Abstract
The Wnts are a family of glycoproteins that regulate cell proliferation, fate decisions, and differentiation. In our study, we examined the contribution of Wnts to the development of ventral midbrain (VM) dopaminergic (DA) neurons. Our results show that β-catenin is expressed in DA precursor cells and that β-catenin signaling takes place in these cells, as assessed in TOPGAL [Tcf optimal-promoter β-galactosidase] reporter mice. We also found that Wnt-1, -3a, and -5a expression is differentially regulated during development and that partially purified Wnts distinctively regulate VM development. Wnt-3a promoted the proliferation of precursor cells expressing the orphan nuclear receptor-related factor 1 (Nurr1) but did not increase the number of tyrosine hydroxylase-positive neurons. Instead, Wnt-1 and -5a increased the number of rat midbrain DA neurons in rat embryonic day 14.5 precursor cultures by two distinct mechanisms. Wnt-1 predominantly increased the proliferation of Nurr1+ precursors, up-regulated cyclins D1 and D3, and down-regulated p27 and p57 mRNAs. In contrast, Wnt-5a primarily increased the proportion of Nurr1+ precursors that acquired a neuronal DA phenotype and up-regulated the expression of Ptx3 and c-ret mRNA. Moreover, the soluble cysteine-rich domain of Frizzled-8 (a Wnt inhibitor) blocked endogenous Wnts and the effects of Wnt-1 and -5a on proliferation and the acquisition of a DA phenotype in precursor cultures. These findings indicate that Wnts are key regulators of proliferation and differentiation of DA precursors during VM neurogenesis and that different Wnts have specific and unique activity profiles.
The development of midbrain dopaminergic (DA) neurons requires a complex combination of transcriptional regulators and diffusible signals to control both the acquisition and maintenance of a neurotransmitter-specific phenotype. The orphan nuclear receptor-related factor 1 (Nurr1, also known as NR4A2) is the only factor known to be required by midbrain precursor cells for the acquisition of a midbrain DA phenotype (1–4). Null mutations in other transcriptional regulators expressed in DA neurons, such as the homeodomain proteins Lmx1b and Ptx3, result in the loss of midbrain DA neurons after their birth (5–7). With regard to soluble diffusible signals, intersections of Shh (ventrally) and FGF8 (in the isthmus) create sites for the induction of DA neurons (8). Members of the Wnt family of secreted glycoproteins are also expressed in the midbrain (9) and are known to regulate precursor proliferation (10–12), fate decisions (13–17), and neuronal differentiation (18–20) in the nervous system. Interestingly, deletion of Wnt-1 results in the loss of DA neurons (21) and of the entire midbrain–hindbrain junction (22, 23). Another mutant mouse with a similar phenotype in the midbrain is the LRP6 (low-density lipoprotein receptor-related protein 6) null (24), which lacks a receptor necessary for Wnt signaling. Combined, these findings suggest an important role for Wnts during the development of midbrain DA neurons. Thus, the aim of our study has been to establish whether Wnts regulate the development of DA neurons and, if so, to characterize the mechanisms involved.
Materials and Methods
In Situ Hybridization and Immunohistochemistry on Brain Sections. Male and female wild-type CD-1 mice (25–35 g; Charles River Breeding Laboratories) were housed, bred, and treated according to the guidelines of the European Communities Council (directive 86/609/EEC) and the Society for Neuroscience (available at www.sfr.org/handbook), and all experiments were approved by the local ethical committee. Mice from embryonic day (E) 10.5 and E11.5 were removed and rapidly frozen in OCT (optimal cutting temperature) compound at –70°C. Serial sagittal sections (14 μm thick) were collected on microscope slides (StarFrost, Friedrichsdorf, Germany). In situ hybridization was performed on fresh frozen tissue with 35S-labeled RNA probes as described (25). Immunohistochemistry was performed on 4% paraformaldehyde-postfixed slides. Incubations were carried out at 4°C overnight with mouse anti-β-catenin (1:250 dilution; BD Biosciences) and rabbit anti-Nurr1 (1:200 dilution; Santa Cruz Biotechnology) in dilution buffer (PBS, pH 7.3/1% BSA/0.3% Triton X-100). After washes with 0.2% Tween 20/PBS, the sections were blocked for 30 min in dilution buffer and then incubated for 2 h with a secondary antibody [Cy2 (cyanine)-coupled horse anti-mouse IgG, Cy2-coupled horse anti-rabbit IgG, or rhodamine-coupled horse anti-mouse IgG; 1:200 dilution; Jackson ImmunoResearch). Immunostaining was visualized by using a Zeiss HBO100 microscope.
Tcf-Optimal Promoter β-Galactosidase (TOPGAL) Transgenic Mice. E10.5 TOPGAL transgenic mice (26) were fixed in 4% paraformaldehyde, cryoprotected in 20% sucrose, and frozen. Next, 14-μm sections through the ventral midbrain (VM) were cut on a cryostat and stored at –20°C. Sections were air dried at room temperature for 15 min, washed with PBS, and incubated for 16–24 h at 37°C in the dark with the following histochemistry solution: 0.1 M phosphate buffer, pH 7.3–7.4/3.1 mM potassium ferricyanide/3.1 mM potassium ferrocyanide/1 mM magnesium chloride/0.4 mg/ml 5-bromo-4-chloro-3-indolyl β-d-galactoside substrate. Slides were washed in PBS and mounted in 4:1 glycerol/PBS. The sections were observed by using a Zeiss Axiovert 100M microscope, and photographs were taken with a Kodak MDS 290 camera.
Real-Time RT-PCR and Quantification of Gene Expression. These methods are described in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org. The PCR started with 94°C for 2 min and then continued with 35–40 cycles of 30 s at 94°C, 30 s at 60°C, 45 s at 72°C, and 15 s at 80°C (for SYBR green detection) on the ABI Prism 5700 detection system (Applied Biosystems). A melting curve was obtained for each PCR product after each run to confirm that the signal corresponded to a unique amplicon of the predicted size. The specificity of the PCR product was verified by sequencing. Expression levels were obtained by subtracting the value for each sample in the absence of reverse transcriptase from the corresponding value in the presence of reverse transcriptase and then normalizing to the housekeeping gene encoding 18S rRNA, obtained for every sample in parallel assays in two to four independent experiments.
Partial Purification of Wnt and the Frizzled-8 Cysteine-Rich Domain (Fz8-CRD) Conditioned Medium. Rat B1a fibroblast lines stably overexpressing hemagglutinin-tagged Wnt-1a, -3a, or -5a (26) or mouse Fz8-CRD IgG were generated as described in Supporting Text. Serum-free N2 medium was conditioned by fibroblasts for 24 h and concentrated with Centricon Plus-80 columns (Millipore) according to the manufacturer's instructions. Protein content was examined by Western blot detection of the hemagglutinin tag (see Fig. 6, which is published as supporting information on the PNAS web site), and aliquots with equivalent concentrations were prepared and stored at –80°C. All experiments were performed with 10 μl/ml partially purified Wnts or 5 μl/ml partially purified Fz8-CRD, unless stated otherwise.
Precursor Cultures and Treatments. VM from E14.5 embryos obtained from timed-mated Sprague–Dawley rats were dissected, mechanically dissociated, and plated at a final density of 1 × 105 cells per cm2 on poly-d-lysine-coated 12- or 24-well plates in serum-free N2 medium, consisting of a 1:1 mixture of F12 and DMEM with 10 ng/ml insulin/100 μg/ml transferrin/100 μM putrescine/20 nM progesterone/30 nM selenium/6 mg/ml glucose/1 mg/ml BSA. All factors were added once (at the initiation of culture), and BrdUrd was added 6 h before fixation.
Immunocytochemical Analysis. Cultures were fixed with 4% paraformaldehyde, washed in PBS, and incubated at 4°C overnight or at room temperature for 1 h in dilution buffer with one of the following antibodies: mouse anti-BrdUrd, 1:50 dilution (DAKO); mouse anti-β-tubulin type III (TuJ1), 1:250 dilution (Sigma); rabbit anti-Adh2, 1:4,000 dilution (a gift from R. Lindahl, University of South Dakota, Vermillion, SD); mouse anti-tyrosine hydroxylase (TH), 1:10,000 dilution (Incstar, Still-water, MN); rabbit anti-TH, 1:250 dilution (Pel-Freez Biologicals); rabbit anti-Nurr1, 1:2,000 dilution (a gift from T. Perlmann, Karolinska Institute, Stockholm); rabbit anti-Nurr1, 1:1,000 dilution (Santa Cruz Biotechnology); or mouse anti-Nurr1, 1:250 dilution (BD Biosciences). After washing, cultures were incubated for 1–3 h in dilution buffer with the appropriate secondary anti-mouse or anti-rabbit IgG antibody or antibodies (biotinylated, 1:500 dilution; FITC- or rhodamine-coupled IgG, 1:100 dilution; Vector Laboratories). Bright-field immunostaining was visualized with the ABC (avidin–biotin complex) immunoperoxidase kit (Vector Laboratories) by using either NovaRED or AEC (3-amino-9-ethylcarbazole) (red), SG (substrate gray) or 3,3′-diaminobenzidine tetrahydrochloride (DAB) 0.5 mg/ml/nickel chloride 1.6 mg/ml (gray/black), or VIP (violet precipitate) substrates. Double staining was performed by sequential single staining as described above. The order of staining was critical only for BrdUrd, in which case the labeling was always performed second. Control experiments, in which primary or secondary antibodies were omitted, demonstrated little to no crossreactivity. Only clearly stained cells were counted as positive cells. Spheres were considered positive if they contained one or more positive cell. Quantitative immunocytochemical data represent means ± SEs of counts obtained by a blinded observer from 10–20 nonoverlapping fields (cells) or entire wells (clusters) in three or four wells per condition from three or four separate experiments, unless stated otherwise. Photographs were taken with a Zeiss Axiovert 100M microscope and collected with a Hamamatsu C4742-95 camera and qed camera plug-in software (QED Imaging, Pittsburgh). Images in Fig. 5I were collected with a Zeiss MC80 camera with Kodak Ektachrome 64T tungsten film, developed, and digitized from slides.
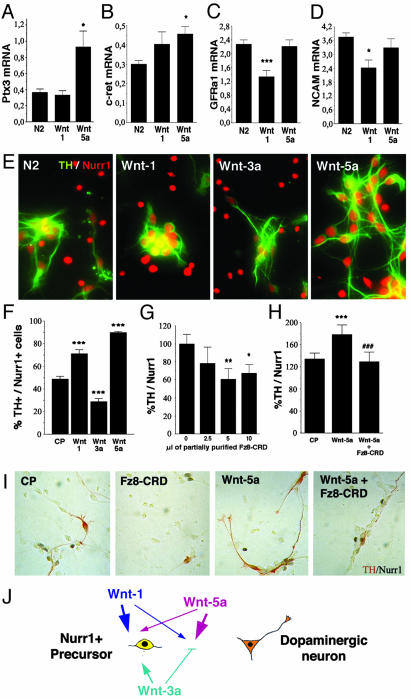
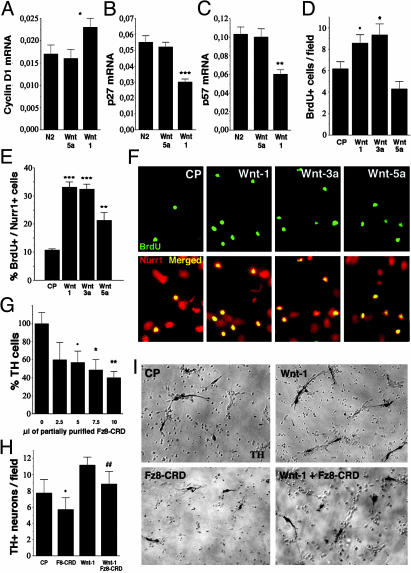
Fig. 5.
Wnts differentially control the development of DA neurons by regulating precursor proliferation and the acquisition of a DA phenotype. Wnt-5a, but not Wnt-1, up-regulated the expression of Ptx3 mRNA (A) and c-ret mRNA (B) and maintained the expression of GDNF family receptor α1 (GFRα1) mRNA (C) and NCAM mRNA (D) at 3 days in vitro, as assessed by real-time RT-PCR in E14.5 rat VM precursor cultures. (E and F) Double immunocytochemistry revealed that Wnt-5a increased the percentage of TH+/Nurr1+ cells from 50% to 90%. Wnt-1 was less efficient than Wnt-5a, and Wnt-3a actually decreased the proportion of TH+ cells from 50% to 30%. (G and I) Fz8-CRD decreased, in a dose-dependent manner, the proportion of Nurr1+ cells that acquired TH expression in E14.5 VM precursor cultures, indicating that Wnt signaling is required for the acquisition of a DA phenotype. (H and I) Treatment of rat E14.5 VM precursor cultures with Fz8-CRD decreased the percentage of TH+/Nurr1+ cells after treatment with Wnt-5a. Statistical analysis and concentrations as in Fig. 4. (J) Model of the mechanisms by which Wnt-1, -3a, and -5a regulate the development of VM DA neurons. Wnt-3a, which is mainly expressed in the dorsal midbrain, enhances the proliferation of Nurr1-expressing precursors and decreases the proportion of neurons that acquire a DA phenotype. Wnt1, probably derived from the midbrain–hindbrain organizer, controls the proliferation of Nurr1-expressing precursors and increases the number of VM neurons. Finally, Wnt-5a increases the number of VM DA neurons specifically by regulating the acquisition of a DA phenotype in Nurr1-expressing precursors. Note that the size of the arrows correlates with the intensity of the effects.
Results and Discussion
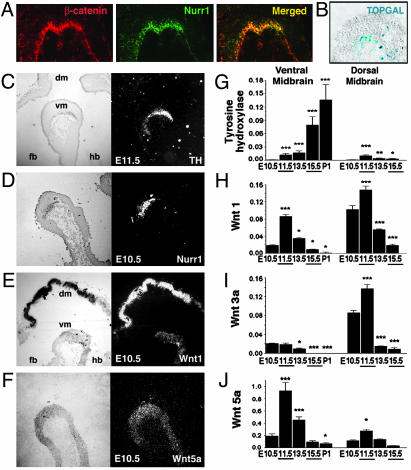
Expression and Transcriptional Activity of β-Catenin and Wnts in the Developing VM. To establish whether Wnts play a physiological role in the development of midbrain DA neurons in vivo, we first examined DA precursor cells for the expression and transcriptional activity of β-catenin, a central signaling component of the Wnt canonical pathway. Double immunohistochemistry showed that β-catenin is expressed in the same domain as the orphan nuclear receptor Nurr1 (Fig. 1A), a marker for DA precursor cells at E10.5 in mice. Moreover, the activation of the Wnt canonical signaling pathway was examined at a transcriptional level by analyzing the VM of TOPGAL reporter mice (27). In these mice, β-galactosidase expression is under the control of an enhancer containing multimerized consensus binding sites for the TCF/LEF family of DNA-binding proteins. The enhancer is activated in cells that both express a member of the TCF/LEF family and receive a canonical Wnt signal to stabilize β-catenin. In control mice, no β-galactosidase-positive cells were detected in the VM (data not shown). However, when the β-catenin-positive VM domain of TOPGAL mice was examined, several β-galactosidase-positive cells were found at E10.5 (Fig. 1B), suggesting that canonical Wnt signaling takes place in the developing VM in vivo before the birth of DA neurons.
Fig. 1.
Detection of β-catenin transcriptional activity (A and B) and Wnt expression (C–J) in the developing midbrain in vivo. (A) Sagittal sections through the developing VM revealed that β-catenin and Nurr1 are expressed in the same domain at E10.5. (B) Analysis of the Nurr1+ domain in TOPGAL reporter mice at E10.5 revealed the presence of β-galactosidase-positive cells, indicating that the Wnt canonical signaling pathway is active in the developing VM before the birth of TH+ neurons. (C–F) In situ hybridization on sagittal brain sections including forebrain (fb), dorsal midbrain (dm), VM (vm), and hindbrain (hb). Wnt-1 (E) and Wnt-5a (F) mRNA expression domains coincide with those of Nurr1+ (D) and TH+ (C) cells. (G–J) Real-time RT-PCR analysis revealed that Wnt-1 is expressed at high levels in the VM and dorsal midbrain, whereas Wnt-3a expression predominates in the dorsal midbrain and Wnt-5a predominates in the VM. *, P < 0.05; **, P < 0.001; and ***, P < 0.0001 (compared with the first stage analyzed for every brain region by one-way ANOVA with Fisher's post hoc test).
Next, the expression of Wnts, Nurr1, and TH, a marker of DA neurons, was examined in the developing VM. TH mRNA was clearly detected by in situ hybridization at E11.5 in mice and was first detected by real-time RT-PCR at E11.5 in rats (Fig. 1 C and G), coinciding with the birth of DA neurons (28), which takes place after the onset of Nurr1 expression (Fig. 1D) and continues until E16 in rat VM. Interestingly, Wnt-1 and -5a were highly expressed in the VM (Fig. 1 E and F), peaking at E11.5 in rats (Fig. 1 H and J). A similar peak was detected in E11.5 rat dorsal midbrain for Wnt-1 and -3a (Fig. 1 H and I) but not for Wnt-5a (Fig. 1J), reinforcing the argument that Wnt-5a has a predominant role in ventral development (29).
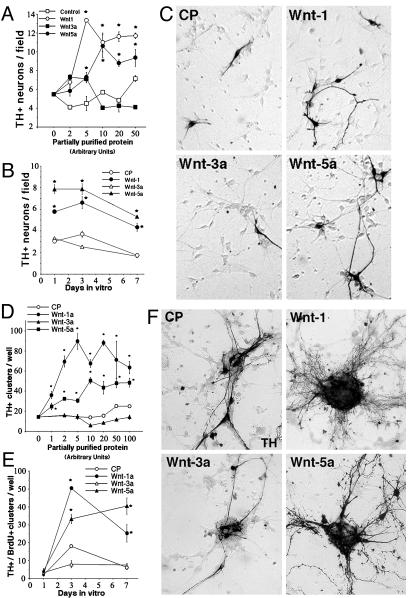
Wnt-1 and -5a, but Not Wnt-3a, Increase the Number of DA Neurons Derived from VM Precursors. To directly assess the role of Wnts in the development of the VM, conditioned medium from mock- or stably transfected fibroblast cell lines engineered to secrete Wnts (27) was concentrated (≈250-fold) and partially purified by size-exclusion-based filtration. Cultures of VM precursors obtained from E14.5 rats were treated with increasing concentrations of partially purified Wnt-1, -3a, -5a, or control medium for 3 days (Fig. 2A). Treatment with Wnt-1 or -5a resulted in robust dose-dependent increases in numbers of TH+ cells, whereas Wnt-3a and control treatment had no significant effect at doses ≤50 units. These results suggested that the increases in TH+ cells induced by Wnt-1 or -5a treatment were specific to those ligands. Interestingly, the effects of 10 units of Wnt-1 or -5a were detected as early as 1 day in vitro, were clearly higher than of those of CP and Wnt-3a at 3 days in vitro (Fig. 2 B and C), and started to diminish after 7 days (Fig. 2B).
Fig. 2.
Wnt-1 and -5a, but not Wnt-3a, increase the number of DA neurons (A–C) and proliferating cell clusters containing DA neurons (D–F) in rat E14.5 VM precursor cultures. Partially purified Wnts were normalized to each other based on the density of Western blot product bands and expressed in arbitrary units, equivalent to 1 μl of normalized partially purified product. Control partially purified medium (CP) was diluted to achieve the same protein concentration as the partially purified Wnts. (A and D) Dose dependency at 3 and 7 days in vitro, respectively. (B and E) Time-course analysis (10 units). (C and F) Comparison of the effect of 10 units of Wnts with CP. TH-immunostained cultures show that Wnt-1 and -5a induce a very dramatic increase in the number of DA neurons and TH+ cell clusters. *, P < 0.01 [compared with CP, by one-way ANOVA with Fisher's post hoc test (n = 4–6)]. A field is 3.14 mm2 and a well is 4 cm2.
In addition to effects on individual DA neurons, it was observed early in the study that Wnt treatment induced the appearance of large spherical cell clusters. These clusters were initially small but increased in size over time in culture, and virtually all clusters were BrdUrd positive after an acute BrdUrd pulse before fixation (data not shown). Clusters of TH+ neurons, more than five cells wide, were counted at various times in vitro (these clusters were excluded from the previous quantification). Strikingly, 10 units of Wnt-1 and -5a, but not Wnt-3a, treatment produced a 3- to 8-fold increase in the number of these clusters observed at 7 days in vitro (Fig. 2 D–F). Moreover, the clusters that did appear in response to Wnt-1 and -5a were larger, and the vast majority consisted almost entirely of TH+ neurons. These results suggested that Wnts may have two different functions in the VM: controlling the number of proliferating precursor cells and/or the number of TH+ neurons.
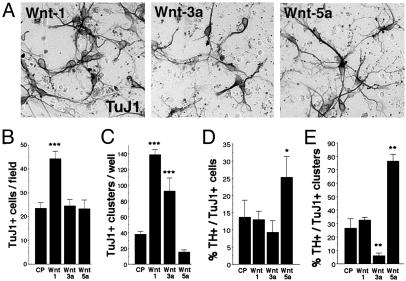
Differential Generation and Differentiation of Neurons by Wnt-1, -3a, and -5a. We next determined whether the observed effects of Wnts were general to all VM neurons or specific to VM DA neurons. We found that Wnt-1 increased the number of cells positive for the early neuronal marker TuJ1 in the cultures by 2-fold (Fig. 3 A and B) and the number of TuJ1+ clusters by 3.5-fold (Fig. 3C). In contrast, Wnt-5a did not increase the number of TuJ1+ cells or TuJ1+ clusters, and Wnt-3a increased only the number of proliferating clusters containing TuJ1+ cells. Interestingly, when we examined the proportion of TH+ neurons of the total of TuJ1+ neurons in the culture, we found that Wnt-1 did not increase this proportion (Fig. 3D) or the proportion of TuJ1+ clusters containing TH+ cells (Fig. 3E), suggesting that the effects of Wnt-1 are general to VM neurons. Wnt-3a showed a similar effect on the proportion of neurons but actually decreased the proportion of DA clusters from 30% to <10% (Fig. 3E), suggesting that Wnt-3a increases the number of TuJ1+ clusters but decreases their content of DA neurons. In contrast, Wnt-5a did not alter the number of proliferative clusters containing neurons (Fig. 3C) but selectively enriched for both TH+ neurons and TH+ cell clusters ≈2-fold (Fig. 3 D and E), suggesting that Wnt-5a promotes the acquisition of a DA phenotype. Thus, our results show that the three Wnts analyzed differentially regulate both neuron number and DA phenotype. Moreover, despite the similar effects of Wnt-1 and -5a on TH+ neurons and TH+ cell clusters, our results indicate that Wnt-1 and -5a may act on the DA lineage by two partially distinct mechanisms.
Fig. 3.
Wnts promote neurogenesis and increase the number of midbrain DA neurons in E14.5 rat VM precursor cultures by two different mechanisms. (A and B) Wnt-1, but not Wnt-3a or -5a, increased the number of TuJ1+ neurons outside cell clusters in VM cultures. A field is 3.14 mm2.(C) Wnt-1 and -3a, but not Wnt-5a, increased the number of proliferating clusters that contained TuJ1+ neurons. A well is 4 cm2.(D) Despite the increase in neurons outside the clusters, Wnt-1 did not increase the proportion of DA neurons. Instead, treatments that did not change the numbers of Tuj1+ neurons either decreased (Wnt-3a) or increased (Wnt-5a) the proportion of DA neurons. (E) The proportion of DA neurons in proliferating clusters containing neurons did not change after Wnt-1 treatment, was decreased by Wnt-3a, and was increased by treatment with Wnt-5a. Concentrations were 10 units/μl partially purified media. *, P < 0.01; **, P < 0.001; and ***, P < 0.0001 [compared with CP, by one-way ANOVA with Fisher's post hoc test (n = 3–5)].
Wnt-1 Increases the Number of TH+ Neurons by Regulating Nurr1+ Precursor Proliferation. Because Wnts are known to play a fundamental role in controlling proliferation, cell fate decisions, and differentiation in the nervous system, we investigated the possibility that Wnts increased the number of TH+ neurons in VM cultures by increasing proliferation and/or promoting the acquisition of a DA phenotype in precursor cells. We first examined by real-time RT-PCR whether cyclin D1, a target of β-catenin (30, 31) that mediates cell cycle progression by Wnt-1 (12) but not by Wnt-5a (32), or other cyclins were regulated by Wnt-1 and -5a. Whereas cyclin D2 mRNA was not affected by any of the treatments (data not shown), cyclin D1 and D3 mRNAs were up-regulated by Wnt-1 but not by Wnt-5a (Fig. 4A; data not shown). Similarly, analysis of cell-cycle inhibitors revealed that Wnt-1 but not Wnt-5a down-regulated p27 and p57 mRNA (Fig. 4 B and C), whereas neither Wnt-1 nor -5a affected p21 mRNA expression (data not shown). Thus, our results suggest that Wnt-1, by regulating G1 → S progression at a transcriptional level, may act to counterbalance cell-cycle arrest induced by Nurr1 (33). We therefore examined whether Wnts regulated proliferation of DA precursor cells in VM cultures. Interestingly, we did not observe BrdUrd incorporation in newborn DA neurons (Nurr1+/TH+ cells), indicating that proliferating cells are not differentiating into TH+ cells during the 6-h labeling period at the end of the experiment. We next examined whether Nurr1-expressing neuronal precursors in the midbrain (Nurr1+/BrdUrd+ cells) were a potential target for Wnts. BrdUrd incorporation was increased by Wnt-1 and -3a but not by Wnt-5a treatment (Fig. 4D), indicating that Wnt-1 and -3a may work as mitogens, promoting general precursor proliferation in the cultures. The effects of Wnt-1 and -3a were even greater (3-fold increase) when Nurr1 and BrdUrd double-positive precursor cells were examined after 1 day in vitro. Partially purified Wnt-5a also increased mitosis in the Nurr1+ population but to a lesser extent (Fig. 4 E and F). The clear increase in proliferation induced by Wnt-1 and -3a correlates well with their ability to stabilize β-catenin (27), with the role of β-catenin in promoting cell cycle reentry in stem cells and neural precursors (10, 11), and with the role of Wnt-3a in the self-renewal of hematopoietic stem cells (34). Thus, the effects of Wnt-1 on precursor proliferation and the regulation of genes controlling G1 → S progression indicate that Wnt-1 primarily enhances the number of TH+ cells by regulating mitosis and proliferation in precursor cells. However, Wnt-3a was as efficacious as Wnt-1 at enhancing mitosis in Nurr1+ cells, but unlike Wnt-1, it decreased the proportion of DA neurons in proliferating clusters (Fig. 3E), suggesting a role of Wnt-3a in maintaining the precursor population. These results indicate that the numbers of TH+ and proliferating cells in the VM are independently regulated by Wnts.
Fig. 4.
Wnt-1 regulated the expression of cyclin D1 and the cell cycle inhibitors p27 and p57 and increased the proliferation of VM precursors. Real-time RT-PCR showed that Wnt-1, but not Wnt-5a, increased the expression of cyclin D1 mRNA (A) and decreased the expression of the cell cycle inhibitors p27 (B) and p57 (C) at 3 days in vitro. (D) Wnt-1 and -3a, but not Wnt-5a, increased the proliferation of VM precursors at 3 days in vitro.(E and F) Wnt-5a was less efficient than Wnt-1 and -3a at increasing BrdUrd incorporation in VM Nurr1+ precursor cells after 1 day in vitro.(G) Increasing units of partially purified Fz8-CRD, a Wnt blocking reagent, reduced the number of TH+ neurons in VM cultures dose-dependently after 3 days in vitro, indicating that endogenous Wnts are required for DA development. (H and I) The increase in the number of TH+ neurons by Wnt-1 was partially blocked by Fz8-CRD, suggesting that the effects of Wnts are specific. •, P < 0.05; *, P < 0.01; **, P < 0.001; ***, P < 0.0001, compared with control; and ##, P < 0.0001 [compared Wnt treatment alone, by one-way ANOVA with Fisher's post hoc test (n = 3–5; for Fz8-CRD, n = 2 or 3)]. Concentrations are 10 units/μl Wnts or CP and 5 units/μl Fz8-CRD. A field (in D and H) is 3.14 mm2.
We also examined whether the effects of Wnt-1 could be blocked by the Fz8-CRD, a Wnt inhibitor (35, 36). Partially purified conditioned medium from fibroblasts overexpressing Fz8-CRD was added to control (Fig. 4G) and Wnt-1-treated cultures (Fig. 4 H and I), and the number of TH+ neurons was examined. Fz8-CRD decreased the number of TH+ neurons in both conditions, indicating that endogenous Wnts are necessary for the development of DA neurons in the cultures and that the effects of Wnts on TH+ cells are specific.
Wnt-5a Increases the Number of TH+ Neurons by Promoting the Maturation of Nurr1+ Precursors into DA Neurons. All of the results presented until now suggest that Wnt-1, although increasing proliferation and neurogenesis, does not increase the proportion of DA neurons. Instead, Wnt-5a, although not increasing general precursor proliferation and having only a minor effect on DA precursor proliferation, does increase the proportion of TH+ neurons within and outside proliferating clusters by an additional mechanism. We, therefore, examined whether Wnt-5a treatment could be involved in promoting the differentiation of midbrain DA neurons by regulating the expression of genes characteristic of midbrain DA neurons. We first examined the expression of the bicoid-related homeodomain gene Ptx3 (37) by real-time RT-PCR and found that Wnt-5a but not Wnt-1 treatment increased Ptx3 mRNA (Fig. 5A). This finding is interesting because Ptx3 is required for the development of DA neurons (6, 7) and Ptx2, another homeodomain gene of the same family, is directly regulated by Wnt signaling (32, 38). We next examined whether the expression of the protooncogene c-ret (39) and the two other glial cell line-derived neurotrophic factor (GDNF) coreceptors, GDNF family receptor α1 (GFRα1) (40) and neural cell adhesion molecule (NCAM) (41), were also regulated by Wnts. Interestingly, both c-ret and NCAM are known to be regulated by Wnt signaling (42, 43), and their ligand, GDNF, is required for the postnatal development of DA neurons (44). We found that all of the GDNF receptors analyzed were differentially regulated by Wnt-1 and -5a. Although c-ret expression was up-regulated by Wnt-5a but not by Wnt-1 (Fig. 5B), the expression of GFRα1 and NCAM was maintained by Wnt-5a and repressed by Wnt-1 (Fig. 5 C and D). Thus, our results suggested a model in which Wnt-5a increases the number of DA neurons by a mechanism that involved the acquisition of Ptx3 and GDNF receptor expression, two distinctive features of midbrain DA neurons. We therefore examined whether Wnt-5a influenced the conversion of ventral mesencephalic Nurr1+ neuronal precursors (Nurr1+/TH–cells) into DA neurons (Nurr1+/TH+ cells). In control conditions ≈50% of all Nurr1+ cells expressed TH, whereas Wnt-3a treatment decreased this expression to 30%. In contrast, Wnt-1 increased the proportion of Nurr1+/TH+ cells to 70% and Wnt-5a increased this population to 90% (Fig. 5 E and F). These results were also confirmed by double ADH-2+/TH+ immunostaining (data not shown) because ADH-2 labels DA precursors and neurons (45). Thus, our data suggest that Wnt-5a increases the number of DA neurons by a mechanism that predominantly involves promoting the maturation of DA precursors and the acquisition of a neuronal DA phenotype.
We next examined whether partially purified Fz8-CRD could block the normal development of DA precursors into DA neurons in E14.5 VM cultures. Dose–response analysis indicated that Fz8-CRD partially blocked the acquisition of a DA phenotype in untreated precursor cultures (Fig. 5G,%TH+/Nurr1+), indicating that endogenous Wnts are required for DA development. Moreover, Fz8-CRD decreased the proportion of TH+/Nurr1+ cells induced by Wnt-5a (Fig. 5 H and I), suggesting that Wnts are partially required for the acquisition of a neuronal DA phenotype in precursor cells.
Final Comments. Our work suggests that Wnts are essential regulators of two crucial aspects of neurogenesis in the VM: the proliferation and differentiation of VM precursors (Fig. 5J). Wnt-1 promoted neurogenesis by increasing the proliferation of precursors and affected both DA and non-DA VM neurons. Wnt-3a promoted the proliferation and maintenance of Nurr1+ precursors and decreased the number of DA neurons. In contrast, Wnt-5a was a weak mitogen; it increased the expression of Ptx3 and GDNF receptors and efficiently promoted the acquisition of a DA phenotype in Nurr1-expressing precursors. These results clearly show that DA precursors respond to Wnts in a very specific manner. We report that Wnt-1, -3a, and -5a differentially regulate the development of midbrain DA neurons by partially overlapping mechanisms, which include promoting the proliferation of DA precursors (Wnt-1 ≥ Wnt-3a > Wnt-5a) and the differentiation of DA precursors into DA neurons (Wnt-5a > Wnt-1). Thus, our results suggest that the two ventrally expressed Wnts, Wnt-1 and -5a, may work sequentially and probably in concert with other signals such as Shh and FGF8 (8) to promote DA neuron development. Future work could address the role of other Wnts in DA neuron development, the function of Wnts in vivo, and whether Wnts contribute to instructing a DA phenotype in stem cells.
Thus, we hereby identify Wnts as key cell-extrinsic signals that specifically regulate partially overlapping and distinct aspects of DA neuron development, and we suggest that activation of Wnt signaling may find a therapeutic application in diseases affecting DA neurons, including Parkinson's disease.
Supplementary Material
Acknowledgments
We thank Drs. Anita Hall, Helena Mira, Carmen Saltó, and Gunnar Schulte for critical reading of the manuscript; Lotta Skoog and Lena Amaloo for secretarial help; Annika Ahlsén and Claudia Tello for additional assistance; Dr. Jen Chih Hsieh for the Fz8-CRD plasmid; and Dr. Andy McMahon for the Wnt-1, -3a, and -5a probes. Financial support was obtained from the Swedish Foundation for Strategic Research, the Swedish Royal Academy of Sciences, the Knut and Alice Wallenberg Foundation, the Swedish Research Council, the Michael J. Fox Foundation, the European Commission, the Juvenile Diabetes Research Foundation, the Karolinska Institute, and the Petrus and Augusta Hedlunds Foundation. G.C.-B. was supported by the Portuguese Praxis XXI program of the Fundação para a Ciência e a Tecnologia. F.J.R. was supported by the Karolinska Institute and the Marie Curie program of the European Union. J. Kele was supported by the Swedish National Neuroscience Network. J. Kitajewski was supported by National Institutes of Health Grant R01 CA75353.
Abbreviations: CP, control partially purified medium; DA, dopaminergic; En, embryonic day n; Fz8-CRD, Frizzled-8 cysteine-rich domain; GDNF, glial cell line-derived neurotrophic factor; NCAM, neural cell adhesion molecule; Nurr1, nuclear receptor-related factor 1; TOPGAL, Tcf-optimal promoter β-galactosidase; TH, tyrosine hydroxylase; TuJ1, β-tubulin type III; VM, ventral midbrain.
References
- 1.Zetterstrom, R. H., Solomin, L., Jansson, L., Hoffer, B. J., Olson, L. & Perlmann, T. (1997) Science 276 248–250. [DOI] [PubMed] [Google Scholar]
- 2.Saucedo-Cardenas, O., Quintana-Hau, J. D., Le, W. D., Smidt, M. P., Cox, J. J., De Mayo, F., Burbach, J. P. & Conneely, O. M. (1998) Proc. Natl. Acad. Sci. USA 95 4013–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castillo, S. O., Baffi, J. S., Palkovits, M., Goldstein, D. S., Kopin, I. J., Witta, J., Magnuson, M. A. & Nikodem, V. M. (1998) Mol. Cell. Neurosci. 11 36–46. [DOI] [PubMed] [Google Scholar]
- 4.Le, W., Conneely, O. M., Zou, L., He, Y., Saucedo-Cardenas, O., Jankovic, J., Mosier, D. R. & Appel, S. H. (1999) Exp. Neurol. 159 451–458. [DOI] [PubMed] [Google Scholar]
- 5.Smidt, M. P., Asbreuk, C. H., Cox, J. J., Chen, H., Johnson, R. L. & Burbach, J. P. (2000) Nat. Neurosci. 3 337–341. [DOI] [PubMed] [Google Scholar]
- 6.Van Den Munckhof, P., Luk, K. C., Ste-Marie, L., Montgomery, J., Blanchet, P. J., Sadikot, A. F. & Drouin, J. (2003) Development (Cambridge, U.K.) 130 2535–2542. [DOI] [PubMed] [Google Scholar]
- 7.Nunes, I., Tovmasian, L. T., Silva, R. M., Burke, R. E. & Goff, S. P. (2003) Proc. Natl. Acad. Sci. USA 100 4245–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye, W., Shimamura, K., Rubenstein, J. L., Hynes, M. A. & Rosenthal, A. (1998) Cell 93 755–766. [DOI] [PubMed] [Google Scholar]
- 9.Parr, B. A., Shea, M. J., Vassileva, G. & McMahon, A. P. (1993) Development (Cambridge, U.K.) 119 247–261. [DOI] [PubMed] [Google Scholar]
- 10.Taipale, J. & Beachy, P. A. (2001) Nature 411 349–354. [DOI] [PubMed] [Google Scholar]
- 11.Chenn, A. & Walsh, C. A. (2002) Science 297 365–369. [DOI] [PubMed] [Google Scholar]
- 12.Megason, S. G. & McMahon, A. P. (2002) Development (Cambridge, U.K.) 129 2087–2098. [DOI] [PubMed] [Google Scholar]
- 13.Dorsky, R. I., Moon, R. T. & Raible, D. W. (1998) Nature 396 370–373. [DOI] [PubMed] [Google Scholar]
- 14.Baker, J. C., Beddington, R. S. & Harland, R. M. (1999) Genes Dev. 13 3149–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson, S. I., Rydstrom, A., Trimborn, T., Willert, K., Nusse, R., Jessell, T. M. & Edlund, T. (2001) Nature 411 325–330. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Castro, M. I., Marcelle, C. & Bronner-Fraser, M. (2002) Science 297 848–851. [DOI] [PubMed] [Google Scholar]
- 17.Muroyama, Y., Fujihara, M., Ikeya, M., Kondoh, H. & Takada, S. (2002) Genes Dev. 16 548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall, A. C., Lucas, F. R. & Salinas, P. C. (2000) Cell 100 525–535. [DOI] [PubMed] [Google Scholar]
- 19.Patapoutian, A. & Reichardt, L. F. (2000) Curr. Opin. Neurobiol. 10 392–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krylova, O., Herreros, J., Cleverley, K. E., Ehler, E., Henriquez, J. P., Hughes, S. M. & Salinas, P. C. (2002) Neuron 35 1043–1056. [DOI] [PubMed] [Google Scholar]
- 21.Danielian, P. S. & McMahon, A. P. (1996) Nature 383 332–334. [DOI] [PubMed] [Google Scholar]
- 22.McMahon, A. P. & Bradley, A. (1990) Cell 62 1073–1085. [DOI] [PubMed] [Google Scholar]
- 23.Thomas, K. R. & Capecchi, M. R. (1990) Nature 346 847–850. [DOI] [PubMed] [Google Scholar]
- 24.Pinson, K. I., Brennan, J., Monkley, S., Avery, B. J. & Skarnes, W. C. (2000) Nature 407 535–538. [DOI] [PubMed] [Google Scholar]
- 25.Åkerud, P., Canals, J. M., Snyder, E. Y. & Arenas, E. (2001) J. Neurosci. 21 8108–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu, H., Julius, M. A., Giarre, M., Zheng, Z., Brown, A. M. & Kitajewski, J. (1997) Cell Growth Differ. 8 1349–1358. [PubMed] [Google Scholar]
- 27.DasGupta, R. & Fuchs, E. (1999) Development (Cambridge, U.K.) 126 4557–4568. [DOI] [PubMed] [Google Scholar]
- 28.Foster, G. A., Schultzberg, M., Kokfelt, T., Goldstein, M., Hemmings, H. C., Jr., Ouimet, C. C., Walaas, S. I. & Greengard, P. (1988) Int. J. Dev. Neurosci. 6 367–386. [DOI] [PubMed] [Google Scholar]
- 29.Saneyoshi, T., Kume, S., Amasaki, Y. & Mikoshiba, K. (2002) Nature 417 295–299. [DOI] [PubMed] [Google Scholar]
- 30.Tetsu, O. & McCormick, F. (1999) Nature 398 422–426. [DOI] [PubMed] [Google Scholar]
- 31.Shtutman, M., Zhurinsky, J., Simcha, I., Albanese, C., D'Amico, M., Pestell, R. & Ben-Ze'ev, A. (1999) Proc. Natl. Acad. Sci. USA 96 5522–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kioussi, C., Briata, P., Baek, S. H., Rose, D. W., Hamblet, N. S., Herman, T., Ohgi, K. A., Lin, C., Gleiberman, A., Wang, J., et al. (2002) Cell 111 673–685. [DOI] [PubMed] [Google Scholar]
- 33.Castro, D. S., Hermanson, E., Joseph, B., Wallen, A., Aarnisalo, P., Heller, A. & Perlmann, T. (2001) J. Biol. Chem. 276 43277–43284. [DOI] [PubMed] [Google Scholar]
- 34.Reya, T., Duncan, A. W., Ailles, L., Domen, J., Scherer, D. C., Willert, K., Hintz, L., Nusse, R. & Weissman, I. L. (2003) Nature 423 409–414. [DOI] [PubMed] [Google Scholar]
- 35.Hsieh, J. C., Rattner, A., Smallwood, P. M. & Nathans, J. (1999) Proc. Natl. Acad. Sci. USA 96 3546–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dann, C. E., Hsieh, J. C., Rattner, A., Sharma, D., Nathans, J. & Leahy, D. J. (2001) Nature 412 86–90. [DOI] [PubMed] [Google Scholar]
- 37.Smidt, M. P., van Schaick, H. S., Lanctot, C., Tremblay, J. J., Cox, J. J., van der Kleij, A. A., Wolterink, G., Drouin, J. & Burbach, J. P. (1997) Proc. Natl. Acad. Sci. USA. 94 13305–13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baek, S. H., Kioussi, C., Briata, P., Wang, D., Nguyen, H. D., Ohgi, K. A., Glass, C. K., Wynshaw-Boris, A., Rose, D. W. & Rosenfeld, M. G. (2003) Proc. Natl. Acad. Sci. USA 100 3245–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trupp, M., Arenas, E., Fainzilber, M., Nilsson, A. S., Sieber, B. A., Grigoriou, M., Kilkenny, C., Salazar-Grueso, E., Pachnis, V. & Arumae, U. (1996) Nature 381 785–789. [DOI] [PubMed] [Google Scholar]
- 40.Airaksinen, M. S. & Saarma, M. (2002) Nat. Rev. Neurosci. 3 383–394. [DOI] [PubMed] [Google Scholar]
- 41.Paratcha, G., Ledda, F. & Ibanez, C. F. (2003) Cell 113, 867–879. [DOI] [PubMed] [Google Scholar]
- 42.Zheng, S., Ramachandran, B., Haigh, J. R., Palos, T. P., Steger, K. & Howard, B. D. (1996) Oncogene 12 555–562. [PubMed] [Google Scholar]
- 43.Conacci-Sorrell, M. E., Ben-Yedidia, T., Shtutman, M., Feinstein, E., Einat, P. & Ben-Ze'ev, A. (2002) Genes Dev. 16 2058–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Granholm, A. C., Reyland, M., Albeck, D., Sanders, L., Gerhardt, G., Hoernig, G., Shen, L., Westphal, H. & Hoffer, B. (2000) J. Neurosci. 20 3182–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner, J., Åkerud, P., Castro, D. S., Holm, P. C., Canals, J. M., Snyder, E. Y., Perlmann, T. & Arenas, E. (1999) Nat. Biotechnol. 17 653–659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.