Abstract
In Escherichia coli, division site selection is regulated in part by the Min-protein system. Oscillations of the Min proteins from pole to pole every ≈40 sec have been revealed by in vivo studies of GFP fusions. The dynamic oscillatory structures produced by the Min proteins, including a ring of MinE protein, compact polar zones of MinD, and zebra-striped oscillations in filamentous cells, remain unexplained. We show that the Min oscillations, including mutant phenotypes, can be accounted for by in vitro-observed interactions involving MinD and MinE, with a crucial role played by the rate of nucleotide exchange. Recent discoveries suggest that protein oscillations may play a general role in proper chromosome and plasmid partitioning.
In Escherichia coli, two systems are known to regulate the placement of the division site: nucleoid occlusion (1, 2) and the Min proteins (3). Both systems interfere with the formation of a ring of FtsZ protein believed to define the division site. The three Min proteins, MinC, MinD, and MinE, are required to prevent minicelling (3), asymmetric cell divisions that produce one small daughter cell lacking a chromosome and hence nonviable. In contrast, overexpression of MinC results in filamentous growth (4) by inhibiting polymerization of FtsZ on the cell membrane (5), a necessary first step in cell division (6). MinC is recruited to the membrane by MinD (7), which is membrane-associated only in its ATP-bound form (MinD:ATP) (8). Like MinC, MinE is naturally cytoplasmic and is recruited to the membrane by MinD:ATP (8).
In vivo observations of GFP fusions in living cells reveal spatial oscillations of the three Min proteins (9–11). In each oscillation period, the cell's entire complement of MinD accumulates in the cell membrane in a “polar zone” at one end of the cell. This polar zone then shrinks toward the end of the cell as a new accumulation forms at the opposite pole. MinC follows the same pattern as MinD, and the two proteins form complexes on the membrane. In contrast, MinE forms a ring at the boundary of the MinD polar zone with some MinE dispersed throughout the polar zone. The MinE ring moves toward the end of the cell as the MinD polar zone shrinks. The oscillation period is approximately proportional to the amount of MinD and inversely proportional to the amount of MinE in the cell, with a minimum period of ≈30 sec (9). Both MinD and MinE, but not MinC, are required for oscillations (9).
The average spatial distribution of the Min proteins naturally prevents minicelling without blocking normal cell division. The polar zones of MinD and MinC are believed to block FtsZ ring formation at the ends of the cell. At the same time, the concentrations of MinD and MinC remain low near the center of the cell, so an FtsZ ring can form there to establish the site of cell division.
Several models have been put forward to account for Min protein oscillations. All of these models successfully generate oscillations, but none can be meaningfully compared with in vivo observations. The model of Meinhardt and de Boer (12) requires newly synthesized protein to form both the MinD polar zones and MinE ring, because the proteins disappear (are degraded?) from the simulation on dissociation from the membrane. However, Min oscillations have been observed to persist for at least 45 min after protein synthesis was blocked, demonstrating Min-protein stability. Furthermore, to obtain a MinE ring, the Meinhardt and de Boer model assumes ad hoc that MinE attaches preferentially to an intermediate concentration of MinD.
The model of Howard, Rutenberg, and de Vet (HRdV) (13) produces oscillations only if MinE is driven onto the membrane by cytoplasmic MinD, despite evidence that MinE is recruited to the membrane by membrane-associated MinD (8). Moreover, the oscillations in the HRdV model have the opposite dependence of frequency on MinD concentration than is observed, and MinD forms a medial band that moves toward the end of the cell, contrary to the experimental observation that MinD forms directly as a polar zone. The model of Kruse (14) avoids ad hoc assumptions and produces a MinD polar zone by a MinD concentration-dependent slowing of MinD diffusion on the membrane, a natural assumption in light of recent experimental evidence for MinD polymer formation (8). However, the Kruse model requires unrealistically rapid membrane diffusion of MinD and fails to produce a MinE ring without ad hoc modification.
A Model Using Only Reported in Vitro Molecular Interactions Leads to Oscillations
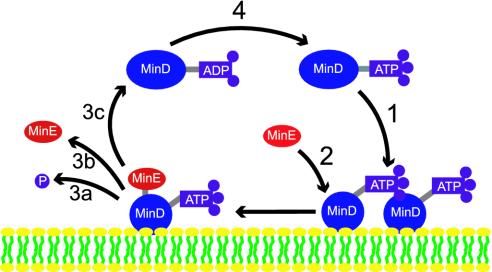
Here we show that a model including only the reported in vitro interactions of MinD and MinE can fully account for the Min oscillations, including formation of compact MinD polar zones and a MinE ring. The specific interactions are shown schematically in Fig. 1. The oscillations are driven by a cycle in which MinD:ATP first associates with the membrane, preferentially where other MinD:ATP is located. MinE then attaches to the MinD:AT P, activates AT P hydrolysis, and MinE and MinD:ADP reenter the cytoplasm in a 1:1 ratio. Evidence for this cycle comes from in vitro experiments by Lutkenhaus and colleagues (8). In particular, MinD bound to ATP reshapes spherical phospholipid vesicles into tubes by forming helical polymers, demonstrating self-association of membrane-bound MinD:ATP. Moreover, MinE cosediments with MinD in the membrane and activates hydrolysis and disassembly of MinD polymers (8). Recent in vivo experiments by Rothfield and colleagues (15) also indicate formation of MinD polymers on the membrane.
Fig. 1.
Model MinD,E cycle driven by ATP hydrolysis. 1, Cytoplasmic MinD:ATP complex attaches to the membrane, preferentially where other MinD:ATP is bound. 2, MinE in the cytoplasm attaches to a membrane-associated MinD:ATP complex. 3, MinE activates ATP hydrolysis by MinD, breaking apart the complex and releasing phosphate (a), MinE (b), and MinD:ADP (c) into the cytoplasm. 4, MinD:ADP is converted back into MinD:ATP by nucleotide exchange. In wild-type cells, MinE is likely active as a homodimer (25).
Reaction–Diffusion Equations
The equations describing the time evolution of MinD and MinE concentrations in a cylindrical cell (see schematic of reaction cycle in Fig. 1) are
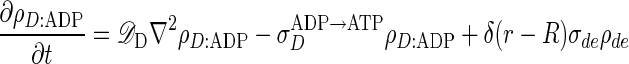 |
[1] |
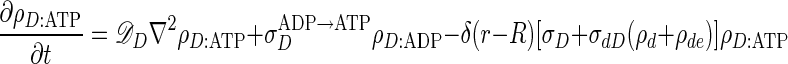 |
[2] |
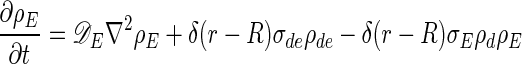 |
[3] |
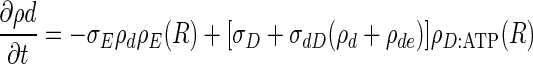 |
[4] |
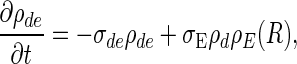 |
[5] |
where ρD:ADP, ρD:ATP, ρE are the concentrations in the cytoplasm of MinD:ADP complexes, MinD:ATP complexes, and MinE, and ρd, ρde are the concentrations on the membrane of MinD: ATP complexes and MinE:MinD:ATP complexes. We have verified that introducing an intermediate free cytoplasmic MinD species, thereby converting the single rate constant 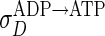 into two sequential decay rates, does not introduce any significant changes.
into two sequential decay rates, does not introduce any significant changes.
The cell radius is R = 0.5 μm, and results are given for cells of length L = 4 μm and 10 μm. The δ functions, δ(r – R), represent local exchange of proteins between membrane and cytoplasm; additional δ functions, δ(z) and δ(z – L), are implemented for the end caps of the cylinder. The total concentrations of MinD and MinE are 1,000/μm and 350/μm, respectively (assuming MinE is active as a homodimer, this implies 700 monomers per μm) (16). The diffusion constants are
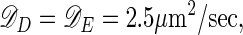 |
as measured for the cytoplasmic diffusion of a maltose-binding protein in the E. coli cytoplasm (17), and the reaction rates are
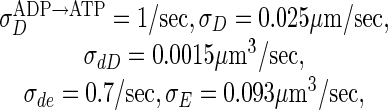 |
unless otherwise indicated. We discretize and solve Eqs. 1–5 on a 3D lattice in cylindrical coordinates, with grid spacing dr = dz = 0.05 μm.
Periodic Oscillations with MinD Polar Zones and a MinE Ring
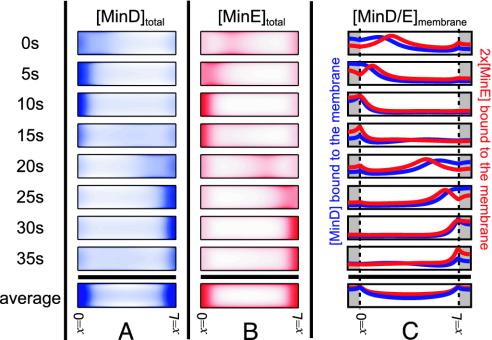
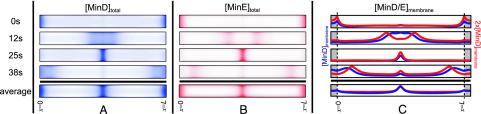
The results of numerical integration in time of our model equations (see Fig. 1) for a 4-μm cylindrical cell are shown in Fig. 2. Periodic oscillations that are independent of initial conditions occur for a wide range of parameters. The oscillations have the same spatial character as those observed in experiment, including the formation and shrinkage of the MinD polar zones and the appearance of a MinE ring. These structures form spontaneously without special targets for MinD at the cell ends, without MinE–MinE interactions, and with no new protein synthesis. MinD:ATP and MinE dwell in one-half of the cell membrane during a period of polar zone compaction, before a brief cytoplasmic burst results in rapid reformation of a new MinD polar zone and MinE ring at the opposite end of the cell. The bottom row of Fig. 2 shows the time-averaged concentrations of MinD and MinE. The minimum for membrane-bound MinD:ATP occurs at the center of the cell.
Fig. 2.
Time slices in 5-sec increments of one complete MinD, MinE oscillation in a 4-μm cell. To mimic experimental observations of GFP fluorescence, we show 2D projections of the concentrations of MinD (A) and MinE (B) inside a 3D cylindrical cell, with the concentrations assumed rotationally symmetric about the axis of the cylinder. In A, the MinD polar zone shrinks toward the end of the cell and reforms at the opposite pole. In B, MinE forms a ring near the boundary of the MinD polar zone. Except during brief cytoplasmic-burst phases (15 sec, 35 sec), both MinD and MinE are primarily membrane-bound. (C) The membrane-associated concentrations, MinD:ATP in blue and MinE in red. The vertical dashed lines and gray shading indicate the caps of the cylindrical cell membrane. The final row shows the time average of each quantity over a complete cycle.
The basic mechanism of the oscillations is that a typical MinD, once released from the membrane, diffuses farther in the cytoplasm than does a typical MinE before reattaching to the membrane. The delay before MinD becomes competent to reattach to the membrane stems from its need to release ADP and rebind ATP, allowing the formation of a new polar zone of MinD:ATP, whereas MinE progressively hydrolyzes the old polar zone.
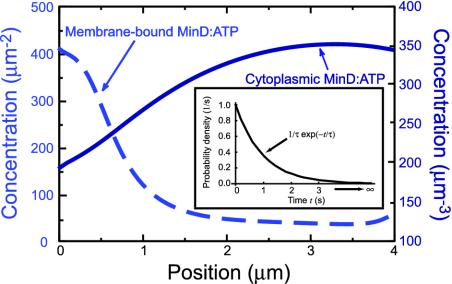
But why does MinD:ATP accumulate at the far pole rather than reattaching uniformly throughout the cell? The reason is that MinD:ATP in the cytoplasm sticks rapidly to the old polar zone from which it was released. Indeed, a MinD protein typically recycles ≈4 times through the old polar zone in each half cycle before finally lodging in the new polar zone. Each such recycling of MinD requires the hydrolysis of one molecule of ATP. The “stickiness” of the old polar zone creates a distribution of MinD:ATP in the cytoplasm that is peaked at the opposite end of the cell (Fig. 3). Thus MinD:ATP accumulates in a new polar zone, with a profile sharpened by the tendency of MinD:ATP in the membrane to self-associate (8).
Fig. 3.
Concentration of ATP-bound MinD (MinD:ATP) in the cytoplasm corresponding to time t = 5 sec in Fig. 2. The distribution is peaked at the opposite end of the cell from the existing MinD:ATP polar zone, indicated by the peak of the dashed curve, leading to the accumulation of MinD:ATP in a new polar zone. (Inset) Waiting-time distribution for recovery of MinD:ATP, assuming a nucleotide-exchange rate 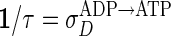 of 1/sec.
of 1/sec.
Proper formation of the new MinD:ATP polar zone requires that MinD have time to diffuse throughout the cytoplasm before rebinding ATP. The nucleotide exchange rate we use, 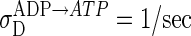 , is well within the observed range, which is known to span more than five orders of magnitude for guanine nucleotide exchange (18). If the exchange rate is too large, cytoplasmic MinD:ATP reappears mainly near the old polar zone, eliminating the peaked distribution of cytoplasmic MinD:ATP (Fig. 3) that is responsible for the formation of a new polar zone. The delay of MinD:ATP recovery due to nucleotide exchange has been ignored in previous models.
, is well within the observed range, which is known to span more than five orders of magnitude for guanine nucleotide exchange (18). If the exchange rate is too large, cytoplasmic MinD:ATP reappears mainly near the old polar zone, eliminating the peaked distribution of cytoplasmic MinD:ATP (Fig. 3) that is responsible for the formation of a new polar zone. The delay of MinD:ATP recovery due to nucleotide exchange has been ignored in previous models.
Once MinE completes the hydrolysis of the old polar zone, it begins to diffuse through the cytoplasm. Because MinE sticks rapidly to available MinD:ATP in the membrane, most of the MinE attaches to the edge of the new MinD:ATP polar zone. This is the origin of the MinE ring. The subsequent movement of the MinE ring toward the end of the cell reflects release from the membrane, diffusion, and reattachment of individual MinE molecules (or homodimers). Because the diffusion length of MinE before reattaching is generally smaller than the ≈1 μm transverse dimension of the cell, it is important to model the oscillations in a fully 3D cell rather than in a 1D approximation as used in the models from refs. 12–14. We neglect membrane diffusion as too slow to affect the dynamics of the oscillations.
The Oscillation Period Depends Linearly on the Ratio of MinD to MinE Concentration
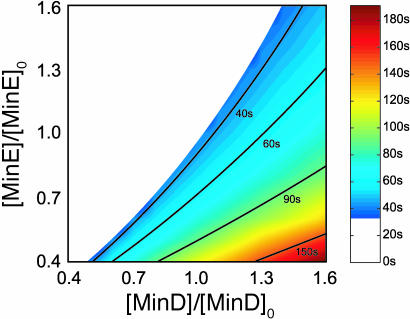
Fig. 4 shows the period of oscillation for a range of protein concentrations. Consistent with experiment (9), the oscillation period is proportional to the total amount of MinD and inversely proportional to the total amount of MinE in the cell. The period is simply determined by the rate at which the MinE ring hydrolyzes the polar zone of MinD:ATP. This period increases almost linearly with the amount of MinD in the cell and decreases inversely with both the amount of MinE and the hydrolysis rate σde. We chose σde = 0.7/sec to match the observed wild-type period of ≈40 sec (9). Varying the total amounts of MinD and MinE in the cell, we find a minimum oscillation period of ≈33 sec, also consistent with experiment (9). Increasing the amount of MinE beyond this limit forces a majority of the MinD into the cytoplasm and eliminates oscillations. In contrast, decreasing the concentration of MinE results in slower and slower oscillations with no apparent limit, consistent with in vivo observations (9).
Fig. 4.
Dependence of the oscillation period on the average concentration of MinD and MinE in a 4-μm cell. Wild-type values are [MinD]0 ≈ 1,000/μm and [MinE]0 ≈ 350/μm, respectively (16). Isoperiod curves from 40 s to 150 s are shown.
Filamentous Cells Have “Doubled” Oscillation Patterns
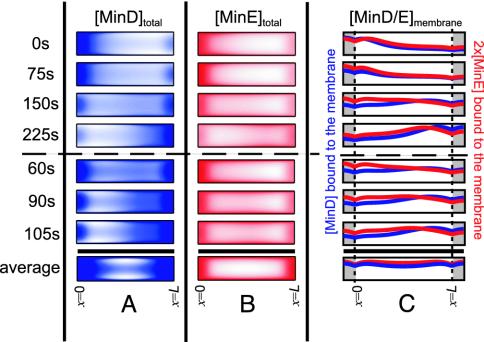
A critical test of the model is the striking behavior of Min oscillations in long cells. Filamentous cells can be obtained by using temperature-sensitive FtsZ mutants, which cannot undergo cell division at the nonpermissive temperature. For cells longer than ≈10 μm, the number of wavelengths of Min oscillations present in the cell increases; zebra-striped cells with as many as eight half-wavelengths have been observed (9). Fig. 5 shows the oscillation pattern obtained from our model for a 10-μm cell. Consistent with experimental observations, the oscillation pattern “doubles;” the oscillatory dynamics in each half of the cell mimics the dynamics of the normal 4-μm cell in Fig. 2. We find that this doubled oscillation pattern is stable for a 3D cell but collapses back to an undoubled pattern (Fig. 2) in the 1D approximation. In addition, a nucleotide exchange rate of ≈1/sec or slower is necessary for stability of the doubled oscillation pattern. The bottom row of Fig. 5 shows the time-averaged concentrations of MinD and MinE. The minima for membrane-bound MinD:ATP occur at 1/4 and 3/4 of the cell length, as observed by Gullbrand and Nordström (19) for FtsZ ring placement in long cells. Oscillations in a 20-μm cell closely correspond to a “doubled” 10-μm cell (not shown).
Fig. 5.
Time slices in 12.5-sec increments of one complete MinD, MinE oscillation in a 10-μm cell. The same quantities are shown as in Fig. 2. The zebra-striped oscillation pattern now includes two half-wavelengths. The system exhibits two separate MinE rings and an alternation between two MinD polar zones and a central MinD tube. The final row shows the time average of each quantity over a complete cycle.
MinE “Mutants”: Slow Oscillations and No MinE Ring Formation
Another striking experimental observation is that cells with the wild-type MinE1–88 protein replaced by the fragment MinE1–53 display oscillations with a period of ≈10 min with no detectable MinE ring and with diffuse MinD polar zones (20). Recent experiments with MinE mutants have correlated a weak or absent MinE ring with extended MinD polar zones and an increased minicelling probability (16). Within our model, the oscillation period varies inversely with the hydrolysis rate σde, and ring formation depends on the attachment probability of MinE to available MinD:ATP in the membrane. In Fig. 6, we show oscillations of MinD and MinE with a reduced hydrolysis rate, σde = 0.07/sec, which slows the oscillations, and a reduced MinE attachment coefficient σE = 0.047 μm/sec. With this value of σE, the MinE ring fails to form, because MinE diffuses well into the new polar zone before reattaching to MinD:ATP (increasing the MinE diffusion constant has a similar effect). Importantly, this invasion of the MinD:ATP polar zone by MinE results in less accumulation of MinD:ATP and a more diffuse MinD:ATP polar zone, similar to experimental observations.
Fig. 6.
An oscillation period for a “MinE mutant” with reduced hydrolysis rate, σde = 0.07/sec, and reduced MinE sticking coefficient, σE = 0.047 μm/sec. The upper half portrays time slices of half an oscillation cycle in 75-sec increments, whereas the lower half focuses on the time interval from 60 to 105 sec, during which the MinE finishes hydrolyzing the old MinD polar zone, diffuses across the cell, and reforms on the opposite half. The same quantities are shown as in Fig. 2. Note the long oscillation period and the suppression of a MinE ring.
Discussion
Some aspects of Min protein oscillations are not captured by our minimal model. For example, the MinE ring is occasionally observed to reverse direction or “stutter” (16), likely reflecting the persistent presence of helical accumulations of MinD (15). Although the model cannot predict the fine structure of MinD polymerization (15), nevertheless, the agreement between model and experiment is close enough to indicate some fundamental properties of the real Min system. The model oscillations represent a limit cycle; that is, the same oscillation develops from any set of initial conditions. Moreover, the pattern of oscillation, particularly the formation of compact polar zones of MinD:ATP capped by a ring of MinE, is insensitive to fluctuations in the amounts of MinD and MinE. A linear-stability analysis around the uniform solution yields instability only for half-wavelengths >2 μm. This minimum wavelength guarantees that oscillations can develop only in the cell's long dimension. Interestingly, the period of oscillations is sensitive to protein number fluctuations (Fig. 4), but periods of up to ≈120 sec (9) appear to yield a normal division phenotype.
Recent observations suggest a general role for protein oscillators in chromosome and plasmid partitioning (21, 22). Nevertheless, it remains an open question why bacteria use such oscillators. The Gram-positive bacterium Bacillus subtilis uses homologs to MinC and MinD to prevent minicelling. However, these proteins form static polar zones in B. subtilis. (B. subtilis also undergoes highly asymmetric cell division during sporulation.) In the absence of Min proteins in E. coli, nucleoid occlusion results in division sites either near the cell ends, resulting in minicelling, or medially, resulting in essentially normal divisions, but with considerably less division accuracy than in wild type (2). In anucleate cells without nucleoid occlusion but with Min proteins, medial FtsZ ring placement is favored but again with less accuracy than in wild type (2). It is unknown whether Min protein oscillations alone can be sufficient for accurate FtsZ ring placement, or whether cooperation and possibly direct interaction between the Min system and nucleoid occlusion are required. In B. subtilis, the nonoscillatory MinCD homologs are not required for medial division accuracy (23). One possible advantage of an oscillator for determining the cell center is that in each half cycle, essentially the same amount of MinD protein accumulates in each polar zone. Thus, the time-averaged minimum of MinD, and hence the minimum of the division-site blocker MinC, will occur at the cell center independent of protein number fluctuations. Furthermore, recent indications (24) that MinC and MinE may competitively bind to MinD and that MinD forms dimers in the cytoplasm may enhance the FtsZ ring placement accuracy of the Min oscillator. Finally, the observation of FtsZ ring formation at the 1/4 and 3/4 points in long cells (19) very likely reflects the doubling of the Min-oscillation pattern. The multiwavelength Min oscillations in long cells may play a role in proper fragmentation and reduce the likelihood of internal minicelling during recovery from filamentous growth induced by the SOS response.
Acknowledgments
We thank Joe Lutkenhaus for valuable suggestions and Bonnie Bassler, Laura Garwin, and Tom Silhavy for critical readings of the manuscript. Partial funding for this research was provided by the Materials Research Science and Engineering Centers Program of the National Science Foundation under Grant DMR-0213282.
References
- 1.Woldringh, C. L., Mulder, E., Valkenburg, J. A., Wientjes, F. B., Zaritsky, A. & Nanninga, N. (1990) Res. Microbiol. 141 39–49. [DOI] [PubMed] [Google Scholar]
- 2.Yu, X.-C. & Margolin, W. (1999) Mol. Microbiol. 32 315–326. [DOI] [PubMed] [Google Scholar]
- 3.de Boer, P. A. J., Crossley, R. E. & Rothfield, L. I. (1989) Cell 56 641–649. [DOI] [PubMed] [Google Scholar]
- 4.de Boer, P. A. J., Crossley, R. E. & Rothfield, L. I. (1992) J. Bacteriol. 174 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi, E. & Lutkenhaus, J. (1993) J. Bacteriol. 175 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi, E. & Lutkenhaus, J. (1991) Nature 354 161–164. [DOI] [PubMed] [Google Scholar]
- 7.Huang, J., Cao, C. & Lutkenhaus, J. (1996) J. Bacteriol. 178 5080–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu, Z., Gogol, E. P. & Lutkenhaus, J. (2002) Proc. Natl. Acad. Sci. USA 99 6761–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raskin, D. M. & de Boer, P. A. J. (1999) Proc. Natl. Acad. Sci. USA 96 4971–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu, Z. & Lutkenhaus, J. (1999) Mol. Microbiol. 34 82–90. [DOI] [PubMed] [Google Scholar]
- 11.Fu, X., Shih, Y.-L., Zhang, Y. & Rothfield, L. I. (2001) Proc. Natl. Acad. Sci. USA 98 980–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meinhardt, H. & de Boer, P. A. J. (2001) Proc. Natl. Acad. Sci. USA 98 14202–14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howard, M., Rutenberg, A. D. & de Vet, S. (2001) Phys. Rev. Lett. 87 278102-1–278102-4. [DOI] [PubMed] [Google Scholar]
- 14.Kruse, K. (2002) Biophys. J. 82 618–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shih, Y.-L., Le, T. & Rothfield, L. (2003) Proc. Natl. Acad. Sci. USA 100 7865–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shih, Y.-L., Fu, X., King, G. F., Le, T. & Rothfield, L. (2002) EMBO J. 21 3347–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elowitz, M. B., Surette, M. G., Wolf, P.-E., Stock, J. B. & Leibler, S. (1998) J. Bacteriol. 181 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenzen, C., Cool, R. H., Prinz, H., Kuhlmann, J. & Wittinghofer, A. (1998) Biochemistry 37 7420–7430. [DOI] [PubMed] [Google Scholar]
- 19.Gullbrand, B. & Nordström, K. (2000) Mol. Microbiol. 36 1349–1359. [DOI] [PubMed] [Google Scholar]
- 20.Rowland, S. L., Fu, X., Sayed, M. A., Zhang, Y., Cook, W. R. & Rothfield, L. I. (2000) J. Bacteriol. 182 613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamaichi, Y. & Niki, H. (2000) Proc. Natl. Acad. Sci. USA 97 14656–14661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebersbach, G. & Gerdes, K. (2001) Proc. Natl. Acad. Sci. USA 98 15078–15083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Migocki, M. D., Freeman, M. K., Wake, R. G. & Harry, E. J. (2002) EMBO Rep. 3 1163–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu, Z., Saez, C. & Lutkenhaus, J. (2003) J. Bacteriol. 185 196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pichoff, S., Vollrath, B., Touriol, C. & Bouché, J.-P. (1995) Mol. Microbiol. 18 321–329. [DOI] [PubMed] [Google Scholar]