Abstract
The crystal structure of the Escherichia coli lactose permease at 3.5 Å with a bound substrate has been reported recently. The structure reveals the sugar–protein contacts, which include hydrophobic stacking between the galactopyranosyl ring of substrate and the indole side chain of Trp-151, as proposed previously. The nature of this interaction is studied here by exploiting the luminescence properties of Trp-151 in a mutant devoid of other tryptophan residues. The following phenomena are observed. (i) The fluorescence emission spectrum of Trp-151 and fluorescence-quenching experiments with water-soluble quenchers demonstrate that Trp-151 is in a hydrophilic environment. (ii) Substrate binding leads to a blue shift in the emission spectrum and reduction in accessibility to polar quenchers, indicating that Trp-151 becomes less exposed to aqueous solvent. (iii) The phosphorescence spectrum of Trp-151 is red-shifted in the presence of substrate, indicating charge separation of the triplet state due to a direct stacking interaction between the galactopyranosyl and indole rings. The spectroscopic data fully complement the x-ray structure and demonstrate the feasibility of fluorescence spectroscopy for studying sugar–protein interactions.
Keywords: bioenergetics, transport, fluorescence/phosphorescence, hydrophobic stacking, sugar binding
The lactose permease (LacY) of Escherichia coli is a paradigm for the major facilitator superfamily (1) of membrane transport proteins, many of which transduce free energy stored in an electrochemical proton gradient into a concentration gradient of substrate and vice versa (2). LacY contains 12 transmembrane helices connected by hydrophilic loops with both the N and C termini on the cytoplasmic face of the membrane (3–6), and it is functionally and structurally a monomer (6–11).
LacY is selective for disaccharides containing a d-galactopyranosyl ring as well as d-galactose (12–14) but does not interact with d-glucopyranosides or d-glucose (14–16). The specificity of LacY is clearly directed toward the galactopyranosyl ring of substrate, and galactose is the most specific substrate but has very low affinity (14). Although the C4 OH group is by far the most important for specificity, the C2, C3, and C6 OH groups are also important for binding (C4 OH ≫ C3 OH ≃ C6 OH > C2 OH) (14, 16).
The use of site-directed mutagenesis, and Cys-scanning mutagenesis in particular (reviewed in ref. 17), has led to the identification of side chains that are irreplaceable for substrate binding and proton translocation. Moreover, the combination of various site-directed biophysical and biochemical techniques has allowed the formulation of a hypothesis for the mechanism of lactose/H+ symport (18).
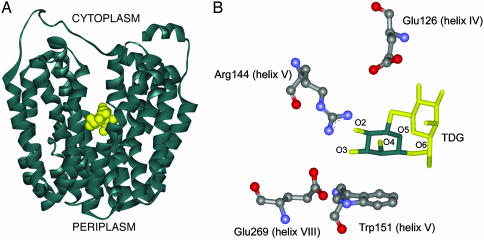
The x-ray structure of the inward-facing conformation of a LacY mutant (C154G) with a bound lactose homologue, β-d-galactopyranosyl-1-thio-β-d-galactopyranoside (TDG), has been solved recently (6). The overall structure reveals pseudo-twofold symmetry between the N- and C-terminal six-helix domains, as proposed (19) for other members of the major facilitator superfamily transporters. Similar symmetry has also been found recently in the x-ray structure of GlpT (phosphate/glycerol phosphate antiporter), another member of the major facilitator superfamily transporters (20). Importantly, the sugar-binding site is found at the twofold axis of symmetry, which is located approximately in the middle of the membrane, facing a large water-filling internal cavity open to the cytoplasm (Fig. 1A). This finding is consistent with the idea that LacY has only one binding site that has alternating accessibility to each side of the membrane.
Fig. 1.
Overall x-ray structure of LacY and binding-site interactions of the galactopytanosyl ring. (A) Ribbon representation of the crystal structure of the C154G LacY mutant in an inward-facing conformation with a bound lactose homologue, TDG (yellow). The sugar-binding site is present in a hydrophilic cavity in the center of the molecule. (B) Detail of the important binding-site interactions between TDG and LacY. The bottom of one galactopyranosyl ring (green) is within van der Waal distance of Trp-151 (helix V), and Arg-144 (helix V) forms H-bonds with the O3 and O4 atoms of the galactopyranosyl ring. Also shown are Glu-269 (helix VIII), which appears to form a salt bridge with Arg-144 as well as an H-bond with Trp-151, and Glu-126 (helix IV), which may interact with the O4, O5, and/or O6 atoms of the galactopyranosyl ring via water molecules. The data shown are derived from Abramson et al. (6).
The main residues involved in substrate binding are shown in Fig. 1B (6). Arg-144 (helix V) and Glu-126 (helix IV) are the major determinants. Arg-144 forms H-bonds with the O3 and O4 atoms of the galactopyranosyl ring, whereas Glu-126 may interact with the O4, O5, or O6 via water molecules. Although not shown in the structure, biochemical evidence indicates that these residues form a charge pair in the absence of the substrate (21–25). Recent experiments (26) using carbodiimide labeling in the absence or presence of ligand in conjunction with mass spectroscopy indicate that Glu-269 (helix VIII) may be implicated in sugar binding, possibly interacting with the C3 OH of the galactopyranosyl ring. The x-ray crystal structure reveals that this residue forms a salt bridge with Arg-144 as well as an H-bond with Trp-151 (6). Because all replacements for Glu-269, with the exception of aspartic acid, do not bind sugar and because all modes of translocation are abrogated, it is likely that Glu-269 may be a key residue, coupling H+ translocation and sugar binding.
Trp-151 (helix V) also plays an important role in substrate binding and specificity, as evidenced by studies (27) demonstrating that the W151F and W151Y mutants exhibit markedly decreased affinity for substrate. These observations led to a model for the binding site in which Trp-151 lies perpendicular to helix V with the indole side chain stacking hydrophobically at the bottom of the galactopyranosyl. The x-ray crystal structure is in excellent agreement, showing directly that the galactopyranosyl group of TDG faces the indole group of Trp-151 at a distance sufficient for a hydrophobic and/or van der Waals interaction (Fig. 1B). In this manner, the galactopyranosyl ring is held in such a position that important H-bonds between the ring and specific side chains in LacY can be realized. These types of interactions are common in the carbohydrate-binding sites of >50 proteins of known structure (28–32).
Here we present spectroscopic evidence for an interaction between LacY substrates and Trp-151 by exploiting its natural luminescence. For this purpose, a fully functional single-Trp-151 mutant of LacY was constructed by replacing the other five tryptophan residues with tyrosine. An additional mutation, C154G, was also introduced, which renders the protein defective with respect to sugar translocation but has little or no effect on binding affinity and increases the stability of LacY in dodecyl-β-d-maltopyranoside (DDM) (33). The findings demonstrate that Trp-151 is in a hydrophilic environment despite its location in the middle of the membrane, which is consistent with the x-ray crystal structure where Trp-151 faces the internal aqueous cavity (6). Moreover, the presence of LacY substrates clearly protects Trp-151 from solvent exposure, as judged by a blue shift in the emission spectrum and the decreased ability of hydrophilic collisional quenchers to quench Trp-151 fluorescence. Finally, the phosphorescence spectrum of Trp-151 is red-shifted in the presence of the substrate, thereby providing direct evidence for the hydrophobic stacking.
Experimental Procedures
Materials. Allyl-α-d-galactopyranoside, melibiose, d-glucose, sucrose, TDG, KI, CsCl, and acrylamide were purchased from Sigma. DDM was purchased from Calbiochem. Synthetic deoxyoligonucleotides were purchased from Genosys (The Woodlands, TX); restriction endonucleases, T4 DNA ligase, and Vent DNA polymerase were purchased from New England Biolabs. We purchased 2-(4-maleimidylanilino)naphthalene-6-sulfonic acid (MIANS) from Molecular Probes. All other materials were reagent grade and obtained from commercial sources.
Construction of LacY Mutants. The gene encoding wild-type LacY was subcloned from plasmid pT7-5/cassette lacY (EMBL accession no. X56095) into the expression vector pCS19 (34) after NcoI and BglII sites were introduced by PCR at the 5′ and 3′ ends, respectively, resulting in plasmid pCS19/cassette lacY. Wild-type LacY is expressed from the T5 promoter with an Asp in place of the Tyr at position 2, a 6-histidine tag at the C terminus, and two additional residues (Arg and Ser) between the 6-histidine tag and the native C-terminal Ala residue, respectively. By applying PCR, plasmid pCS19/single-Trp-151-encoding tyrosine residues in place of five native tryptophan residues at positions 10, 33, 78, 171, and 223 were constructed. In this background, the codon for Cys-154 was replaced with the codon for glycine by PCR, yielding plasmid pCS19/single Trp-151/C154G. All constructs were confirmed by DNA sequencing.
Bacterial Strains, Growth, and LacY Purification. E. coli T184 (lacZ–Y–) transformed with pCS19 plasmid encoding single-W151/C154G LacY with a 6-histidine tag at the C terminus was grown in 6 liters of Luria–Bertani broth at 30°C containing ampicillin (100 μg/ml) to an OD600 of 0.6 and induced with 0.5 mM isopropyl 1-thio-β-d-galactopyranoside. Cells were disrupted by passage through a French pressure cell, and the membrane fraction was harvested by ultracentrifugation. Membranes were solubilized by adding DDM to a final concentration of 2%, and LacY was purified by Co(II) affinity chromatography using Talon Superflow (BD Biosciences Clontech) as described (33, 35). Protein eluted with 150 mM imidazole was dialyzed against 20 mM Tris·HCl, pH 7.5/0.008% DDM, concentrated by using a Vivaspin 20 concentrator (30-kDa cutoff; Vivascience, Hannover, Germany), and stored on ice. As determined by SDS/12% polyacrylamide gel electrophoresis followed by silver nitrate staining, the preparations contained only a single band with a Mr of ≈36 kDa. Protein was assayed by using a MicroBCA kit (Pierce).
Fluorescence Spectroscopy. Fluorescence was recorded by a Fluorolog 3 spectrofluorimeter (Spex Industries, Edison, NJ), equipped with a double-grating monochromator, Glan Thompson polarizers, and a cuvette holder thermostated at 25°C. All experiments were carried out in 20 mM Tris·HCl buffer (pH 7.5) containing 0.008% DDM at a given final protein concentration.
Time courses of MIANS labeling were conducted with 1 μM protein in 20 mM Tris·HCl (pH 7.5), containing 0.008% DDM in 1 × 1-cm quartz cuvettes with constant stirring at 25°C. Reactions were initiated by adding MIANS to a final concentration of 5 μM, and fluorescence was recorded over time. Excitation and emission wavelengths were 332 and 415 nm, respectively; slits were 2 nm in both monochromators.
Fluorescence-quenching experiments were performed by adding appropriate aliquots of stock solutions of 5 M CsCl, KI, or acrylamide to 200-μl samples containing 5 μM LacY. The final concentration of the quencher was as indicated. The data were analyzed by using Stern–Volmer plots for collisional quenching,
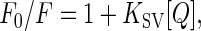 |
[1] |
where F0 and F correspond to the fluorescence emission of the protein in the absence and presence of quencher at a given concentration [Q]. The slope of the plot F0/F over [Q] is the Stern–Volmer constant (KSV), which is defined by the product of the bimolecular rate constant of the quenching process (kq) and the fluorophore lifetime in the absence of quencher (τ) (36). Because acrylamide absorbs weakly at 295 nm, fluorescence intensity was corrected as described (37).
Steady-state f luorescence emission and f luorescence-quenching studies were performed in 0.3 × 1-cm quartz cuvettes. Tryptophan was exited at 295 nm, and emission spectra were recorded at 1-nm intervals between 310 and 450 nm by using an integration time of 0.5 s. In all cases, polarizers were used in a magic-angle position (54.7°) to correct for polarization effects on intensity, reduce direct contribution of light scattering, and eliminate polarization effects in monochromator transmittance (38). Each spectrum is the average of two scans.
Phosphorescence measurements were conducted at 77 K by using a xenon-flash lamp as an excitation source. The excitation wavelength (280 nm) was selected with a monochromator (12-nm band pass), and the spectra were recorded between 390 and 490 nm at 1-nm intervals. Data were acquired with an initial delay of 0.1 ms, and every data point corresponds to the accumulation of 200 flashes. Measurements were made in a 5 × 5-mm quartz cuvette immersed in liquid nitrogen.
Results
Properties of Single-W151/C154G LacY. Previous studies (39) describe the construction of a functional permease in which each native tryptophan residue was replaced with phenylalanine. However, expression of single-Trp-151 LacY with phenylalanine in place of the remaining native tryptophan residues is poor. Because phenylalanine residues are essentially randomly distributed throughout transmembrane helical regions, whereas tyrosine residues like tryptophan tend to be distributed near the membrane–water interface (40), each tryptophan residue except Trp-151 was replaced with tyrosine instead of phenylalanine, which markedly increases expression with vector pCS19 (L.G. and H.R.K., unpublished data).
The mutant C154G binds ligand at least as well as wild-type LacY but is severely defective with respect to all modes of sugar translocation across the membrane (33, 41, 42). In addition, the mutant undergoes conformational changes associated with substrate binding at a negligible rate, indicating that the molecule favors a particular conformation (6, 33). The C154G mutant also exhibits little tendency to aggregate in DDM and is thermostable (33). In this study, the C154G mutation was introduced into single-Trp-151 LacY to study a well expressed, stable LacY molecule containing a single tryptophan residue in the sugar-binding site.
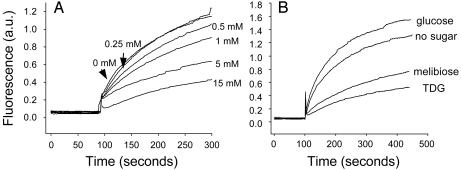
MIANS is a particularly convenient probe because it is essentially nonfluorescent until the maleimide moiety reacts covalently with a Cys residue (43), when a time-dependent increase in fluorescence is observed. At a low MIANS/LacY ratio, the increase in fluorescence is due primarily to reaction with Cys-148, as shown by mass spectrometry (44), and the reaction is inhibited as a function of substrate concentration. Similar behavior is observed with single Trp-151/C154G (Fig. 2A); increasing concentrations of TDG progressively protect against MIANS labeling of this mutant in much the same manner as observed previously for the mutant C154G (33).
Fig. 2.
Substrate binding by single-W151/C154G LacY. Time courses of MIANS labeling of purified detergent-solubilized LacY in the absence or presence of different sugars are shown. Reactions were initiated by adding 5 μM MIANS to 1 μM LacY. Fluorescence was recorded as described in Experimental Procedures. (A) Protection against MIANS labeling of single-W151/C154G LacY by given concentrations of TDG. (B) Effect of TDG, melibiose, or glucose (15 mM final concentration) on the rate of MIANS labeling.
Similar MIANS protection is observed in the presence of another LacY substrate, melibiose (Fig. 2B), whereas glucose, which is not a substrate for LacY, causes a mild increase in the rate and extent of MIANS fluorescence in all likelihood because of a nonspecific increase in viscosity. The findings demonstrate that purified single-W151/C154G LacY in DDM retains high affinity and specificity.
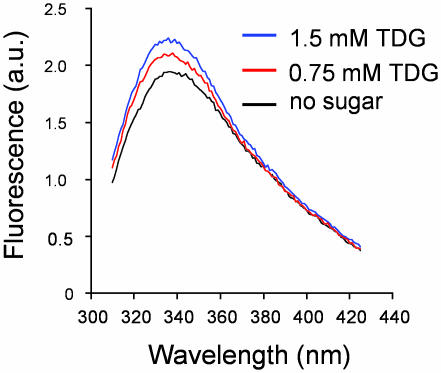
Steady-State Emission Properties of Trp-151. In the absence of ligand, the Trp-151 emission spectrum is relatively broad, with a maximum at ≈340 nm (Fig. 3), indicating that the indole ring is solvated by water (38) although it is located approximately in the middle of the membrane (45). The emission properties clearly agree with the position of Trp-151 in the crystal structure (6). Importantly, addition of TDG shifts the emission maximum to a shorter wavelength (≈336 nm) in a highly reproducible fashion and at the same time narrows the spectrum, indicating that the indole ring becomes less accessible to water. However, the mobility of Trp-151, as reflected by steady-state emission anisotropy, does not change significantly after addition of TDG. It is unlikely, therefore, that reorientation of the aromatic ring causes the observed blue shift. Rather, the blue shift likely represents a change in polarity of the local environment of Trp-151, resulting from hydrophobic stacking between the galactopyranosyl ring of the sugar and the indole ring of Trp-151, thereby excluding water molecules from the side chain.
Fig. 3.
Effect of TDG on fluorescence emission of Trp-151. The emission spectrum of purified detergent-solubilized single-W151/C154G LacY (final concentration 5 μM) was measured in the absence (bottom curve) or presence (top and middle curves) of given concentrations of TDG as described in Experimental Procedures with an excitation wavelength of 295 nm. The results shown are typical of ≈20 independent experiments.
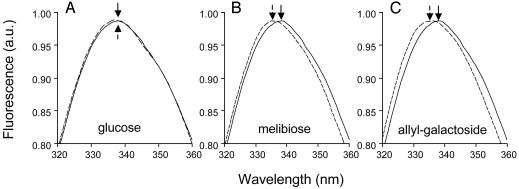
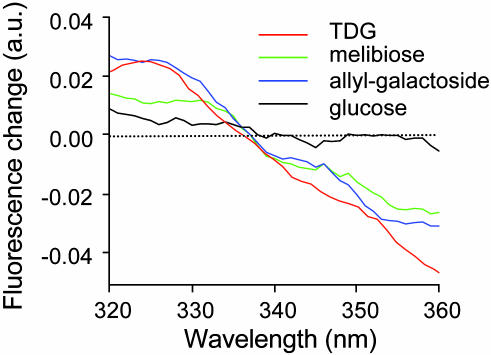
Glucose does not shift the normalized Trp-151 emission spectra (Fig. 4A). However, similar small but reproducible blue shifts are observed with other LacY substrates, such as melibiose (Fig. 4B) and particularly allyl-α-d-galactopyranoside (Fig. 4C). For clarity, Fig. 5 shows the difference in the Trp-151 emission spectra determined in the presence or absence of given sugars. The negative difference above 340 nm represents the displacement of Trp-151 maximum emission wavelength to shorter wavelengths in the presence of LacY substrates. The data provide strong evidence that the galactopyranosyl moiety of the sugar is solely responsible for the shift in Trp-151 emission.
Fig. 4.
Effect of various sugars on the emission spectrum of Trp-151. For each experiment, 5 μM purified detergent-solubilized single-W151/C154G LacY was incubated in the absence (solid line) or presence (broken line) of the indicated sugar at a final concentration of 15 mM before recording the emission spectrum of Trp-151. The excitation wavelength was 295 nm. For clarity, the emission spectra were normalized to the emission maximum in each experiment. The arrows indicate the positions of the maximum emission wavelengths. The results shown are typical of ≈20 independent experiments.
Fig. 5.
Changes of Trp-151 emission spectrum in the presence of various sugars. The curves were generated by using the data from Figs. 3 and 4 and subtracting the normalized Trp-151 emission spectrum in the absence of sugars from the normalized emission spectrum in the presence of different sugars. Experimental conditions and data collection are as described for Figs. 3 and 4.
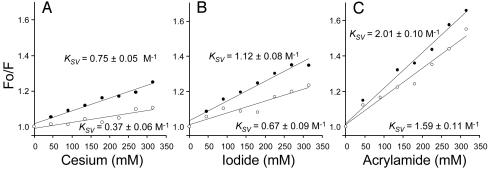
Quenching of Trp-151 Fluorescence. Changes in polarity of the environment surrounding Trp-151 were studied further by fluorescence quenching. The water-soluble ions Cs+ (Fig. 6A) and I– (Fig. 6B) were used as collisional quenchers to probe the solvent accessibility of Trp-151 in the absence or presence of TDG. Acrylamide, another collisional quencher, which is neutral, was also tested (Fig. 6C) to investigate possible electrostatic effects of I– or Cs+. All experimental data fit linear Stern–Volmer plots for collisional (dynamic) quenching, thereby excluding a contribution from static quenching (46). The extent of exposure of Trp-151 to a given quencher is related to the slope of the plot, which yields the collisional KSV (see Experimental Procedures). In the absence of TDG, the KSV values obtained for all three quenchers reflect moderate accessibility of Trp-151 to solvent (36). Moreover, the accessibility of the three quenchers is proportional to their size, indicating that Trp-151 is in a relatively neutral environment. Importantly, there is a significant reduction of the KSV values for all three quenchers in the presence of TDG (50%, 60%, and 30% for Cs+, I–, and acrylamide, respectively), providing further evidence that Trp-151 is shielded from the surrounding solvent by substrate.
Fig. 6.
The effect of sugar on collisional quenching of Trp-151 fluorescence. Stern–Volmer plots of Trp-151 fluorescence quenching by Cs+ (Cesium) (A), I– (Iodide) (B), or acrylamide (C) in the absence (filled circles) or presence (open circles) of 15 mM TDG are shown. Purified detergent-solubilized single-W151/C154G LacY (5 μM) was titrated with small aliquots of stock solutions of 5 M CsCl, KI, or acrylamide. The data were fit to linear Stern–Volmer plots (see Experimental Procedures), and KSV values were calculated from the slope of the plots. Fluorescence spectra were recorded as described previously.
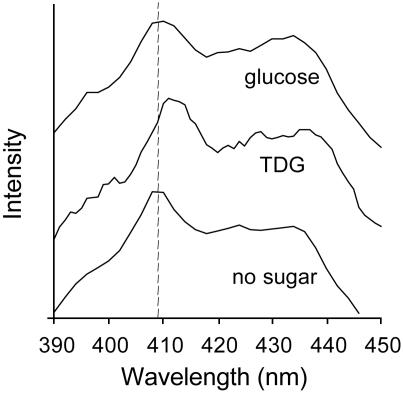
Phosphorescence. Trp-151 phosphorescence studies provide further support for hydrophobic stacking between the indole side chain and the galactopyranosyl moiety of substrate (Fig. 7). In the absence of sugar, the characteristic vibronic 0,0 band of tryptophan appears at 408 nm, indicating that the indole group is exposed to a polar environment (47, 48). Significantly, the 0,0 band of Trp-151 is reproducibly red-shifted 4 nm in the presence of TDG, but no change is observed in the presence of glucose. The spectral shift observed in the presence of TDG is consistent with hydrophobic stacking between the indole group of Trp-151 and the galactopyranosyl ring of the lactose analog.
Fig. 7.
Effect of TDG or glucose on Trp-151 phosphorescence. Phosphorescence spectra at 77 K of 10 μM purified detergent-solubilized single-W151/C154G LacY in the absence or presence of a given sugar are shown (final concentration 15 mM). The excitation wavelength was 280 nm, and the spectra were recorded as described in Experimental Procedures.
Discussion
Until very recently, crystal structures of sugar-binding proteins were the only atomic-resolution structures that provide insight into the nature of sugar–protein interactions (29–32). The x-ray structure of LacY reveals sugar–protein interactions that are similar to those observed in many other sugar-binding proteins (6). Evidently, both affinity and specificity are driven by H-bonding interactions between the OH groups of the pyranose ring and polar side chains of the protein, sometimes mediated by water molecules. Although energetically less important, crystal structures commonly reveal stacking interactions between aromatic amino acid residues and the pyranose ring of the sugar, and their proximity (3–4 Å) suggests that the binding interactions are driven primarily by hydrophobic and van der Waals interactions.
The sugar-binding site of LacY observed in the x-ray crystal structure shows that the galactopyranosyl group of TDG is oriented toward the indole group of Trp-151, which strongly suggests a hydrophobic stacking interaction, as in the case of soluble sugar-binding proteins. The importance of the aromaticity in position 151 regarding substrate binding has been revealed by studying binding (27). Replacement of the Trp-151 by either phenylalanine or tyrosine results in a decrease in substrate affinity by 50- and 20-fold, respectively, with relatively little change in the kinetics of lactose transport. Moreover, the interaction between Trp-151 and the galactopyranosyl ring may orient the substrate in such a position that important H-bonds can be formed (Fig. 1B).
As an indirect method, the luminescence properties of Trp-151 provide a powerful tool to understand the nature of this interaction fully, and single-Trp-151/C154G LacY was constructed for this purpose. The emission properties of Trp-151 reveal that the indole side chain is partially solvated by water and are totally consistent with the crystal structure. Interestingly, Trp-151 has restricted mobility, which is reflected by a high steady-state anisotropy value of 0.17 (the limiting anisotropy value of tryptophan is ≈0.25). Furthermore, moderate accessibility of Trp-151 to solvent and the neutral character of the local environment are consistent with fluorescence-quenching experiments using Cs+, I–, or acrylamide as collisional-quenching agents.
Hydrophobic interaction of the galactopyranosyl ring with Trp-151 is revealed by observations of (i) a blue shift in the fluorescence emission spectrum in the presence of LacY substrates, indicating that Trp-151 becomes less solvated by water, and (ii) marked reduction in solvent accessibility as revealed by decreased KSV values in the presence of ligand. These observations are consistent with the interpretation that the galactopyranosyl ring is in close proximity to Trp-151 and replaces the water molecules solvating the indole ring.
As revealed by the structure, the distance between the galactopyranosyl ring and the center of the indole ring of Trp-151 is ≈4 Å. However, is the galactopyranosyl ring sufficiently close to Trp-151 for hydrophobic stacking and/or van der Waal interactions? Drawing from several examples in chemistry and biology, interactions between the π electrons of aromatic side chains and the C—OH-bonds present in hexopyranoses are a common motif (49). The O and OH groups of sugars have an inductive effect on the C—H-bond H atoms, making them more positively charged and enhancing the possibility for interaction with electron-rich aromatic rings (50). Consequently, the electron distribution of the aromatic residue becomes more diffuse, which is reflected in the shift of its phosphorescence spectrum to longer wavelengths (47, 51). Tryptophan phosphorescence spectroscopy has been used (52–54) to study stacking interactions of nucleic acids with various trytophan-containing peptides and proteins. The results presented in Fig. 7 demonstrate clearly that the phosphorescence spectrum of Trp-151 is red-shifted in the presence of TDG but not in the presence of glucose. This finding is consistent with the conclusion that Trp-151 stacks hydrophobically with TDG.
Finally, it is apparent that the x-ray crystal structure and the findings presented here are highly complementary and, as such, provide strong evidence for the conclusion that the galactopyranosyl moiety of LacY substrates stacks with Trp-151. This strong correlation demonstrates the feasibility of luminescence spectroscopic tools for studying sugar–protein interactions.
Acknowledgments
We thank Michael Ehrmann for generously providing plasmid pCS19. We also thank Ruth Gordillo, Miguel Prieto, Adam Weinglass, Shushi Nagamori, Ernest Wright, and Douglas Rees for advice and helpful suggestions and Vladimir Kasho for molecular graphics. This work was supported in part by National Institutes of Health Grant DK 5113108 (to H.R.K.).
Abbreviations: DDM, dodecyl-β-d-maltopyranoside; KSV, Stern–Volmer quenching constant; LacY, lactose permease; MIANS, 2-(4-maleimidylanilino)naphthalene-6-sulfonic acid; TDG, β-d-galactopyranosyl-1-thio-β-d-galactopyranoside.
References
- 1.Saier, M. H., Jr. (2000) Mol. Microbiol. 35 699–710. [DOI] [PubMed] [Google Scholar]
- 2.Konings, W. N., Kaback, H. R. & Lolkema, J. S. (1996) Handbook of Biological Physics: Transport Processes in Eukaryotic and Prokaryotic Organisms (Elsevier, Amsterdam).
- 3.Foster, D. L., Boublik, M. & Kaback, H. R. (1983) J. Biol. Chem. 258 31–34. [PubMed] [Google Scholar]
- 4.Calamia, J. & Manoil, C. (1990) Proc. Natl. Acad. Sci. USA 87 4937–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaback, H. R. & Wu, J. (1997) Q. Rev. Biophys. 30 333–364. [DOI] [PubMed] [Google Scholar]
- 6.Abramson, J., Smirnova, I., Kasho, V., Verner, G., Kaback, H. R. & Iwata, S. (2003) Science 301 610–615. [DOI] [PubMed] [Google Scholar]
- 7.Costello, M. J., Escaig, J., Matsushita, K., Viitanen, P. V., Menick, D. R. & Kaback, H. R. (1987) J. Biol. Chem. 262 17072–17082. [PubMed] [Google Scholar]
- 8.Sahin-Tóth, M., Lawrence, M. C. & Kaback, H. R. (1994) Proc. Natl. Acad. Sci. USA 91, 5421–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun, J. & Kaback, H. R. (1997) Biochemistry 36 11959–11965. [DOI] [PubMed] [Google Scholar]
- 10.Guan, L., Murphy, F. D. & Kaback, H. R. (2002) Proc. Natl. Acad. Sci. USA 99 3475–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ermolova, N., Guan, L. & Kaback, H. R. (2003) Proc. Natl. Acad. Sci. USA 100 10187–10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandermann, H., Jr. (1977) Eur. J. Biochem. 80 507–515. [DOI] [PubMed] [Google Scholar]
- 13.Olsen, S. G. & Brooker, R. J. (1989) J. Biol. Chem. 264 15982–15987. [PubMed] [Google Scholar]
- 14.Sahin-Tóth, M., Akhoon, K. M., Runner, J. & Kaback, H. R. (2000) Biochemistry 39 5097–5103. [DOI] [PubMed] [Google Scholar]
- 15.Wu, J. & Kaback, H. R. (1994) Biochemistry 33 12166–12171. [DOI] [PubMed] [Google Scholar]
- 16.Sahin-Tóth, M., Lawrence, M. C., Nishio, T. & Kaback, H. R. (2001) Biochemistry 43 13015–13019. [DOI] [PubMed] [Google Scholar]
- 17.Frillingos, S., Sahin-Tóth, M., Wu, J. & Kaback, H. R. (1998) FASEB J. 12 1281–1299. [DOI] [PubMed] [Google Scholar]
- 18.Kaback, H. R., Sahin-Tóth, M. & Weinglass, A. B. (2001) Nat. Rev. Mol. Cell Biol. 2 610–622. [DOI] [PubMed] [Google Scholar]
- 19.Hirai, T., Heymann, J. A., Maloney, P. C. & Subramaniam, S. (2003) J. Bacteriol. 185 1712–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, Y., Lemieux, M. J., Song, J., Auer, M. & Wang, D. N. (2003) Science 301 616–620. [DOI] [PubMed] [Google Scholar]
- 21.Frillingos, S., Gonzalez, A. & Kaback, H. R. (1997) Biochemistry 36 14284–14290. [DOI] [PubMed] [Google Scholar]
- 22.Venkatesan, P. & Kaback, H. R. (1998) Proc. Natl. Acad. Sci. USA 95 9802–9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahin-Tóth, M., le Coutre, J., Kharabi, D., le Maire, G., Lee, J. C. & Kaback, H. R. (1999) Biochemistry 38 813–819. [DOI] [PubMed] [Google Scholar]
- 24.Zhao, M., Zen, K. C., Hubbell, W. L. & Kaback, H. R. (1999) Biochemistry 38 7407–7412. [DOI] [PubMed] [Google Scholar]
- 25.Wolin, C. D. & Kaback, H. R. (2000) Biochemistry 39 6130–6135. [DOI] [PubMed] [Google Scholar]
- 26.Weinglass, A. B., Whitelegge, J. P., Hu, Y., Verner, G. E., Faull, K. F. & Kaback, H. R. (2003) EMBO J. 22 1467–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan, L., Hu, Y. & Kaback, H. R. (2003) Biochemistry 42 1377–1382. [DOI] [PubMed] [Google Scholar]
- 28.Vyas, N. K., Vyas, M. N. & Quiocho, F. A. (1988) Science 242 1290–1295. [DOI] [PubMed] [Google Scholar]
- 29.Vyas, N. K., Vyas, M. N. & Quiocho, F. A. (1991) J. Biol. Chem. 266 5226–5237. [PubMed] [Google Scholar]
- 30.Sixma, T. K., Pronk, S. E., Kalk, K. H., van Zanten, B. A., Berghuis, A. M. & Hol, W. G. (1992) Nature 355 561–564. [DOI] [PubMed] [Google Scholar]
- 31.Merritt, E. A., Sarfaty, S., Feil, I. K. & Hol, W. G. (1997) Structure (London) 5 1485–1499. [DOI] [PubMed] [Google Scholar]
- 32.Quicho, F. A. & Vyas, N. K. (1999), ed. Hecht, S. M. (Oxford Univ. Press, Oxford), pp. 441–457.
- 33.Smirnova, I. N. & Kaback, H. R. (2003) Biochemistry 42 3025–3031. [DOI] [PubMed] [Google Scholar]
- 34.Spiess, C., Beil, A. & Ehrmann, M. (1999) Cell 97 339–347. [DOI] [PubMed] [Google Scholar]
- 35.le Coutre, J., Narasimhan, L. R., Patel, C. K. & Kaback, H. R. (1997) Proc. Natl. Acad. Sci. USA 94 10167–10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eftink, M. R. & Ghiron, C. A. (1976) Biochemistry 15 672–680. [DOI] [PubMed] [Google Scholar]
- 37.Coutinho, A. & Prieto, M. (1993) J. Chem. Educ. 70 425–428. [Google Scholar]
- 38.Lakowicz, J. R. (1999) Principles of Fluorescence Spectroscopy (Academic, New York).
- 39.Menezes, M. E., Roepe, P. D. & Kaback, H. R. (1990) Proc. Natl. Acad. Sci. USA 87 1638–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White, S. H. & Wimley, W. C. (1999) Annu. Rev. Biophys. Biomol. Struct. 28 319–365. [DOI] [PubMed] [Google Scholar]
- 41.Menick, D. R., Sarkar, H. K., Poonian, M. S. & Kaback, H. R. (1985) Biochem. Biophys. Res. Commun. 132 162–170. [DOI] [PubMed] [Google Scholar]
- 42.van Iwaarden, P. R., Driessen, A. J., Lolkema, J. S., Kaback, H. R. & Konings, W. N. (1993) Biochemistry 32 5419–5424. [DOI] [PubMed] [Google Scholar]
- 43.Haugland, R. P. (2002) Handbook of Fluorescent Probes and Research Products (Molecular Probes, Eugene, OR).
- 44.le Coutre, J. & Kaback, H. R. (2000) Biopolymers 55 297–307. [DOI] [PubMed] [Google Scholar]
- 45.Sorgen, P. L., Hu, Y., Guan, L., Kaback, H. R. & Girvin, M. E. (2002) Proc. Natl. Acad. Sci. USA 99 14037–14040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dewey, G. (1991) Biophysical and Biochemical Aspects of Fluorescence Spectroscopy (Plenum, New York).
- 47.Geacintov, N. E. & Brenner, H. C. (1989) Photochem. Photobiol. 50 841–858. [DOI] [PubMed] [Google Scholar]
- 48.Papp, S. & Vanderkooi, J. M. (1989) Photochem. Photobiol. 49 775–784. [DOI] [PubMed] [Google Scholar]
- 49.Brandl, M., Weiss, M. S., Jabs, A., Suhnel, J. & Hilgenfeld, R. (2001) J. Mol. Biol. 307 357–377. [DOI] [PubMed] [Google Scholar]
- 50.Meyer, E., Castellano, R. & Diederich, F. (2003) Angew. Chem. Int. Ed. Engl. 42 1210–1250. [DOI] [PubMed] [Google Scholar]
- 51.Co, T. & Maki, A. H. (1978) Biochemistry 17 182–186. [DOI] [PubMed] [Google Scholar]
- 52.Khamis, M. I., Casas-Finet, J. R. & Maki, A. H. (1987) J. Biol. Chem. 262 1725–1733. [PubMed] [Google Scholar]
- 53.Misra, A., Ozarowski, A., Casas-Finet, J. R. & Maki, A. H. (2000) Biochemistry 39 13772–13780. [DOI] [PubMed] [Google Scholar]
- 54.Maki, A. H., Ozarowski, A., Misra, A., Urbaneja, M. A. & Casas-Finet, J. R. (2001) Biochemistry 40 1403–1412. [DOI] [PubMed] [Google Scholar]