–95. [DOI] [PubMed] [Google Scholar]
- 43.Kisker, C., Hinrichs, W., Tovar, K., Hillen, W. & Saenger, W. (1995) J. Mol. Biol. 247 260–280. [DOI] [PubMed] [Google Scholar]
- 44.Uchida, A., Sugasawa, K., Masutani, C., Dohmae, N., Araki, M., Yokoi, M., Ohkuma, Y. & Hanaoka, F. (2002) DNA Repair 1 449–461. [DOI] [PubMed] [Google Scholar]
- 45.Wagner, G. (1997) Nat. Struct. Biol. NMR 4 Suppl., 841–844. [PubMed] [Google Scholar]
- 46.Bertolaet, B. L., Clarke, D. J., Wolff, M., Watson, M. H., Henze, M., Divita, G. & Reed, S. I. (2001) J. Mol. Biol. 313 955–963. [DOI] [PubMed] [Google Scholar]
- 47.Bax, A., Kontaxis, G. & Tjandra, N. (2001) Methods Enzymol. 339, 127–174. [DOI] [PubMed] [Google Scholar]
- 48.Kleijnen, M. F., Shih, A. H., Zhou, P., Kumar, S., Soccio, R. E., Kedersha, N. L., Gill, G. & Howley, P. M. (2000) Mol. Cell 6 409–419. [DOI] [PubMed] [Google Scholar]
- 49.Walters, K. J., Goh, A. M., Wang, Q., Wagner, G. & Howley, P. M. (2003) Biochim. Biophys. Acta, in press. [DOI] [PubMed]
- 50.Chen, L., Shinde, U., Ortolan, T. G. & Madura, K. (2001) EMBO Rep. 2 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicholls, A. J. (1993) GRASP Manual (Columbia Univ., New York).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.