Abstract
Hepatocyte growth factor/scatter factor (HGF/SF), acting through the Met receptor, plays an important role in most human solid tumors, and inappropriate expression of this ligand–receptor pair is often associated with poor prognosis. The molecular basis for the malignant potential of the HGF/SF–Met signal in cancer cells has mostly been attributed to its mitogenic and invasive properties. However, HGF/SF also induces angiogenesis, but the signaling mechanism has not been fully explained, nor has this activity been directly associated with HGF/SF–Met-mediated tumorigenesis. It is known that HGF/SF induces in vitro expression of vascular endothelial growth factor (VEGF), a key agonist of tumor angiogenesis; by contrast, thrombospondin 1 (TSP-1) is a negative regulator of angiogenesis. Here, we show that, in the very same tumor cells, in addition to inducing VEGF expression, HGF/SF dramatically down-regulates TSP-1 expression. We show that TSP-1 shut-off plays an important, extrinsic role in HGF/SF-mediated tumor development, because ectopic expression of TSP-1 markedly inhibits tumor formation through the suppression of angiogenesis. Interestingly, although VEGF-induced expression is sensitive to inhibitors of several pathways, including mitogen-activated protein kinase, phosphoinositide 3-kinase, and signal transducer and activator of transcription 3, TSP-1 shut-off by HGF/SF is prevented solely by inhibiting mitogen-activated protein kinase activation. These studies identify HGF/SF as a key switch for turning on angiogenesis. They suggest that TSP-1 is a useful antagonist to tumor angiogenesis and that it may have therapeutic value when used in conjunction with inhibitors of VEGF.
Hepatocyte growth factor/scatter factor (HGF/SF) and its tyrosine kinase receptor, Met, have been associated with most types of human cancers, and their expression often correlates with poor prognosis and metastases (1, 2). Constitutively active mutations in Met, either sporadic or inherited, have been found in human cancers, providing strong genetic evidence for the role of Met in human malignancies (1). Multiple biological outcomes of HGF/SF–Met signaling account for its role in cancer, among which the most critical are cell proliferation, tumor cell invasion, and angiogenesis (1).
Angiogenesis is an essential component for tumor development (3) regulated by both proangiogenic and antiangiogenic factors (4). Vascular endothelial growth factor (VEGF) is a potent agonist of angiogenesis that activates both endothelial cell proliferation and migration (5). By contrast, thrombospondin 1 (TSP-1) suppresses angiogenesis by inhibiting endothelial cell proliferation and inducing endothelial cell apoptosis (6, 7). It has been previously shown that TSP-1 expression is positively regulated by the p53, PTEN, Smad, and APC tumor suppressor proteins (8–11). It can also be down-regulated by activation of oncoproteins such as ras, src, myc, and c-jun as well as by a novel metastasis-associated gene product, MTS-1 (12–15). Many cells express TSP-1, and low levels of TSP-1 expression have been associated with increased recurrence rates and decreased overall survival in several human cancers (6), suggesting that the loss of TSP-1 is critical for tumor development. Overexpression of TSP-1 in human skin carcinoma, mammary carcinoma, glioblastoma, hemangioblastoma, and colorectal cancer cell lines has been shown to block angiogenesis and suppress tumor progression (16–20). Thus, VEGF and TSP-1 can contribute to an angiogenic switch, with the type of angiogenic effector that becomes dominant, determining angiogenesis or vascular quiescence (4). HGF/SF is angiogenic: the ligand–receptor interaction stimulates endothelial cells to proliferate and migrate in vitro, induces blood vessel formation in vivo (21–23), and induces the VEGF expression in human cancer cells (24, 25). Here, we show that HGF/SF–Met signaling operates as a key switch, in turning on VEGF and turning off TSP-1 expression, thus invoking angiogenesis.
Materials and Methods
Cell Lines and Growth Conditions. The human leiomyosarcoma cell line SK-LMS-1 and the breast cancer cell line MDA-MB-231 were obtained from American Type Culture Collection. The autocrine SK-LMS-1 cell line stably transfected with human HGF/SF cDNA (designated SK/HGF hereafter) has been described (26). The SK/HGF cell line that stably transfected with signal transducer and activator of transcription 3 (Stat3)β (designated SK/HGF-Stat3β) is described elsewhere (27). The series of SK-LMS-1 cell lines were grown in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin solution (Invitrogen). The MDA-MB-231 cells were grown in RPMI medium 1640 supplemented with 5% FBS and 1% penicillin/streptomycin (Invitrogen).
RNA Preparation. Total RNA was isolated from cells with or without treatment with recombinant HGF (200 units/ml) by using TRIzol reagent (Invitrogen). For inhibitor treatments, cells were administered with different inhibitors for 1 h before HGF stimulation. All inhibitors were dissolved in DMSO. The working concentrations of individual inhibitors used in this study were 80 μM PD98059, 40 μM U0126, and 40 μM LY294002 (Cell Signaling Technology, Beverly, MA). As a control, the same amount of DMSO was used to treat cells for the same amount of time before HGF treatment.
Northern Blot Analyses and RT-PCR. Total RNA (10–20 μg per sample) was separated in 1% agarose-formaldehyde gel and was transferred onto a Hybond-XL nylon membrane (Amersham Biosciences). Northern blot analyses were performed according to the manufacturer's protocol. The DNA probes used in this study were generated by RT-PCR amplification from RNAs isolated from SK-LMS-1 cells by using SuperScript One-Step RT-PCR with the Platinum Taq System (Invitrogen). The locations of RT-PCR products were nucleotides 141–803 of human TSP-1 (GenBank accession no. XM_007606), nucleotides 803-1260 of human VEGF (accession no. AF022375), nucleotides 92–1090 of human GAPDH (accession no. NM_002046), and nucleotides 1617–2311 of human HGF (accession no. X16323). All RT-PCR products were confirmed by sequencing.
Expression Plasmids and Transfection. The expression vector pcDNA3/Hygro-TSP1 was constructed by insertion of the TSP-1 coding sequence released from pcDNA1-TSP1 into HindIII and XbaI sites of the pcDNA3.1/Hygro vector (Invitrogen), which carries a hygromycin-resistant gene. For establishment of stable cell lines with or without ectopic expression of TSP-1, the SK/HGF cells were transfected with either pcDNA3.1/Hygro empty vector or pcDNA3/Hygro-TSP1 by using FuGENE 6 reagent (Roche Applied Science). Forty-eight hours after transfection, cells were subjected to drug selection by the addition of 400 μg/ml Hygromycin B (Invitrogen). Two weeks after Hygromycin B addition, individual colonies were picked and screened for positive clones (i.e., those having TSP-1 expression) by RT-PCR or Northern blot.
Immunoprecipitation and Western Blot Analyses. Cells were treated with different inhibitors 1 h before HGF stimulation. After treatment with HGF for 15 min, cells were lysed in RIPA buffer consisting of 20 mM Tris·HCl (pH 7.5), 150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 1 mM EDTA, 50 mM NaF, 1 mM sodium orthovanadate, and Complete Proteinase Inhibitor Mixture Tablets (Roche Applied Science). Protein concentrations were quantified by using the DC Protein Assay (Bio-Rad). For immunoprecipitation, 400 μg of protein was incubated overnight with anti-h-Met antibody (c-28, Santa Cruz Biotechnology) in 1 ml of RIPA buffer at 4°C. Immune complexes were collected by using protein A-agarose beads (Invitrogen) and washed three times in the same buffer. Immune complexes were eluted in 20 μl of Laemmli sample buffer (Sigma) and boiled for 5 min before being loaded onto a gel. For tissue samples, 100 mg of tumor tissues were homogenized in 1 ml of RIPA buffer. For Western blot analyses, 40 μg of protein extracts was separated in a Tris-glycine gel (Invitrogen) and transferred onto a polyvinylidene difluoride membrane. Western blot analyses were performed as described (27). The antibodies used for detection were anti-thrombospondin (Ab-3, Oncogene Research Products), anti-phosphotyrosine (clone 4G10, Upstate Biotechnology, Lake Placid, NY), anti-h-Met (C-28), anti-phospho-p44/42 mitogen-activated protein kinase (MAPK) (Thr-202/Tyr-204), anti-p44/42 MAPK, anti-Phospho-Akt (Ser-473), and anti-Akt (Cell Signaling Technology).
Tumorigenicity Study. Female athymic nude mice (Ncr nu/nu; Animal Production Area, National Cancer Institute–Frederick Cancer Research and Development Center, Frederick, MD) at 4 weeks old were used for this study. For each mouse, 1 × 106 cells suspended in 100 μl of serum-free DMEM were s.c. injected into the back. Tumor growth was monitored and measured twice a week. At the end of the experiment, tumors were removed from each mouse and frozen in liquid nitrogen or fixed in formalin.
Immunohistochemistry. Immunohistochemical staining of blood vessels was performed on 5-μm paraffin-embedded tissue sections by using rat anti-mouse CD31 antibody (Pharmingen) and was detected by using the rat ABC Staining System (Santa Cruz Biotechnology). Sections were counterstained with hematoxylin. After staining, the section was observed under a light microscope and three CD31-positive (“hot”) fields were photographed under ×10 magnification for each tumor section. The representative number of CD31-positive vessels was counted based on the total from the three digitized images from each tumor section.
Results and Discussion
Gene expression analysis studies performed on a human leiomyosarcoma cell line (SK-LMS-1, data not shown) showed that VEGF expression increased after HGF/SF treatment, as was previously reported for other tumor cells lines (24, 25). We performed Northern analysis on SK-LMS-1 cells and showed that HGF/SF induced the expression of VEGF, which persisted for at least 48 h (Fig. 1A). VEGF was also elevated in long-term cultures of the SK/HGF cell line (SK-LMS-1 cells having autocrine regulation of HGF/SF; ref. 26). We also examined MDA-MB-231 cells, a human breast cancer cell line, and, as with SK-LMS-1, after HGF/SF treatment the levels of VEGF increased and persisted for 48 h (Fig. 1B).
Fig. 1.
HGF/SF up-regulates VEGF and down-regulates TSP-1 expression in SK-LMS-1 cells (A) and MDA-MB-231 cells (B). Total RNA was prepared from SK-LMS-1 cells or MDA-MB-231 cells treated with recombinant human HGF/SF (200 units/ml) at the indicated time points. Total RNA was also prepared from the SK/HGF cell line, a long-term culture derivative of SK-LMS-1 cells that is autocrine for human HGF/SF (26). Northern blots were probed with 32P-radiolabeled TSP-1, VEGF, or GAPDH cDNA fragments, respectively.
Using gene expression profiling, we also observed that the antiangiogenic factor TSP-1 was decreased in response to HGF/SF stimulation. This observation was confirmed in SK-LMS-1 cells by Northern blot analyses (Fig. 1 A) after HGF/SF treatment. This effect was seen as early as 6 h after HGF treatment and continued to 48 h. Moreover, TSP-1 expression was completely shut down in the SK/HGF cell line. The down-regulation of TSP-1 by HGF/SF was also observed in MDA-MB-231 cells 24 and 48 h after HGF treatment (Fig. 1B).
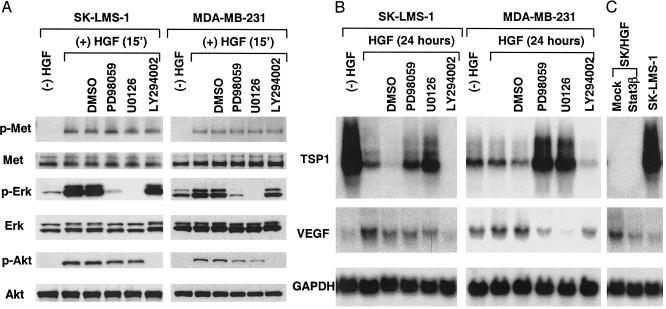
HGF/SF, acting through its tyrosine kinase receptor, Met, is known to activate several intracellular signaling pathways, including MAPK, phosphoinositide 3-kinase (PI3-kinase), and Stat3 (1, 28). We asked which of these pathways might be involved in regulating VEGF and TSP-1 expression. We pretreated SK-LMS-1 and MDA-MB-231 cells with several specific kinase inhibitors for 1 h, followed by 15 min of treatment with HGF/SF (Fig. 2A). In both cell types, phosphorylation of the Met receptor on its tyrosine residues in response to HGF/SF was followed by the activation of down-stream targets of extracellular signal-related kinase (ERK or p44/42 MAPK) and Akt/PI3-kinase. PD98059 and U0126 (which are inhibitors of MEK1, the activator of MAPK) blocked the activation of ERK, whereas LY294002, an inhibitor of PI3-kinase, blocked Akt activation (Fig. 2 A). RNA samples from SK-LMS-1 and MDA-MB-231 cells treated with individual inhibitors followed by HGF/SF treatment for 24 h were subjected to Northern blot analysis. We found that the shutdown of TSP-1 by HGF/SF was blocked by PD98059 or U0126 but not by LY294002 (Fig. 2B) or by overexpression of Stat3β (Fig. 2C), a dominant-negative form of Stat3 that blocks Met-mediated tumorigenesis (27). These results indicated that neither Akt/PI3-kinase nor Stat3 is involved in TSP-1 down-regulation by HGF/SF.
Fig. 2.
HGF/SF–Met signaling pathways regulate TSP-1 and VEGF expression. (A) HGF/SF induces activation of the MAPK and PI3-kinase pathways in SK-LMS-1 cells and MDA-MB-231 cells. Serum-starved cells were untreated ([–]HGF); treated with only HGF ([+]HGF, blank column); or treated with DMSO (control), PD98059 (80 μM), U0126 (40 μM), or LY294002 (40 μM) for 1 h, followed by HGF/SF stimulation for 15 min (a good time point for observing the status of all tyrosine phosphorylations). Whole-cell extracts were prepared, and the state of Met phosphorylation was detected by immunoprecipitation with anti-human Met antibody, followed by Western blot with anti-phosphotyrosine (and/or anti-human Met antibody). For detection of extracellular signal-related kinase (Erk) and Akt, Western blots were probed with anti-phospho p44/42 MAPK, anti-p44/42 MAPK, anti-phospho Akt (Ser-473), or anti-Akt antibodies, respectively. (B) Negative regulation of TSP-1 expression occurs primarily through the MAPK pathway, whereas positive regulation of VEGF expression occurs through MAPK and PI3-kinase pathways. Total RNA was prepared from cells with or without HGF/SF treatment and/or inhibitor treatment. Northern blot analyses were performed as described for Fig. 1. Down-regulation of TSP-1 by HGF/SF was inhibited by the MAPK inhibitors PD98059 or U0126 but was not affected by LY294002. Up-regulation of VEGF was inhibited by PD98059 and U0126 as well as LY294002. (C) VEGF (but not TSP-1) expression was regulated by Stat3 signaling. Total RNA was prepared from SK/HGF cells with or without overexpression of a dominant-negative form of Stat3 and Stat3β (27). Overexpression of Stat3β decreased VEGF expression but did not affect TSP-1 expression in SK/HGF cells.
The MAPK inhibitors PD98059 or U0126 dramatically inhibited TSP-1 down-regulation by HGF/SF in both SK-LMS-1 and MDA-MB-231 cells (Fig. 2B); interestingly, TSP-1 expression after treatment was higher than the basal level in MDA-MB-231 cells (Fig. 2B). This latter finding is consistent with the high basal level of MAPK activity in MDA-MB-231 cells (Fig. 2A) contributing to the low basal level of TSP-1 expression (Fig. 2B). Thus, HGF/SF-mediated down-regulation of TSP-1 depends on the MAPK pathway and is independent of PI3-kinase and Stat3 pathways.
In contrast to the TSP-1 case, we found that, whereas in MDA-MB-231 cells PD98059 and U0126 suppressed VEGF expression induced by HGF/SF, in SK-LMS-1 cells the inhibition was marginal (Fig. 2B). However, VEGF expression was suppressed by LY294002 and Stat3β (Fig. 2 B and C). These data are consistent with previous reports showing that MAPK, PI3-kinase, and Stat3 pathways positively regulate VEGF expression (24, 29). Our results suggest that the HGF/SF-induced down-regulation of TSP-1 and the up-regulation of VEGF are subject to differential regulation by distinct intracellular pathways.
To determine whether down-regulation of TSP-1 by HGF/SF had any biological effect on HGF/SF-induced tumor growth, we overexpressed TSP-1 in SK/HGF cells to generate SK/HGF-TSP1 cells (Fig. 3A). Overexpression of TSP-1 has no effect on cell proliferation or anchorage-independent growth compared with the parental SK/HGF cells in vitro (data not shown). To test whether TSP-1 influences tumorigenicity, we s.c. implanted SK/HGF and SK/HGF-TSP1 cells in athymic nude mice (nu/nu) and compared their tumorigenicities (tumor growth rate). At early times (≈5 weeks), no differences were observed in tumor growth between the two groups. However, by 6 weeks, the difference in tumor volumes between these two groups became more apparent (t test; P < 0.025). We found that HGF/SF-dependent tumor growth was partially inhibited by TSP-1 overexpression (Fig. 3 B and C). TSP-1 protein expression was confirmed in the SK/HGF-TSP1 tumor group by Western blot analysis of tumor tissue (Fig. 3D). These results suggest that down-regulation of TSP-1 by HGF/SF contributes to tumor development.
Fig. 3.
TSP-1 inhibits HGF/SF-induced tumor growth in vivo. (A) TSP-1 was ectopically expressed in SK/HGF cells, establishing the SK/HGF-TSP1 cell line. The expression of TSP-1 in SK/HGF-TSP1 cells was confirmed by Northern blot analysis. (B) Tumor growth of SK-LMS-1 cells and the influence of TSP-1 overexpression. SK-LMS-1 control cells, SK/HGF control cells, and SK/HGF-TSP1 cells (clone 26) were s.c. implanted in athymic nude mice. The animals were monitored for tumor growth, and tumor volume was measured twice a week. The tumor volume values represent an average of four mice for each group (P < 0.025). (C) Visualization of the tumors after the mice were killed. (D) TSP-1 protein in tumor xenografts is derived from SK/HGF-TSP1 cells. Cell extracts were prepared from fresh tumors, and TSP-1 protein was detected by anti-TSP-1 antibody under denaturing conditions.
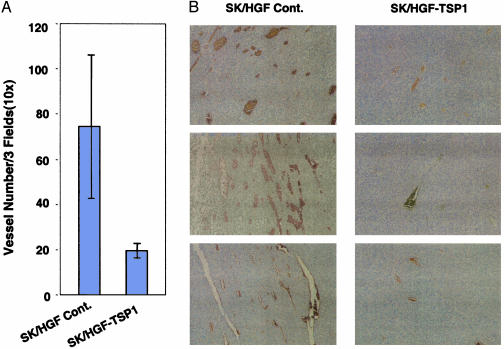
To test whether the inhibition of tumor growth by TSP-1 was due to its extrinsic antiangiogenic activity, we compared microvascular density between these tumor groups. To detect the number of blood vessels in SK/HGF and SK/HGF-TSP1 tumor sections, we used immunohistochemical staining with antibodies against mouse endothelial cell surface marker CD31. The average number of CD31-positive capillary structures in the SK/HGF control tumor group was significantly higher than that in SK/HGF-TSP1 tumor group [Fig. 4 (t test, P < 0.01)]. These results suggest that TSP-1 inhibition of HGF/SF-induced tumor growth occurs through suppression of tumor angiogenesis.
Fig. 4.
TSP-1 inhibits HGF/SF-induced tumor angiogenesis. (A) Decreased neovascularization in SK/HGF-TSP1 tumors: tissue sections prepared from tumors derived from SK/HGF and SK/HGF-TSP1 cells were immunohistochemically stained with anti-mouse CD31 antibody. Three fields (×10 magnification) from each stained tumor section were photographed, and the number of CD31-positive vessels (brown staining) was scored for each field. The values in the graph represent the average number of blood vessels in the three selected sections from each of four tumors for each cell type (P < 0.01). (B) Three representative fields from each group of tumors are displayed.
Our study provides insight into the mechanisms by which HGF induces tumor angiogenesis. The mechanism has two distinct components (Fig. 5). First, HGF/SF acts directly on endothelial cells, inducing proliferation and migration (21–23). Second, HGF/SF acts on tumor cells, in which it triggers angiogenic switching (4) by up-regulating the expression of the proangiogenic factor VEGF (Fig. 1) (24, 25) and down-regulating the expression of TSP-1, an angiogenesis inhibitor (Fig. 1). The distinct components of the angiogenic switch are modulated by means of distinct branches of the HGF signaling pathway; i.e., whereas VEGF is modulated by means of PI3-kinase, Stat3, and, in some cases, MAPK, TSP-1 in these cells is targeted only by means of MAPK (Fig. 5). In addition, it has been shown in vitro that TSP-1 can inhibit HGF/SF activity through direct interaction with the ligand (30). Oncoproteins such as ras and myc have been shown previously to coordinate the expression of VEGF and TSP-1 (12, 31). Recently, Watnick et al. (32) reported that TSP-1 repression by activated H-Ras in human embryonic kidney cells was mediated by the P13-K pathway and not the MAPK pathway that we observe in SK-LMS-1 and MDA-MB-231 cells (Fig. 2). However, they also used the MDA-MB-231 cells in their study, and, similar to our study (Fig. 2), they show that treatment of this cell line with LY294002 did not affect the expression of TSP-1 (32). Cell type differences and variation in the signaling intensity, duration, or downstream molecules induced by the Ras oncoprotein compared to HGF/SF could account for the sensitivity to different signaling inhibitors.
Fig. 5.
Schematic representation of tumor angiogenesis induced by HGF/SF–Met signaling. Intrinsically, HGF/SF activates the Met receptor on the surface of host endothelial cells, inducing proliferation and migration. Extrinsically, HGF/SF–Met signaling turns on the angiogenic switch by simultaneously up-regulating proangiogenic VEGF expression and down-regulating antiangiogenic TSP-1 expression from the tumor cells. We do not exclude the possibility that, in endothelial cells, HGF/SF–Met signaling may have extrinsic activity in addition to intrinsic activity.
The regulation by HGF/SF–Met signaling is systemic and qualifies as a dominant-acting angiogenic switch and would be thus expected to dramatically enhance neovascularization. Hanahan and Folkman (4) have emphasized the importance of angiogenesis for tumor development. Angiogenesis itself depends on tumor growth, and much effort has been directed toward blocking this tumor-associated process. Many angiogenesis inhibitors have been characterized, and >30 are now in clinical trials (33). TSP-1 is an angiogenic factor with potential clinical utility, whereas the neutralizing monoclonal antibody to VEGF, which inhibits tumor angiogenesis, shows promise in clinical trials (34, 35). Given that angiogenic factors like HGF/SF can simultaneously up-regulate VEGF and down-regulate TSP-1 expression (Fig. 1), whether the combination of anti-VEGF neutralizing antibodies plus TSP-1 will provide a therapeutic synergism to inhibit tumor angiogenesis and tumor growth is an important question and needs to be tested. An alternative strategy would be to target the signaling pathways that are responsible for inhibiting TSP-1 shut-off and VEGF expression. Inhibitors of the HGF/SF–Met pathway would therefore have the potential to both block VEGF expression and boost TSP-1 levels while simultaneously interfering with invasion and metastasis.
We have demonstrated that the MAPK pathway plays a dual role in regulating the expression of angiogenic effectors, but it is especially effective in preventing the negative regulation of TSP-1 expression induced by HGF/SF. It is less effective in controlling the up-regulation of VEGF expression in tumor cells such as SK-LMS-1 (Fig. 2B), but the MAPK pathway is an important and intrinsic target in many tumor types (36). Conceivably, a combination of a small molecule MAPK inhibitor coupled with a neutralizing anti-VEGF therapy might be effective. Note that tumor lethal factor (TLF), the anthrax lethal toxin, has been shown to be a potent MAPK inhibitor (37) and also dramatically suppresses tumor angiogenesis (38). The mechanisms underlying the inhibition of tumor angiogenesis by TLF is not clear, but it is possible that TLF can increase the expression of the antiangiogenic factor TSP1 from tumor cells while decreasing VEGF. Obviously, directly targeting HGF/SF and its receptor, Met, could have potent intrinsic and extrinsic antitumor activity. Anti-HGF/SF neutralizing antibodies and the HGF/SF antagonist HGF/NK4, both of which not only inhibit angiogenesis but also inhibit cell proliferation and invasion, have been shown to effectively inhibit tumor growth in animal models (39, 40).
Acknowledgments
We thank Bree Buckner and Bryn Eagleson for technical assistance, Michelle Reed for help with manuscript preparation, David Nadziejka for editing the manuscript, Meg Gustafson for general help, Douglas Hanahan (University of California, San Francisco), Tony Hunter (The Salk Institute for Biological Sciences, La Jolla, CA), and Beatrice Knudsen (Fred Hutchinson Cancer Research Center, Seattle) for reviewing the paper, and the Van Andel Research Institute for financial support.
Abbreviations: HGF/SF, hepatocyte growth factor/scatter factor; VEGF, vascular endothelial growth factor; TSP-1, thrombospondin 1; MAPK, mitogen-activated protein kinase; PI3-kinase, phosphoinositide 3-kinase; Stat3, signal transducer and activator of transcription 3.
References
- 1.Trusolino, L. & Comoglio, P. M. (2002) Nat. Rev. Cancer 4 289–300. [DOI] [PubMed] [Google Scholar]
- 2.Vande Woude, G. F., Jeffers, M., Cortner, J., Alvord, G., Tsarfaty, I. & Resau, J. (1997) CIBA Found. Symp. 212 119–132. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan, D. & Weinberg, R. A. (2000) Cell 100 57–70. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan, D. & Folkman, J. (1996) Cell 86 353–364. [DOI] [PubMed] [Google Scholar]
- 5.Ferrara, N. (2002) Semin. Oncol. 29 10–14. [DOI] [PubMed] [Google Scholar]
- 6.Sargiannidou, I., Zhou, J. & Tuszynski, G. P. (2001) Exp. Biol. Med. 226 726–733. [DOI] [PubMed] [Google Scholar]
- 7.Jimenez, B., Volpert, O. V., Crawford, S. E., Febbraio, M., Silverstein, R. L. & Bouck, N. (2000) Nat. Med. 6 41–48. [DOI] [PubMed] [Google Scholar]
- 8.Dameron, K. M., Volpert, O. V., Tainsky, M. A. & Bouck, N. (1994) Science 265 1582–1584. [DOI] [PubMed] [Google Scholar]
- 9.Wen, S., Stolarov, J., Myers, M. P., Su, J. D., Wigler, M. H., Tonks, N. K. & Durden, D. L. (2001) Proc. Natl. Acad. Sci. USA 98 4622–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarte-Waldhoff, I., Volpert, O. V., Bouck, N. P., Sipos, B., Hahn, S. A., Klein-Scory, S., Luttges, J., Kloppel, G., Graeven, U., Eilert-Micus, C., et al. (2000) Proc. Natl. Acad. Sci. USA 97 9624–9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutierrez, L. S., Suckow, M., Lawler, J., Ploplis, V. A. & Castellino, F. J. (2003) Carcinogenesis 24 199–207. [DOI] [PubMed] [Google Scholar]
- 12.Rak, J., Mitsuhashi, Y., Sheehan, C., Tamir, A., Viloria-Petit, A., Filmus, J., Mansour, S. J., Ahn, N. G. & Kerbel, R. S. (2000) Cancer Res. 60 490–498. [PubMed] [Google Scholar]
- 13.Tikhonenko, A. T., Black, D. J. & Linial, M. L. (1996) J. Biol. Chem. 271 30741–30747. [DOI] [PubMed] [Google Scholar]
- 14.Dejong, V., Degeorges, A., Filleur, S., Ait-Si-Ali, S., Mettouchi, A., Bornstein, P., Binetruy, B. & Cabon, F. (1999) Oncogene 18 3143–3151. [DOI] [PubMed] [Google Scholar]
- 15.Grigorian, M., Andresen, S., Tulchinsky, E., Kriajevska, M., Carlberg, C., Kruse, C., Cohn, M., Ambartsumian, N., Christensen, A., Selivanova, G., et al. (2001) J. Biol. Chem. 276 22699–22708. [DOI] [PubMed] [Google Scholar]
- 16.Bleuel, K., Popp, S., Fusenig, N. E., Stanbridge, E. J. & Boukamp, P. (1999) Proc. Natl. Acad. Sci. USA 96 2065–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyata, Y., Koga, S., Takehara, K., Kanetake, H. & Kanda, S. (2003) Clin. Cancer Res. 9 1734–1740. [PubMed] [Google Scholar]
- 18.Miyanaga, K., Kato, Y., Nakamura, T., Matsumura, M., Amaya, H., Horiuchi, T., Chiba, Y. & Tanaka, K. (2002) Anticancer Res. 22 3941–3948. [PubMed] [Google Scholar]
- 19.Rodriguez-Manzaneque, J. C., Lane, T. F., Ortega, M. A., Hynes, R. O., Lawler, J. & Iruela-Arispe, M. L. (2001) Proc. Natl. Acad. Sci. USA 98 12485–12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo, N., Zabrenetzky, V. S., Chandrasekaran, L., Sipes, J. M., Lawler, J., Krutzsch, H. C. & Roberts, D. D. (1998) Cancer Res. 58 3154–3162. [PubMed] [Google Scholar]
- 21.Bussolino, F., Di Renzo, M. F., Ziche, M., Bocchietto, E., Olivero, M., Naldini, L., Gaudino, G., Tamagnone, L., Coffer, A. & Comoglio, P. M. (1992) J. Cell Biol. 119 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant, D. S., Kleinman, H. K., Goldberg, I. D., Bhargava, M. M., Nickoloff, B. J., Kinsella, J. L., Polverini, P. & Rosen, E. M. (1993) Proc. Natl. Acad. Sci. USA 90 1937–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosen, E. M. & Goldberg, I. D. (1995) Adv. Cancer. Res. 67 257–279. [DOI] [PubMed] [Google Scholar]
- 24.Dong, G., Chen, Z., Li, Z. Y., Yeh, N. T., Bancroft, C. C. & Van Waes, C. (2001) Cancer Res. 61 5911–5918. [PubMed] [Google Scholar]
- 25.Moriyama, T., Kataoka, H., Hamasuna, R., Yokogami, K., Uehara, H., Kawano, H., Goya, T., Tsubouchi, H., Koono, M. & Wakisaka, S. (1998) Biochem. Biophys. Res. Commun. 249 73–77. [DOI] [PubMed] [Google Scholar]
- 26.Jeffers, M., Rong, S. & Vande Woude, G. F. (1996) Mol. Cell. Biol. 16 1115–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang, Y. W., Wang, L. M., Jove, R. & Vande Woude, G. F. (2002) Oncogene 21 217–226. [DOI] [PubMed] [Google Scholar]
- 28.Furge, K. A., Zhang, Y. W. & Vande Woude, G. F. (2000) Oncogene 19 5582–5589. [DOI] [PubMed] [Google Scholar]
- 29.Niu, G., Wright, K. L., Huang, M., Song, L., Haura, E., Turkson, J., Zhang, S., Wang, T., Sinibaldi, D., Coppola, D., et al. (2002) Oncogene 21 2000–2008. [DOI] [PubMed] [Google Scholar]
- 30.Lamszus, K., Joseph, A., Jin, L., Yao, Y., Chowdhury, S., Fuchs, A., Polverini, P., Goldberg, I. D. & Rosen, E. M. (1996) Am. J. Pathol. 149 805–819. [PMC free article] [PubMed] [Google Scholar]
- 31.Baudino, T. A., McKay, C., Pendeville-Samain, H., Nilsson, J. A., Maclean, K. H., White, E. L., Davis, A. C., Ihle, J. N. & Cleveland, J. L. (2002) Genes Dev. 16 2530–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watnick, R. S., Cheng, Y., Rangarajan, A., Ince, T. A. & Weinberg, R. A. (2003) Cancer Cell 3 219–231. [DOI] [PubMed] [Google Scholar]
- 33.Kerbel, R. & Folkman, J. (2002) Nat. Rev. Cancer 2 727–739. [DOI] [PubMed] [Google Scholar]
- 34.Ferrara, N. (2002) Nat. Rev. Cancer 2 795–803. [DOI] [PubMed] [Google Scholar]
- 35.McCarthy, M. (2003) Lancet 361 1959. [DOI] [PubMed] [Google Scholar]
- 36.Lewis, T. S., Shapiro, P. S. & Ahn, N. G. (1998) Adv. Cancer Res. 74 49–139. [DOI] [PubMed] [Google Scholar]
- 37.Duesbery, N. S., Webb, C. P., Leppla, S. H., Gordon, V. M., Klimpel, K. R., Copeland, T. D., Ahn, N. G., Oskarsson, M. K., Fukasawa, K., Paull, K. D., et al. (1998) Science 280 734–737. [DOI] [PubMed] [Google Scholar]
- 38.Duesbery, N. S., Resau, J., Webb, C. P., Koochekpour, S., Koo, H. M., Leppla, S. H. & Vande Woude, G. F. (2001) Proc. Natl. Acad. Sci. USA 98 4089–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao, B., Su, Y., Oskarsson, M., Zhao, P., Kort, E. J., Fisher, R. J., Wang, L. M. & Vande Woude, G. F. (2001) Proc. Natl. Acad. Sci. USA 98 7443–7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuba, K., Matsumoto, K., Date, K., Shimura, H., Tanaka, M. & Nakamura, T. (2000) Cancer Res. 60 6737–6743. [PubMed] [Google Scholar]