Abstract
Ubiquitination of membrane-associated proteins can direct their proteasome-mediated degradation or activation at the endoplasmic reticulum (ER), as well as their endocytosis and intracellular sorting. However, the full spectrum of ubiquitinated membrane proteins has not been determined. Here we combined proteomic analysis with yeast genetics to identify 211 ubiquitinated membrane-associated proteins in Saccharomyces cerevisiae and map >30 precise sites of ubiquitination. Major classes of identified ubiquitinated proteins include ER-resident membrane proteins, plasma membrane-localized permeases, receptors, and enzymes, and surprisingly, components of the actin cytoskeleton. By determining the differential abundance of ubiquitinated proteins in yeast mutated for NPL4 and UBC7, which are major components of ER-associated degradation (ERAD), we furthermore were able to classify 83 of these identified ubiquitinated membrane proteins as potential endogenous substrates of the ERAD pathway. These substrates are highly enriched for proteins that localize to or transit through the ER. Interestingly, we also identified novel membrane-bound transcription factors that may be subject to ubiquitin/proteasome-mediated cleavage and activation at the ER membrane.
The endoplasmic reticulum (ER) is a primary site of synthesis and folding for ER-resident proteins, as well as those destined for secretion, for the plasma membrane, and for other secretory and endocytic organelles. To maintain the proper function and composition of the ER and downstream compartments, the ER-associated degradation (ERAD) pathway targets unfolded proteins in the ER for ubiquitination and degradation by the 26S proteasome in the cytosol (1). The importance of ERAD is underscored by the finding that its misguided function is central to the development of several infectious and genetic human diseases, including viral infection by human cytomegalovirus (HCMV) and HIV, cystic fibrosis, and certain neurodegenerative disorders (2).
Many transacting ERAD components have been identified, including the E2 ubiquitin conjugating enzyme Ubc7p (recruited to the ER by the integral membrane protein Cue1p) and the E3 ubiquitin ligases Hrd1p and Doa10p (1, 3–6). Recent studies have also found that the highly conserved Cdc48p–Npl4p–Ufd1p protein complex is required for the proteasome-mediated degradation of ERAD substrates (7–12). This protein complex is unique among ERAD components in that it functions after ubiquitination, possibly to fully extract ubiquitinated proteins from the membrane and/or to recruit the proteasome to these substrates (Fig. 1a). Interestingly, the ubiquitin/proteasome-dependent processing and activation of ER-localized membrane-bound transcription factors also requires the Cdc48p–Npl4p–Ufd1p protein complex, highlighting the importance of this protein complex in the ubiquitin-mediated regulation of membrane proteins (13–15),
Fig. 1.
NPL4-dependent ubiquitinated ER protein degradation. (a) Simplified model of ERAD. Unfolded and/or damaged ER proteins (a membrane protein is depicted, soluble lumenal proteins are also subject to ERAD) are ubiquitinated by the E2 ubiquitin-conjugating enzyme Ubc7p (recruited to the ER membrane by Cue1p) and the E3 ubiquitin ligases Hrd1p and Doa10p. The E2s Ubc6p and Ubc1p also make minor contributions to ER protein ubiquitination. Once ubiquitinated, an ERAD substrate requires the activity of the Npl4p–Ufd1p–Cdc48p chaperone to be fully extracted from the membrane and degraded by the proteasome. (b) Accumulation of ubiquitinated proteins in npl4-1 membranes. WT and npl4-1 cells expressing 6×His–Myc-tagged ubiquitin were subjected to subcellular fractionation. Equal amounts of total protein (10 μg) from total cell extract (lanes 1 and 2), soluble (lanes 3 and 4), and membrane (lanes 5 and 6) fractions were separated by SDS/PAGE and immunoblotted with anti-Myc antibodies. (c) Extragenic suppression of npl4-1 by deletion of genes encoding ERAD ubiquitination machinery. Yeast strains of the indicated genetic backgrounds were serially diluted, spotted onto rich-media plates, and incubated for 2–3 days at the indicated temperatures. The number of cells spotted in each dilution is indicated on the bottom. (d) Loss of UBC7 abrogates accumulation of ubiquitinated proteins in npl4-1 membranes. WT and npl4-1 cells ±Δubc7 expressing 6×His–Myc-tagged ubiquitin were subjected to subcellular fractionation. Equal amounts of total protein (10 μg) from total cell extract (lanes 1–4), soluble (lanes 5–8), and membrane (lanes 9–12) fractions were separated by SDS/PAGE and immunoblotted with anti-Myc antibodies.
In the current study, we have found that mutation of the essential Npl4p component of the Cdc48p–Npl4p–Ufd1p protein complex causes the accumulation of a large population of Ubc7p-ubiquitinated membrane-associated proteins in Saccharomyces cerevisiae. Through the implementation of a genetic/proteomic technique that took advantage of the differential ubiquitination of proteins in npl4 and ubc7 yeast mutants, we identified 211 ubiquitinated membrane-associated proteins, 83 of which were classified as potential substrates of NPL4- and UBC7-dependent ERAD. Our findings represent the first large-scale identification of endogenous ERAD substrates and suggest that a diverse set of proteins that localize to or transit through the ER are subject to ERAD.
Materials and Methods
Yeast Strains and Manipulations. The genotypes of all strains used in this study are provided in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. Media were prepared according to standard methods (16). SUB280 (a WT yeast strain deleted for all endogenous ubiquitin genes and expressing untagged ubiquitin from a plasmid) and SUB592 (an isogenic strain expressing 6×His–Myc-tagged ubiquitin) have been described (17). PSY2975 (npl4-1 expressing untagged ubiquitin) was generated by integrating the npl4-1 point mutation into SUB280 by two-step gene replacement (16). PSY2976 (npl4-1 expressing 6×His–Myc-tagged ubiquitin) was obtained by plasmid-swap in PSY2975. PSY2977 (Δubc7 expressing 6×His–Myc-tagged ubiquitin) and PSY2978 (npl4-1 Δubc7 expressing 6×His–Myc-tagged ubiquitin) were generated by transforming a Δubc7::kanMX4 PCR product (derived from RG597) into SUB592 and PSY2976, respectively. Proper integration of Δubc7::kanMX4 in both strains was confirmed by PCR. PSY2967 is an FY23 derived WT strain, whereas the npl4-1 strains PSY2968 and PSY2969 are FY23-backcrossed strains derived from PSY825 (13, 18). RG597 (Δubc7::kanMX4), RG850 (Δcue1::kanMX4), RG1704 (Δhrd1::kanMX4), and RG7299 (Δdoa10::kanMX4) were purchased from Research Genetics (Carlsbad, CA). PSY2970 (npl4-1 Δubc7::kanMX4), PSY2971 (npl4-1 Δcue1::kanMX4), PSY2972 (npl4-1 Δhrd1::kanMX4), and PSY2973 (npl4-1 Δdoa10::kanMX4) were generated by crossing the appropriate RG strain to PSY2968. PSY2974 (npl4-1 Δhrd1::kanMX4 Δdoa10::kanMX4) was generated by crossing PSY2972 to a PSY2973 sister spore of the opposite mating type.
npl4–1 Extragenic Suppressor Screen. PSY2968 and PSY2969 cells were mutagenized with ethyl methane sulfonate (EMS) (19). Colonies able to grow at the nonpermissive temperature of 31°C (Ts+ phenotype) were observed at the rate of 1.3 × 10–4. Of 186 Ts+ candidates, 46 were determined to be recessive based on the Ts– phenotype of a heterozygous diploid. Complementation testing led to the classification of 18 complementation groups, 5 of which contained multiple members. The largest complementation group, Group I (17 alleles), was found to be caused by mutation of the chromosome XIII centromere-linked UBC7 gene based on Δubc7 noncomplementation. Group II (5 alleles) was found to be caused by mutation of chromosome XIII-encoded CUE1 based on noncomplementation with a previously cloned transposon insertion allele of CUE1 (K.A., S. Frietze, A.L.H., and P.A.S., unpublished results).
Membrane Fractionation and 6×His–Myc-Tagged Ubiquitinated Protein Purification. Yeast cells were separated into total, soluble, and membrane fractions as described with the exclusion of the final sucrose step gradient (20). To purify 6×His–Myc-tagged ubiquitinated proteins, membranes containing 20 mg of total protein were solubilized in 20 ml of native solubilization/binding buffer [50 mM NaH2PO4/1 M NaCl/2% Triton X-100/3% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS)/10 mM imidazole/20 mM 2-mercaptoethanol], pH 8.0, supplemented with protease inhibitors (2.5 μg/ml each pepstatin A, leupeptin, aprotinin, and chymostatin) for 1 h at 4°C. The solubilized membranes were incubated with 1 ml Ni-NTA Agarose (Qiagen, Valencia, CA) for an additional 1 h at 4°C. The following washes were performed in column format: (i) 20 ml of native wash buffer (50 mM NaH2PO4/1 M NaCl/0.5% Triton X-100/20 mM imidazole/20 mM 2-mercaptoethanol), pH 8.0; (ii) 20 ml of native wash buffer, pH 6.0; (iii) 20 ml of native wash buffer, pH 8.0; (iv) 40 ml of urea wash buffer (100 mM NaH2PO4/8 M urea/0.1% Triton X-100/20 mM imidazole/20 mM 2-mercaptoethanol), pH 8.0; (v) 10 ml of urea wash buffer, pH 6.0; (vi) 40 ml of urea wash buffer, pH 8.0; and (vii) 10 ml of urea wash buffer, pH 8.0 (no Triton X-100). Bound proteins were eluted by four sequential 1-ml elutions in imidazole elution buffer (100 mM NaH2PO4/8 M urea/300 mM imidazole/20 mM 2-mercaptoethanol), pH 8.0. Purified proteins were processed for trypsin digestion as described (21). Briefly, protein-containing elutions were pooled, concentrated, and treated with 50 mM iodoacetamide for 1 h. Proteins were dialyzed into trypsin buffer (10 mM Tris pH 8.5/1 M urea/0.01% SDS) and digested overnight at 37°C with 5 μg of trypsin in the presence of 1 mM CaCl2.
Two-Dimensional Liquid Chromatography and Tandem Mass Spectrometry. The tryptic peptides were acidified with formic acid and separated by strong cation exchange (SCX) chromatography (buffer A: 5 mM potassium phosphate buffer, pH 3.0/25% acetonitrile; buffer B: 5 mM potassium phosphate buffer, pH 3.0/25% acetonitrile/350 mM KCl) by using a 2.1 mm by 20 cm Polysulfoethyl A column (Poly LC, Columbia, MD) at a flow rate of 200 μl/min. Twelve 1-min peptide-containing SCX fractions were reduced in volume and subjected individually (10% each) to reverse-phase chromatography using 100-μm inner diameter by 12 cm self-packed fused silica C18 capillary columns coupled directly to an LCQ-DECA ion trap mass spectrometer (Thermo Finnigan, San Jose, CA). The sample was loaded onto the microcolumn under high pressure and washed offline with 5–10 μl of solvent A (0.1% formic acid/0.005% heptafluorobutyric acid/5% acetonitrile) to remove salt. Peptides were subjected to a 60-min gradient with solvent B (0.1% formic acid/0.005% heptafluorobutyric acid/95% acetonitrile); as peptides eluted, they were ionized, detected, isolated, and selected for fragmentation/sequencing in an automated manner (21).
Data Processing. Each tandem MS (MS/MS) spectrum was searched independently against a database of predicted spectra derived from S. cerevisiae-encoded proteins by using the sequest algorithm (22). The following modifications were permitted (mass change shown in Daltons): carboxyamidomethylated cysteine (+57), oxidized methionine (+16), and ubiquitinated lysine (+114). We required that peptide matches must (i) be fully tryptic, (ii) have cross-correlation (XCorr) values of ≥2.0, 2.2, and 3.75 for 1+, 2+, and 3+ charge-state peptides, respectively, and (iii) have a delta correlation (dCn) score ≥0.1. With these stringent criteria (23), we identified >6,000 peptides corresponding to 527 unique proteins (including ubiquitin itself) among the five data sets. We applied a series of filtering and elimination steps to these 527 proteins to differentiate strong candidate ubiquitinated proteins from weaker candidates and false positives. First, proteins for which ubiquitination sites were identified (29 total) were classified as ubiquitinated proteins. We eliminated 106 false positive proteins purified in the negative control strains and 11 additional proteins that contained endogenous 3×His motifs. We retained seven proteins that were identified by a single peptide in one of the negative control strains but identified by ≥10 peptides in one or more of the tagged-ubiquitin strains. Of the remaining 374 proteins that displayed specificity for the tagged-ubiquitin strains, we retained only those that were identified by ≥2 peptides in at least one of the tagged ubiquitin strains. This resulted in the final classification of 211 proteins as ubiquitinated proteins, not including ubiquitin itself. Approximately 2/3 of these identified ubiquitinated membrane-associated proteins (n = 141) were also identified in a recent study (24) that cataloged >1,000 ubiquitinated proteins from total yeast extracts; the remaining 1/3 (n = 70), in contrast, are unique to our study.
Classification of Candidate ERAD Substrates. We reasoned that an increase or decrease in ubiquitination for a given protein (on one or several sites, on already assembled ubiquitin chains, or as a percentage of the total protein population) would directly influence the relative abundance of that protein in our purifications of ubiquitinated proteins. Therefore, to identify proteins whose ubiquitination is influenced by the ERAD components NPL4 and UBC7, we compared the relative abundance of each protein among the WT, npl4-1, and npl4-1 Δubc7 data sets by using the number of unique peptides identified as a semiquantitative measure of protein abundance (25, 26). The number of peptides sequenced for a given protein, although a function of abundance, is also a complicated reflection of the efficiency of digestion, peptide solubility, extraction, ionization, and fragmentation for each protein and its peptides. Because, however, we compared the same protein among different fractions, the only major influence on relative peptide number for a given protein should be changes in its abundance among the samples. As an important control for this approach, we determined the fluctuations in peptide number for an “internal control” class of proteins that should be equally abundant in each data set: the false positive proteins that bound the nickel beads by virtue of intrinsic histidine-rich sequences or metal binding capabilities. We determined the no. of peptide(npl4-1)/no. of peptide(WT) and no. of peptide(npl4-1)/no. of peptide(npl4-1Δubc7) ratios for 57 proteins identified in both negative control strains (≥2 peptides). In both cases, the resulting ratios showed a bell curve distribution around a mean log(ratio) of –0.1 ± 0.3 (peptide values of 0 were set to 0.1 to allow for inclusion in this analysis). Only in one case was a protein identified in one but not the other data set. This analysis indicated that equally abundant proteins were found with a similar number of peptides among the three data sets. We then determined the no. of peptide(npl4-1)/no. of peptide(WT) and no. of peptide(npl4-1)/no. of peptide(npl4-1Δubc7) ratios for 194 and 193 ubiquitinated proteins, respectively (≥2 peptides in at least one of the two data sets being compared; peptide values of 0 were set to 0.1). A significant responsiveness to NPL4 (npl4-1/WT ratio) or UBC7 (npl4-1/npl4-1Δubc7 ratio) was assigned to proteins whose ratio was ≥2 standard deviations from the mean of the corresponding set of ratios obtained for the internal control proteins.
Transmembrane Domain (TMD) Prediction. TMDs were predicted by the tmhmm (www.cbs.dtu.dk/services/TMHMM) and tmpred (www.ch.embnet.org/software/TMPRED_form.html) algorithms.
Yeast Databases. Subcellular localizations and biological functions of proteins were compiled from summaries of published literature in the Saccharomyces Genome Database (www.yeastgenome.org) and the Yeast Proteome Database (www.incyte.com).
Results and Discussion
To test for a global role of the Cdc48p–Npl4p–Ufd1p complex in membrane protein turnover, we monitored total protein ubiquitination in yeast mutated for NPL4. We integrated the temperature-sensitive npl4-1 point mutation (G323S) into a yeast strain expressing 6×His–Myc-tagged ubiquitin as the only form of ubiquitin in the cell (13, 17). Strikingly, the npl4-1 strain displayed a marked accumulation of ubiquitinated proteins in total cell extracts as compared with the corresponding WT strain (Fig. 1b, compare lanes 1 and 2). Fractionation of these cellular extracts revealed that the accumulating proteins were highly enriched in the membrane fraction of npl4-1 cells (Fig. 1b, lanes 3–6), demonstrating that the Cdc48p–Npl4p–Ufd1p protein complex effects the accumulation of a large population of membrane-associated or otherwise insoluble ubiquitinated proteins.
The importance of Cdc48p–Npl4p–Ufd1p in pathways involving membrane–protein ubiquitination, and ERAD in particular, was further demonstrated by the isolation of UBC7 and CUE1 in a genetic screen for extragenic suppressors of the temperature-sensitive npl4-1 mutation (Fig. 1c). Ubc7p is the primary ERAD E2 enzyme and is recruited to the ER membrane by the Cue1p protein (1, 4). By direct tests, we found that simultaneous deletion of the ERAD E3-encoding genes HRD1 and DOA10 also suppressed npl4-1 to a similar extent (Fig. 1c). As shown in Fig. 1d, the accumulation of ubiquitinated proteins in npl4-1 membranes was completely reversed in cells lacking UBC7. We concluded that the Cdc48p–Npl4p–Ufd1p protein complex is potentially required for the efficient turnover of a large number of Ubc7p-ubiquitinated membrane-associated proteins.
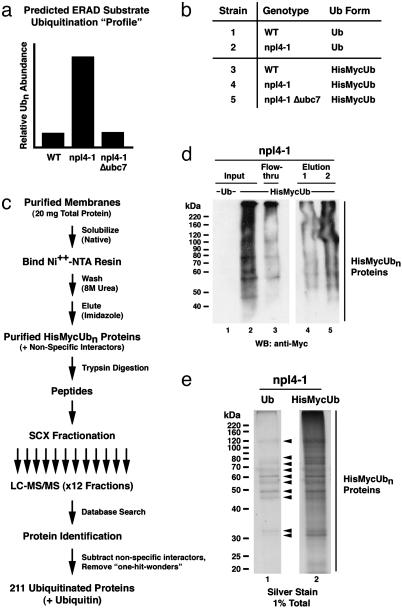
Given the known roles for Cdc48p–Npl4p–Ufd1p and Ubc7p in the ERAD pathway, it is likely that the differentially ubiquitinated proteins in npl4-1 and Δubc7 cells largely represent the endogenous substrates of ERAD. Taking advantage of this differential ubiquitination, we designed and implemented a genetic/proteomic “screen” to identify endogenous ERAD substrates. We first purified 6×His–Myc-ubiquitinated proteins from WT, npl4-1, and npl4-1 Δubc7 membranes (and two negative control strains expressing untagged ubiquitin) by immobilized metal affinity chromatography and identified them by tandem mass spectrometry (24) (Fig. 2 b–e; see Materials and Methods). Applying stringent criteria, we identified 211 candidate ubiquitinated proteins, plus ubiquitin itself, among the three data sets (see Materials and Methods). In addition, we identified 34 precise sites of ubiquitination in 29 of these purified proteins (Fig. 3a and Table 2, which is published as supporting information on the PNAS web site). The distribution of these 211 ubiquitinated proteins among the WT, npl4-1, and npl4-1Δubc7 data sets is depicted in Fig. 4a. Consistent with the increased abundance of ubiquitinated proteins in npl4-1 membranes, the npl4-1 data set contained the largest number of ubiquitinated proteins, and very few ubiquitinated proteins present in the WT and/or npl4-1 Δubc7 data sets were excluded from the npl4-1 data set.
Fig. 2.
A genetic/proteomic screen for endogenous ERAD substrates in S. cerevisiae. (a) Predicted “profile” of ubiquitination for a typical ERAD substrate in WT vs. npl4-1 vs. npl4-1Δubc7 yeast. (b) Yeast strains from which membrane-associated 6×His–Myc-tagged ubiquitinated proteins were purified and identified. WT and npl4-1 strains expressing untagged ubiquitin (strains 1 and 2) served as negative controls for the experimental WT, npl4-1, and npl4-1Δubc7 strains expressing 6×His–Myc-tagged ubiquitin (strains 3–5, respectively). (c) Strategy used for ubiquitinated protein purification and identification. For each of the five strains listed in b, a membrane fraction containing 20 mg of total protein was solubilized under native conditions, and 6×His–Myc-ubiquitin-conjugated proteins were purified by immobilized metal (nickel) affinity chromatography. Columns were washed extensively under highly denaturing conditions (8 M urea) to eliminate copurification of proteins associated with ubiquitin-conjugated proteins. Bound proteins were eluted with imidazole and directly digested with trypsin. The resulting peptide mixture was resolved by SCX chromatography, and 12 peptide-containing SCX fractions were collected. Each SCX fraction was individually subjected to nanoscale microcapillary reverse-phase (RP) chromatography. The RP chromatography was coupled directly to an ion trap tandem mass (MS/MS) spectrometer. Each MS/MS spectrum was searched independently against a database of predicted spectra derived from S. cerevisiae-encoded proteins, leading to the identification of >6,000 peptides corresponding to 527 unique proteins among the five data sets. These identified proteins were subjected to a stringent quality evaluation (see Materials and Methods), and only 211 unambiguously identified candidate ubiquitinated proteins were kept. (d) Example of a representative purification of 6×His–Myc-tagged ubiquitinated proteins (from npl4-1 membranes) as monitored by anti-Myc Western blot. Equivalent amounts (0.025% total) of input (lane 2), flow-through (lane 3), and two protein-containing elution (lanes 4 and 5) fractions were separated by SDS/PAGE and immunoblotted with anti-Myc antibodies to detect ubiquitin conjugates. An equivalent amount of input from the npl4-1-negative control strain expressing untagged ubiquitin (lane 1) was included to indicate the specificity of the anti-Myc antibodies. (e) Example of a representative purification of 6×His–Myc-tagged ubiquitinated proteins (from npl4-1 membranes) as monitored by silver staining. Equivalent amounts (1% total) of purified proteins from npl4-1 cells expressing either untagged (lane 1) or 6×His–Myc-tagged (lane 2) ubiquitin were subjected to SDS/PAGE and were detected by silver staining. Arrowheads indicate the presence of “false positive” proteins purified in the absence of tagged ubiquitin. Many of these false positive proteins contain endogenous histidine-rich sequences and/or have metal binding capabilities.
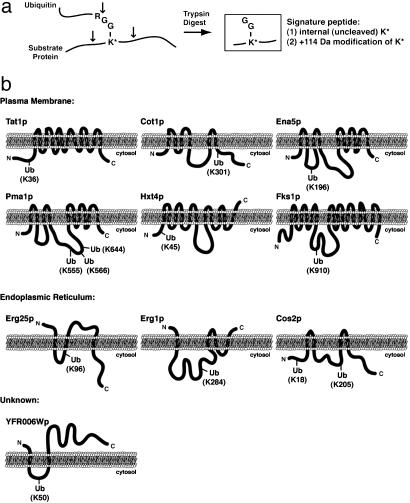
Fig. 3.
Identification of precise sites of substrate protein ubiquitination. (a) Schematic diagram indicating the ability to detect ubiquitinated lysine residues by tandem mass spectrometry. After trypsin digestion, a diglycine ubiquitin remnant remains covalently attached by an isopeptide linkage to the ε-amino side chain of a lysine residue within the substrate protein. The modified lysine, which is resistant to trypsin cleavage, can be identified by tandem mass spectrometry as a 114-Da-modified internal lysine residue. (b) Schematic representations of the predicted membrane topology and identified site(s) of ubiquitination for 15 integral membrane proteins localized to the plasma membrane, ER, and unknown membranes as indicated.
Fig. 4.
Subcellular localization and functional classes of identified ubiquitinated membrane-associated proteins, including candidate ERAD pathway substrates. (a) Venn diagram depiction of the distribution of 211 ubiquitinated proteins among the WT, npl4-1, and npl4-1 Δubc7 data sets. (b) Venn diagram depiction of the overlap between the 108 NPL4-(npl4-1 up-regulated) and 79 UBC7-responsive (Δubc7 down-regulated) ubiquitinated proteins [the small class (<10%) of npl4-1 down-regulated and Δubc7 up-regulated ubiquitinated proteins are not included]. The 68 NPL4- and UBC7-responsive proteins were classified as “strong” candidate endogenous ERAD substrates. Manual inspection of the remaining 51 NPL4-or UBC7-responsive proteins led to the classification of an additional 15 “weak” candidate ERAD substrates (see Materials and Methods). (c) Histogram representation of the membrane associations of 211 identified ubiquitinated proteins (white bars plus black bars) and the ERAD-candidate subpopulation (83 proteins, white bars). A total of 80 proteins (34 ERAD candidates) have known TMDs, and an additional 63 proteins (22 ERAD candidates) contain predicted TMDs. Of the non-TMD containing population, 43 (17 ERAD substrates) have known peripheral associations with cellular membranes, and 25 proteins (10 ERAD substrates) are not known or predicted to associate with membranes. (d) Histogram representation of the distribution of 186 membrane-associated (integral or peripheral) ubiquitinated proteins (73 of which are candidate ERAD substrates) among subcellular membranes. Asterisks indicate cellular membranes that contain an enriched number of ERAD candidate proteins. (e) Histogram representation of the distribution of 211 ubiquitinated proteins (including 83 candidate ERAD substrates) among eight functional classes of proteins. Asterisks indicate functional classes that contain an enriched number of ERAD candidate proteins.
We identified 83 of these 211 ubiquitinated proteins whose relative levels of abundance (i.e., levels of ubiquitination) among the three data sets fit the profile expected for a “typical” ERAD substrate: low levels of ubiquitination in WT, high levels of ubiquitination in npl4-1, and “rescued” low levels of ubiquitination in npl4-1Δubc7. To find these proteins, we first classified proteins as (i) NPL4- and/or (ii) UBC7-responsive based on significant changes in their relative abundance in pair-wise comparisons between (i) the WT vs. npl4-1 data sets and (ii) the npl4-1 vs. npl4-1Δubc7 data sets (see Materials and Methods). In all, 108 proteins displayed a significant increase in abundance in the npl4-1 sample as compared with WT, whereas 79 proteins displayed a significantly decreased abundance in npl4-1Δubc7 when compared with npl4-1. The intersection of NPL4- and UBC7-responsive proteins is shown in Fig. 4b; the 68 proteins that displayed significant responsiveness to both NPL4 and UBC7 are likely to be ubiquitinated as substrates of the ERAD pathway. Based on manual inspection, an additional 15 proteins that just missed the stringent cutoffs used in our classifications (see Materials and Methods) were also included as “weak” candidate ERAD substrates (Fig. 4b). In sum, we identified 211 ubiquitinated membrane-associated ubiquitinated proteins, 83 of which are likely to be ubiquitinated as substrates of the ERAD pathway.
To gain insight into total membrane protein ubiquitination, we categorized the cellular localizations and biological functions of the 211 identified ubiquitinated proteins (Table 3, which is published as supporting information on the PNAS web site). More than 85% of the identified ubiquitinated proteins have known or predicted associations with cellular membranes (Fig. 4c). It is possible that some of the identified ubiquitinated soluble proteins were present because of the mild washing conditions used during the preparation of cellular membranes. As shown in Fig. 4d, membrane-associated ubiquitinated proteins with known localizations fell into two major classes: (i) ER/nuclear membrane proteins and (ii) plasma membrane proteins. The ER/nuclear membrane-localized class of proteins is likely to consist largely of proteins that are ubiquitinated through the ERAD pathway. Indeed, proteins that we classified as ERAD substrates were highly enriched in this category (Fig. 4d; discussed in more detail below). However, we noted that only a small subset of the plasma membrane-localized ubiquitinated proteins were identified as candidate ERAD substrates. This observation suggests that the plasma membrane proteins were mostly ubiquitinated through an ERAD-independent pathway, such as ubiquitin-mediated endocytosis and vacuolar targeting (27, 28). In fact, we identified several plasma membrane proteins that are known to be substrates of ubiquitin-mediated endocytosis, including the amino acid transporters Gap1p and Gnp1p, the glucose transporter Hxt7p, the inositol permease Itr1p, the multidrug resistance transporter Pdr5p, and the α-factor mating pheromone receptor Ste6p. We have additionally identified ≈30 novel substrates of ubiquitination at the plasma membrane, including the amino acid permeases Tat1p, Bap3p, Hip1p, and Lyp1p and the cell wall biosynthetic enzymes Chs1–3p, Fks1p, and Gas1p. Interestingly, we were able to identify many precise sites of ubiquitination for these proteins (Table 2); it is worth noting that the sites of ubiquitination always fell within a predicted cytosolic loop region (Fig. 3b).
Some identified ubiquitinated membrane-associated proteins also localize to other cellular membranes, including the vacuole and transport vesicles, and participate in cellular pathways such as vesicle transport, regulation of the actin cytoskeleton, DNA transcription and replication, and the ubiquitin-proteasome pathway (Fig. 4 d and e). The identification of actin (Act1p) and some of its regulators (Rho3p, Sac6p, Rvs167p) as membrane-associated substrates of ubiquitination may extend our understanding of the suggested role for ubiquitin as a direct regulator of the actin cytoskeleton during the process of endocytosis (29, 30).
We were particularly interested in examining the localizations and biological functions of the 83 identified candidate substrates of the ERAD pathway (listed in Table 4, which is published as supporting information on the PNAS web site). As mentioned earlier, the majority of candidate ERAD substrates localize to the ER/nuclear membrane (Fig. 4d). Many of these candidate ER-localized ERAD substrates function as enzymes in biological processes that take place at the ER, such as ergosterol biosynthesis (Erg1p, Erg9p, Erg27p) and protein glycosylation (Pmt2p, Pmt4p, Alg9p) (Fig. 4e). Additionally, we identified several soluble ER lumenal proteins (Kar2p, Pdi1p) as candidate ERAD substrates; these proteins were likely purified in a ubiquitinated form either before translocation into the ER or during their export from the ER in the process of ERAD. Interestingly, several ER-localized proteins that function in the ERAD pathway itself were also identified as ERAD substrates, including Cdc48p, Ubc6p, Ssm4p/Doa10p, and Cue1p. We identified ubiquitination sites for several ER-localized proteins; as for plasma membrane proteins, ubiquitination sites always fell in predicted cytosolic loops (Fig. 3b). Not all identified ERAD substrates, however, are ER-resident proteins. For example, many candidate ERAD substrate proteins localize to transport vesicles (Erp1p, Sec27p, Hip1p), the vacuole (Pmc1p, Bpt1p, Ccc1p), and the plasma membrane (Snq2p, Dnf2p, Gap1p). It is likely, therefore, that these proteins are not ubiquitinated at their final destination, but rather as they are synthesized and folded in the ER. Overall, these findings suggest that the ERAD pathway of protein ubiquitination and degradation is responsible for the turnover of a diverse set of proteins that localize to or transit through the ER.
Interestingly, two membrane-anchored transcription factors, Mga2p and Spt23p, that are posttranslationally cleaved from the ER membrane by an ubiquitin/proteasome-dependent mechanism (15) were also identified as candidate ERAD substrates in our screen. These transcription factors are known to require the Cdc48p–Npl4p–Ufd1p protein complex for cleavage and/or activation (13, 14). This finding indicates that our screen also identified proteins that are targets of nonclassical ERAD pathways. A very interesting possibility is that the predicted membrane-anchored transcription factor Imp2p (encoded by YIL154C), which we identified as a potential ERAD substrate, might be subject to a similar form of ubiquitin/protesome/Cdc48p–Npl4p–Ufd1p-dependent cleavage and activation (31).
In conclusion, our large-scale proteomic identification of ubiquitinated membrane proteins and many precise sites of ubiquitination has significantly contributed to the understanding of the broad role ubiquitin plays as a posttranslational regulator of membrane proteins. We have found that membrane-associated protein ubiquitination occurs primarily at the plasma membrane and ER/nuclear membrane, likely reflecting two pathways of membrane protein ubiquitination: ubiquitin-mediated endocytosis/vacuolar targeting and ERAD, respectively. Our data also indicate that ubiquitin may be an important but currently unrecognized regulator the actin cytoskeleton. Furthermore, we have described a genetic/proteomic approach that allowed for the large-scale identification of proteins whose ubiquitination is potentially coregulated through the ERAD pathway. We have found that candidate ERAD substrates are largely localized to the ER and function in ER-specific processes. Proteins localized to other membranes, such as the vacuole and plasma membrane, also may be subject to ERAD, presumably as they transit through the ER. This approach also allowed for the identification of nonclassical ERAD substrates, and may have revealed a novel substrate of ubiquitin-mediated membrane-anchored transcription factor activation. We propose that this approach could be adapted to identify the endogenous substrates of other ubiquitin-regulated pathways, such as endocytosis and vacuolar protein targeting.
Supplementary Material
Acknowledgments
We thank D. Finley for plasmids and strains. We especially appreciate the critical comments on the manuscript by G. Adelmant, as well as stimulating discussions and technical assistance from members of the Silver and Gygi laboratories. This work was funded by National Institutes of Health (NIH) grants (to P.A.S. and S.P.G.). A.L.H. and K.A. were supported by NIH training grants to Harvard Medical School and National Cancer Institute grants to the Dana–Farber Cancer Institute.
Abbreviations: ER, endoplasmic reticulum; ERAD, ER-associated degradation; TMD, transmembrane domain; SCX, strong cation exchange.
References
- 1.Kostova, Z. & Wolf, D. H. (2003) EMBO J. 22 2309–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plemper, R. K. & Wolf, D. H. (1999) Trends Biochem. Sci. 24 266–270. [DOI] [PubMed] [Google Scholar]
- 3.Deak, P. M. & Wolf, D. H. (2001) J. Biol. Chem. 276 10663–10669. [DOI] [PubMed] [Google Scholar]
- 4.Biederer, T., Volkwein, C. & Sommer, T. (1997) Science 278 1806–1809. [DOI] [PubMed] [Google Scholar]
- 5.Bays, N. W., Gardner, R. G., Seelig, L. P., Joazeiro, C. A. & Hampton, R. Y. (2001) Nat. Cell Biol. 3 24–29. [DOI] [PubMed] [Google Scholar]
- 6.Swanson, R., Locher, M. & Hochstrasser, M. (2001) Genes Dev. 15 2660–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bays, N. W. & Hampton, R. Y. (2002) Curr. Biol. 12 R366–R371. [DOI] [PubMed] [Google Scholar]
- 8.Bays, N. W., Wilhovsky, S. K., Goradia, A., Hodgkiss-Harlow, K. & Hampton, R. Y. (2001) Mol. Biol. Cell 12 4114–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarosch, E., Taxis, C., Volkwein, C., Bordallo, J., Finley, D., Wolf, D. H. & Sommer, T. (2002) Nat. Cell Biol. 4 134–139. [DOI] [PubMed] [Google Scholar]
- 10.Braun, S., Matuschewski, K., Rape, M., Thoms, S. & Jentsch, S. (2002) EMBO J. 21 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabinovich, E., Kerem, A., Frohlich, K. U., Diamant, N. & Bar-Nun, S. (2002) Mol. Cell. Biol. 22 626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye, Y., Meyer, H. H. & Rapoport, T. A. (2001) Nature 414 652–656. [DOI] [PubMed] [Google Scholar]
- 13.Hitchcock, A. L., Krebber, H., Frietze, S., Lin, A., Latterich, M. & Silver, P. A. (2001) Mol. Biol. Cell 12 3226–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rape, M., Hoppe, T., Gorr, I., Kalocay, M., Richly, H. & Jentsch, S. (2001) Cell 107 667–677. [DOI] [PubMed] [Google Scholar]
- 15.Hoppe, T., Matuschewski, K., Rape, M., Schlenker, S., Ulrich, H. D. & Jentsch, S. (2000) Cell 102 577–586. [DOI] [PubMed] [Google Scholar]
- 16.Adams, A., Gottschling, D. E., Kaiser, C. A. & Stearns, T. (1997) Methods in Yeast Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 17.Spence, J., Gali, R. R., Dittmar, G., Sherman, F., Karin, M. & Finley, D. (2000) Cell 102 67–76. [DOI] [PubMed] [Google Scholar]
- 18.DeHoratius, C. & Silver, P. A. (1996) Mol. Biol. Cell 7 1835–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson, M. K., Kurihara, T. & Silver, P. A. (1993) Genetics 134 159–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latterich, M. & Schekman, R. (1994) Cell 78 87–98. [DOI] [PubMed] [Google Scholar]
- 21.Peng, J. & Gygi, S. P. (2001) J. Mass Spectrom. 36 1083–1091. [DOI] [PubMed] [Google Scholar]
- 22.Eng, J., McKormack, A. L. & Yates, J. R. (1994) J. Am. Soc. Mass Spectrom. 5 976–989. [DOI] [PubMed] [Google Scholar]
- 23.Peng, J., Elias, J. E., Thoreen, C. C., Licklider, L. J. & Gygi, S. P. (2003) J. Proteome Res. 2 43–50. [DOI] [PubMed] [Google Scholar]
- 24.Peng, J., Schwartz, D., Elias, J. E., Thoreen, C. C., Cheng, D., Marsischky, G., Roelofs, J., Finley, D. & Gygi, S. P. (2003) Nat. Biotechnol. 21 921–926. [DOI] [PubMed] [Google Scholar]
- 25.Rappsilber, J., Ryder, U., Lamond, A. I. & Mann, M. (2002) Genome Res. 12 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanders, S. L., Jennings, J., Canutescu, A., Link, A. J. & Weil, P. A. (2002) Mol. Cell. Biol. 22 4723–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hicke, L. (2001) Cell 106 527–530. [DOI] [PubMed] [Google Scholar]
- 28.Bonifacino, J. S. & Traub, L. M. (2003) Annu. Rev. Biochem. 72 395–447. [DOI] [PubMed] [Google Scholar]
- 29.Kaminska, J., Gajewska, B., Hopper, A. K. & Zoladek, T. (2002) Mol. Cell. Biol. 22 6946–6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaw, J. D., Cummings, K. B., Huyer, G., Michaelis, S. & Wendland, B. (2001) Exp. Cell Res. 271 1–9. [DOI] [PubMed] [Google Scholar]
- 31.Masson, J. Y. & Ramotar, D. (1996) Mol. Cell. Biol. 16 2091–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.