Abstract
LTRs of endogenous retroviruses are known to affect expression of several human genes, typically as a relatively minor alternative promoter. Here, we report that an endogenous retrovirus LTR acts as one of at least two alternative promoters for the human β1,3-galactosyltransferase 5 gene, involved in type 1 Lewis antigen synthesis, and show that the LTR promoter is most active in the gastrointestinal tract and mammary gland. Indeed, the LTR is the dominant promoter in the colon, indicating that this ancient retroviral element has a major impact on gene expression. Using colorectal cancer cell lines and electrophoretic mobility-shift assays, we found that hepatocyte nuclear factor 1 (HNF-1) binds a site within the retroviral promoter and that expression of HNF-1 and interaction with its binding site correlated with promoter activation. We conclude that HNF-1 is at least partially responsible for the tissue-specific activation of the LTR promoter of human β1,3-galactosyltransferase 5. We demonstrate that this tissue-specific transcription factor is implicated in the activation of an LTR gene promoter.
Approximately 8% of the human genome is made up of human endogenous retrovirus (HERV) and related sequences (1). Although a few HERVs retain the potential to encode viral proteins (for review see refs. 2 and 3), most HERV ORFs have been degraded by mutation. The majority of HERVs are devoid of all coding regions, having undergone recombination between the two flanking LTRs to produce the elements known as solitary LTRs. In contrast to retroviral coding regions, many HERV LTRs retain functional potential. LTRs contain the viral promoter, enhancer, and polyadenylation signals and can regulate gene expression. Indeed, several reports have demonstrated that LTRs can regulate nearby native (nonretroviral) human genes. In the majority of well-documented cases, the LTR acts as a gene promoter, often as one of multiple alternative promoters for the same gene (4–7).
Many LTR-derived promoters are active primarily in the placenta, such as those of the human endothelin B receptor (EDNRB), Mid1, insulin-like peptide INSL4, pleiotrophin, and aromatase (CYP19) genes (5–9). Other LTRs have different tissue specificities, for example, driving the expression of apolipoprotein C-I and alcohol dehydrogenase 1C in liver cells (5, 10) or of Mid1 in fetal kidney (9). A putative binding site for the placental transcription factor Hand1/E47 was identified in the EDNRB LTR promoter and was shown to be important for placenta-specific promoter activity, but interaction of Hand1/ E47 with this site could not be confirmed (11). In other studies, only ubiquitous transcription factors have been confirmed to bind LTR gene promoters (10, 12). Therefore, the transcription factors that mediate tissue-specific gene expression from LTR promoters remain largely unknown.
Through database searches, we found that the first exon of the β1,3-galactosyltransferase 5 (β3Gal-T5) gene is composed of a HERV LTR sequence. β3Gal-T5 is a member of a family of at least six enzymes, each with a different tissue and substrate specificity (13–18). β3Gal-T5 expression and activity are highest in tissues that produce high levels of carbohydrate sialyl Lewis a antigens upon carcinogenesis, such as the colon, stomach, pancreas, and small intestine (15). β3Gal-T5 is also thought to be responsible for the synthesis of type 1 Lewis antigens in some colorectal and pancreatic cancer cell lines (15, 16). This may be clinically important, as sialyl Lewis a antigen forms an antigenic epitope on the CA19–9 serum marker used to diagnose colorectal, gastric, and pancreatic cancers and may be involved in tumor metastasis (for review, see ref. 19). However, β3Gal-T5 expression and type 1 Lewis antigen synthesis are decreased in colon adenocarcinomas (16). Any relationship between β3Gal-T5 activity and carcinogenesis is clearly complex and may be different at the various stages of tumor development and metastasis.
In this study, we demonstrate that a HERV LTR acts as an alternative tissue-specific promoter for the human β3Gal-T5 gene and is the major promoter for this gene in the colon. Furthermore, we identify a functional binding site for hepatocyte nuclear factor 1 (HNF-1) within the LTR.
Experimental Procedures
Computational Methods. The human RefSeq database (www.ncbi.nlm.nih.gov/RefSeq) was screened for genes that contain transposable elements within their UTRs as described (7). Candidate gene loci were examined by using the University of California, Santa Cruz genome browser (20). Determination of intron–exon boundaries was carried out by using the spidey alignment program (21). Putative transcription factor binding sites were identified by using the TRANSFAC transcription factor database (ref. 22; www.cbil.upenn.edu/tess).
Reverse Transcription, PCR Amplification, and Southern Blotting. Total RNA samples were obtained from Clontech or prepared from cultured cells by using TRIzol (Invitrogen) according to the manufacturer's instructions. Five micrograms of each RNA was treated with DNase I and reverse-transcribed as described (23). RT-PCR was carried out by using 2 ng/μl of each primer in 4 mM MgCl2. All primer sequences are available on request. For Southern blotting, an agarose gel was blotted onto Zeta-Probe GT blotting membrane (Bio-Rad) in 10× SSC. Twenty nanograms of oligonucleotide probe specific for β3Gal-T5 exon 4 was radiolabeled and hybridized to the filter overnight at 61°C in 6× SSC/0.5% SDS/1× Denhardt's solution, and washed three times 10 min in 3× SSC. 5′ RACE. 5′ RACE was carried out by using the FirstChoice RLM-RACE kit (Ambion). Briefly, 10 μg of total brain RNA was treated with calf intestinal phosphatase and tobacco acid pyrophosphatase to ligate an RNA adapter only to the 5′ cap structure of full-length mRNAs. The RNA was reversetranscribed, and two rounds of nested PCR were carried out by using forward primers specific for the 5′-RNA adapter and reverse primers specific for β3Gal-T5 exon 4. The single specific amplification product was cloned into the pGEM-T vector (Promega), and five independent clones were sequenced.
Real-Time RT-PCR. PCR was carried out by using 2 ng/μl of each primer in SYBR green PCR master mix (Applied Biosystems) with 4 mM MgCl2. Reaction conditions on a Bio-Rad iCycler were as follows: initial step 95°C, 3 min, 50 cycles of 95°C, 30 s; 53°C, 30 s; 72°C, 30 s. Dissociation curves were run and demonstrated that each primer pair amplified a single product. Serial dilutions of colon cDNA were used to prepare standard curves for each primer pair, and the relative abundance of the target transcripts in each cDNA was calculated. Levels of total β3Gal-T5 transcripts were normalized to GAPDH and expressed relative to levels detected in kidney. Levels of β3Gal-T5 LTR transcripts were divided by levels of total transcripts. This value was then multiplied by the GAPDH- and kidney-normalized level of total transcripts to determine the contribution of the LTR promoter to total β3Gal-T5 expression.
Plasmid Constructs. ERV-L LTR nucleotides 1–522 were cloned from human genomic DNA by using primers containing terminal restriction sites. The product was digested and cloned into the promoterless luciferase reporter vector pGL3B (Promega). Sequentially 5′ deleted products were amplified by using forward primers based on internal LTR sequences together with the original reverse primer. Putative HNF-1 binding site 1 was disrupted by using a mutagenic forward primer with the original reverse primer. HNF-1 site 2 was mutated by a two-step PCR protocol. A mutagenic reverse primer was used in the first step with the original forward primer, and the resulting product was then used as the forward primer together with the original reverse primer. The identity of all reporter constructs was verified by DNA sequencing of the insert.
Cell Lines and Transient Transfections. The human colorectal cancer cell lines DLD-1 (ATCC CCL-221) and LoVo (ATCC CCL-229) were cultured in MEM, alpha modification with nucleosides supplemented with 10% FCS. Cells were seeded in six-well plates at a density of 2 × 105 cells per well and transfected in duplicate after 24 h with 1 μg of reporter construct DNA and 0.2 μg of pRL-TK vector (Promega) by using 7 μl of Lipofectamine reagent (Invitrogen). Cells were lysed after 24 h, and luciferase activities were measured by using the Dual-Luciferase Reporter Assay System (Promega). Luciferase activities were normalized to the pRL-TK internal control and expressed as fold-activation over the pGL3B promoterless vector.
Electrophoretic Mobility-Shift Assays. Nuclear extracts were prepared from DLD-1 and LoVo cells essentially as described (24). Sense-strand oligonucleotide sequences were as follows: putative HNF-1 site 1, WT, 5′-GTGATGGTTACTTTTAGGTGTCAAC; putative HNF-1 site 2, WT, 5′-TGGCTGGATTAATAAATACCTAGAG; and putative HNF-1 site 2, mutated, 5′-TGGCTGTCTTGATACATACCTAGAG (mutated bases in bold). A sample of nuclear extract containing 5 μg of protein was added to 0.5 μg of poly(dI·dC) in 10 mM Hepes, 4 mM DTT, 0.2 mM EDTA, 100 nM NaCl, 0.1 mg/ml BSA, 4% glycerol. Samples were incubated for 20 min on ice with 100-fold molar excess of unlabeled competitor oligonucleotide or an antibody specific for HNF-1 (H-205X, Santa Cruz Biotechnology). Samples were then incubated for 30 min on ice with 1 ng of radiolabeled oligonucleotide probe. PAGE was carried out at 4°C; gels were fixed for 30 min in 10% acetic acid/10% methanol and dried under vacuum.
Results
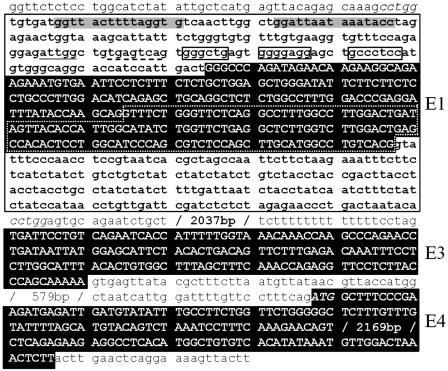
Exon 1 of Human β3Gal-T5 Is Derived from an Endogenous Retroviral LTR. A screen to identify genes containing HERVs within their UTRs revealed that the first exon of the β3Gal-T5 gene is derived from a HERV (Fig. 1). The HERV element is a 650-bp solitary LTR, type MLT2B3, of the ERV-L family and is in the same orientation as the gene. ERV-L is one of the older ERV families, present in the genomes of all placental mammals (25, 26). The β3Gal-T5 gene originally cloned from the Colo205 colorectal cancer cell line has five splicing variants, all with a common transcriptional start site within the LTR and an ORF derived solely from exon 4 (15). The two major splicing variants differ at the 3′ end of exon 1 because of the presence of two alternative splice donor sites, at LTR nucleotide positions 448 (variant 1) and 315 (variant 2, see Fig. 1). The transcriptional start site, as determined by 5′ RACE (15), maps to nucleotide 175 of the ERV-L LTR, suggesting that the LTR acts as a promoter for the β3Gal-T5 gene.
Fig. 1.
Exon 1 of β3Gal-T5 is derived from an endogenous retroviral LTR. Exon (E) sequences are shown in uppercase reverse type, with all other sequences in lowercase. The translation initiation codon at the beginning of exon 4 and the 5-bp direct repeats flanking the LTR are shown in italics. The ERV-L LTR sequence is shown in bold and framed with a solid black line. Dotted white lines frame the portion of exon 1 included in splicing variant 1, but not variant 2; three minor splicing variants, which contain additional exon sequences, are not shown (see ref. 15). Gray boxes, putative HNF-1 binding sites; underlined text, putative CCAAT box; dashed underlined text, putative Jun binding site; unfilled boxes, putative Sp1 binding sites.
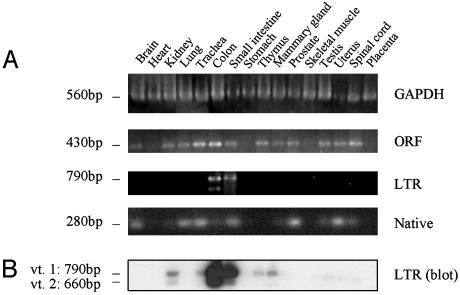
Expression Patterns of β3Gal-T5 Transcripts. Many genes that use a HERV LTR as a promoter also have an additional, native gene promoter(s) (5–7, 9). To determine whether this is also the case for β3Gal-T5, we compared the expression patterns of β3Gal-T5 transcripts containing coding and LTR sequences in normal human tissues by using RT-PCR (Fig. 2A). Total β3Gal-T5 transcripts were detected by using primers specific to the exon 4 ORF. Transcripts were detected in all tissues with the exception of the heart, stomach, skeletal muscle, and placenta (Fig. 2 A). β3Gal-T5 transcripts initiating within the LTR were detected by using primers specific for the LTR-derived exon 1 and exon 4. In contrast to total β3Gal-T5 transcripts, LTR-driven transcripts were detected only in the colon and small intestine (Fig. 2 A). The major splicing variant 1, at 790 bp, and the smaller variant 2 were both detected. However, in contrast to a previous report that these two variants are expressed in equal amounts (15), variant 1 was the dominant isoform in this analysis. This may be caused by the different methods used or represent a difference between normal human tissues and the cell lines used in the previous study. The gel shown in Fig. 2 A was also Southern-blotted and probed with an oligonucleotide specific for β3Gal-T5 exon 4 to enable detection of low levels of LTR-driven transcripts. As shown in Fig. 2B, transcripts initiating from the LTR were detected in several tissues as well as in colon and small intestine. However, the expression pattern of β3Gal-T5 transcripts containing ORF sequences remained broader than that of LTR-containing transcripts. This suggests that β3Gal-T5 has at least one additional promoter with a different tissue specificity to the LTR.
Fig. 2.
Detection of β3Gal-T5 transcripts initiating within the LTR and native promoters. (A) Total cDNAs derived from a range of normal human tissues were used in RT-PCR assays, with primers specific for GAPDH, the β3Gal-T5 ORF, and β3Gal-T5 transcripts initiating within the LTR or native promoter. Approximate molecular weights are indicated on the left. (B) Southern blot of the gel shown in the third panel in A. The blot was probed with a labeled oligonucleotide specific for β3Gal-T5 exon 4.
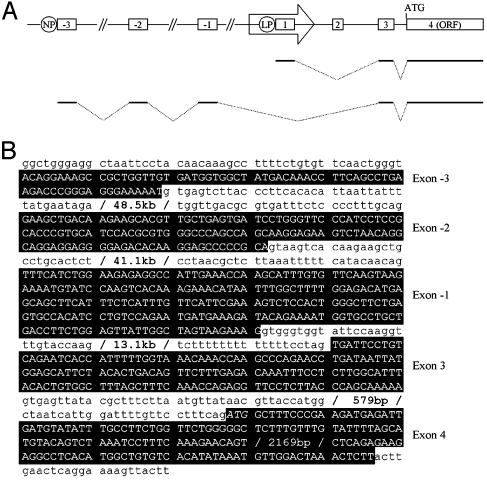
Identification of a Native Promoter for β3Gal-T5. To identify additional promoters, 5′ RACE was carried out on cDNA derived from the brain, which was shown to express transcripts containing sequences from the β3Gal-T5 ORF but not the LTR (Fig. 2). A single specific product was obtained, and the sequence of five clones was determined. The final two exons of the sequenced transcript (exons 3 and 4) are identical to those of transcripts initiating within the LTR (Fig. 3). Three upstream exons were also identified: a first exon (named exon –3) of 69 bp, a second exon (–2) of 132 bp, and a third exon (–1) of 231 bp. All intron–exon boundaries conformed to the GT/AG rule of splice donor and acceptor sites. The exons did not contain any HERV sequences or long ORFs, suggesting that transcripts initiating from the LTR and native promoters encode identical proteins from the ORF within exon 4. As shown in Fig. 2 A Lower, transcripts initiating within the native promoter were detected in the brain, lung, trachea, small intestine, mammary gland, prostate, testis, uterus, and spinal cord. LTR and/or native promoter-driven transcripts were therefore detected in all tissues where β3Gal-T5 coding sequences were present, suggesting that these two promoters may account for all β3Gal-T5 expression. However, as the analysis shown here is not quantitative, the presence of further alternative promoters for β3Gal-T5 cannot be ruled out.
Fig. 3.
Genomic structure and nucleotide sequence of transcripts initiating from the native promoter of β3Gal-T5. (A) Structure of the β3Gal-T5 locus. Exons are boxed and numbered. The ERV-L LTR is represented by an arrow. β3Gal-T5 transcripts initiating from the native promoter (NP) and LTR promoter (LP-splicing variant 1 only) are shown schematically below. The diagram is not to scale. (B) Nucleotide sequence of transcripts initiating from the β3Gal-T5 native promoter. Exon sequences are shown in uppercase reverse type, with all other sequences in lowercase. The translation initiation codon is shown in italics.
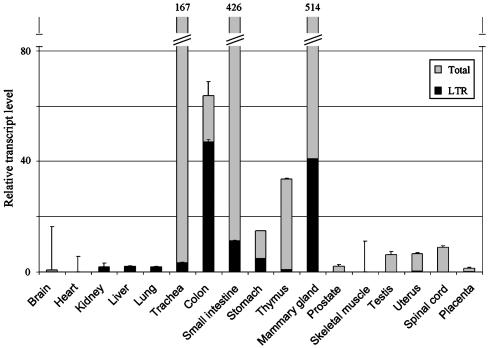
Quantitative Analysis of β3Gal-T5 Expression and LTR Promoter Activity in Human Tissues. Real-time RT-PCR was used to determine the relative levels of β3Gal-T5 transcription and LTR promoter activity in normal human tissues (Fig. 4). Primers annealing to exon 3 and exon 4, common to all β3Gal-T5 transcripts, were used to determine the level of total gene expression. This value was normalized to GAPDH and expressed relative to that obtained for the kidney, arbitrarily chosen on the basis of its low level of β3Gal-T5 expression. Levels of transcripts initiating from the LTR were determined by using exon 1 and exon 3 primers. The contribution of the LTR promoter to total β3Gal-T5 expression was calculated for each tissue (see Experimental Procedures). As the forward primer was designed to anneal to the portion of exon 1 contained in splicing variant 1, but not the less abundant variant 2 (see Fig. 1), the values obtained are an underestimate. As shown in Fig. 4, β3Gal-T5 expression was highest in the mammary gland, small intestine, trachea, colon, thymus, and stomach. Low levels of expression were also detected in a range of other tissues. The LTR promoter had a distinct tissue specificity, with highest activity in the colon, mammary gland, small intestine, and stomach. LTR transcripts were also detected at lower levels in the kidney, liver, lung, trachea, thymus, and uterus. Of the tissues with high overall levels of β3Gal-T5, the LTR promoter made the highest contribution to gene expression in the colon (74% of total transcripts). These results are broadly consistent with those obtained with RT-PCR (Fig. 2), with the exception that β3Gal-T5 transcripts were not detected in the stomach by RT-PCR. The real-time PCR results shown in Fig. 4 are consistent with previous reports of the expression pattern of β3Gal-T5 (15, 18).
Fig. 4.
Contribution of the LTR promoter to β3Gal-T5 expression in human tissues. Primers were used in real-time PCR assays to amplify transcripts specific for GAPDH, total β3Gal-T5, and β3Gal-T5 transcripts initiating within the LTR promoter from cDNAs derived from normal human tissues. Gray bars represent the relative abundance of total β3Gal-T5 transcripts normalized to GAPDH levels ± SEM. Black bars depict the contribution of the LTR promoter to total β3Gal-T5 transcription ± SEM. In some cases, the error bars are too small to see. Relative levels of transcripts are indicated above the bars where necessary. Assays were carried out in duplicate and repeated twice.
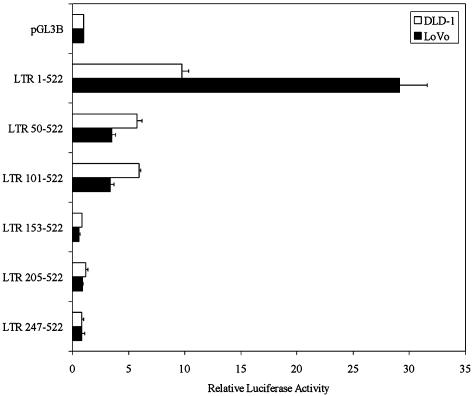
Functional Analysis of the β3Gal-T5 LTR Promoter. As LTR promoter activity was highest in the colon (Fig. 4), we chose to analyze the β3Gal-T5 LTR promoter in human colorectal cancer cell lines. High levels of β3Gal-T5 have been reported in some, but not all, of such cell lines (15, 18). Using RT-PCR assays as before, total β3Gal-T5 transcripts were detected in both the DLD-1 and LoVo colorectal cancer cell lines, whereas LTR transcripts were detected only in LoVo cells (data not shown). Plasmids containing LTR nucleotides 1–522, which includes all of β3Gal-T5 exon 1, and 5′ deleted derivatives were cloned upstream of a luciferase reporter gene. These constructs were transiently transfected into DLD-1 and LoVo cells. The longest LTR construct displayed high levels of promoter activity in both cell lines (Fig. 5). However, promoter activity was ≈3-fold higher in LoVo cells than in DLD-1. Deletion of the first 49 bp of the LTR reduced promoter activity in both cell lines, but to a much greater degree in LoVo cells. This finding suggests that the transcription factor(s) responsible for the high specific activity of the LTR promoter in LoVo cells bind within this region. Deletion of a further 51 bp from the 5′ end of the LTR had no effect on promoter activity. However, deletion of the region from nucleotides 101–152 of the LTR reduced promoter activity to background levels in both cell types. This finding suggests that transcription factors responsible for the low-level basal activity of the promoter bind this region. Continued 5′ deletion of the LTR did not further decrease promoter activity.
Fig. 5.
5′ deletion analysis of the LTR promoter in human colorectal cancer cell lines. ERV-L LTR nucleotides 1–522 and five sequentially 5′ deleted LTRs were cloned into the pGL3B luciferase reporter vector. Plasmids were transiently transfected into DLD-1 and LoVo cells. Luciferase activities are shown relative to the internal control and then to pGL3B to demonstrate fold activation. Bars represent the mean of four independent transfections ± SEM.
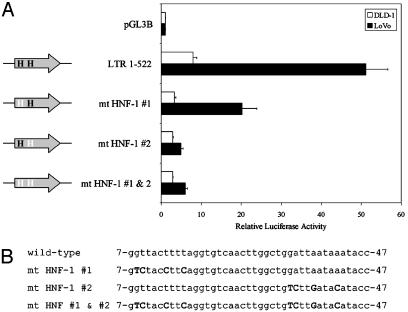
LTR positions 1–49 and 101–152 were screened for putative transcription factor binding sites. Two predicted sites for HNF-1 were identified at nucleotide positions 7 and 33; these sequences were named HNF-1 sites 1 and 2, respectively (see Fig. 1 for position and sequence of all putative binding sites). HNF-1 is expressed in tissues where the LTR promoter is active, including the intestine, stomach, kidney, liver, and thymus (27). HNF-1 therefore represents an excellent candidate transcription factor responsible for tissue-specific activation of the LTR promoter. The LTR sequence from nucleotide positions 101–152 was found to contain a consensus reverse-orientation CCAAT box, a consensus Jun binding site, and three G/C-rich regions (sites 1, 2, and 3), which may act as Sp1 binding sites. These factors are all ubiquitously expressed and are good candidates for the low-level basal activation of the LTR promoter.
The two putative HNF-1 binding sites were disrupted by site-directed mutagenesis to introduce mutations predicted to prevent factor binding (28). Disruption of site 1 decreased promoter activity by ≈2.5-fold in both DLD-1 and LoVo cells (Fig. 6), whereas mutation of site 2 caused a decrease of ≈2.7-fold in DLD-1 cells and >10-fold in LoVo. Simultaneous disruption of both sites did not cause a further decrease in promoter activity. These results suggest that whereas both regions identified as potential HNF-1 binding sites are involved in LTR promoter activity, only site 2 is important for the high LTR promoter activity specifically observed in LoVo cells.
Fig. 6.
Site-directed mutagenesis of putative HNF-1 binding sites within the LTR. (A) Mutation of two putative HNF-1 binding sites within the LTR 1–522 reporter construct. Plasmids were transiently transfected into DLD-1 and LoVo cells. Luciferase activities are shown as fold activation as before. Bars represent the mean of four independent transfections ± SEM. H, putative HNF-1 binding site. White type represents mutation of the putative binding site. (B) The sequence of LTR nucleotides 7–47 in each construct is indicated, with mutated nucleotides in uppercase bold type.
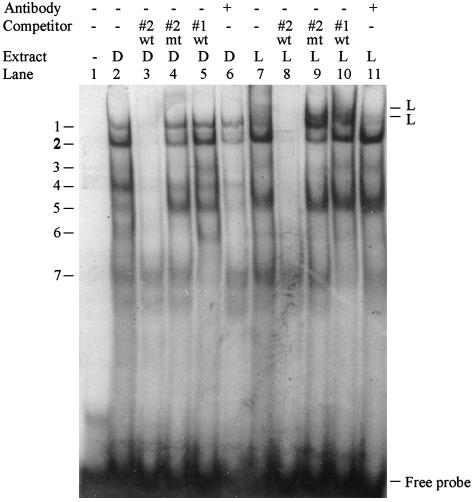
Identification of Transcription Factors Binding to the β3Gal-T5 LTR Promoter. Electrophoretic mobility-shift assays were used to identify the transcription factors binding to the β3Gal-T5 LTR promoter. Only HNF-1 site 2 was important for specific activation of the LTR promoter in LoVo cells (Fig. 6) so a double-stranded oligonucleotide containing this site was used as the probe. Multiple complexes bound the probe in DLD-1 and LoVo cells (Fig. 7, lanes 2 and 7, bands labeled 1–7). Additional bands were observed specifically in LoVo extracts (Fig. 7, lane 7, bands labeled L). Formation of bands 1–6 and the LoVo-specific bands was prevented by preincubation with an excess of unlabeled WT competitor oligonucleotide in both cell types (Fig. 7, lanes 3 and 8). However, oligonucleotides containing the mutated site 2 did not compete for these complexes (Fig. 7, lanes 4 and 9), indicating that they bound specifically to HNF-1 binding site 2. It is important to note that this competitor oligonucleotide contained the same mutation that decreased promoter activity specifically in LoVo cells (see Fig. 6). Band 7 was not disrupted by the WT competitor, suggesting that this complex bound the probe in a nonspecific manner. Addition of an HNF-1 antibody did not affect formation of any of the specific complexes seen in DLD-1 extracts (Fig. 7, lane 6; bands 1–6 all appear less intense, but no change in the relative intensities of the bands was observed). In contrast, addition of HNF-1 antibody to LoVo extracts specifically decreased the intensity of band 1 and the two LoVo-specific bands (Fig. 7, lane 11). The HNF-1 antibody used in this assay was previously shown to prevent protein–DNA complex formation rather than cause a supershift effect (29). HNF-1 therefore interacts with its predicted binding site 2 specifically in LoVo cells.
Fig. 7.
HNF-1 binds the LTR promoter specifically in the LoVo cell line. A radiolabeled oligonucleotide containing putative HNF-1 binding site 2 was incubated with nuclear extracts containing 5 μg of protein from DLD-1 (D) or LoVo (L) cells. Extracts were preincubated with a 100-fold molar excess of WT or mutated competitor oligonucleotides, or with an antibody reactive with HNF-1, as indicated.
None of the bands observed in DLD-1 and LoVo extracts were competed by an oligonucleotide containing putative HNF-1 binding site 1 (Fig. 7, lanes 5 and 10). Unlike site 2, therefore, this site is not bound by HNF-1. This finding is consistent with the different effects of mutation of the two sites on LoVo-specific promoter activity (Fig. 6).
We also determined that Sp1 bound its predicted binding site 2, but not 1 and 3, whereas the putative CCAAT box was bound by NF-YA. Mutations in these sites prevented transcription factor binding and decreased LTR promoter activity in DLD-1 and LoVo cells, whereas mutation of the putative Jun binding site had no significant effect on transcription (data not shown).
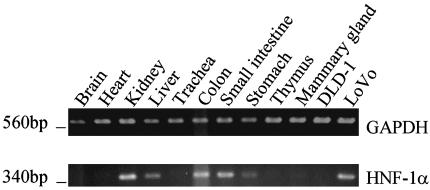
Detection of HNF-1α Transcripts in Human Tissues and Colorectal Cancer Cell Lines. As described above, HNF-1 is known to be expressed in some tissues where the β3Gal-T5 LTR promoter is active (27). However, expression of the factor in other tissues and in colorectal cancer cell lines has not been well characterized. RT-PCR was therefore carried out on RNA derived from normal human tissues, DLD-1, and LoVo cells, using primers specific for the HNF-1α isoform (Fig. 8). HNF-1α transcripts were detected in the kidney, liver, colon, small intestine, and stomach, as expected. However, no transcripts were detected in the thymus, trachea, or mammary gland, where the LTR promoter is also active (see Figs. 2 and 4). HNF-1α transcripts were also absent from the brain and heart, where the LTR promoter is not activated. HNF-1α expression was detected in LoVo cells, but not in DLD-1. Expression of HNF-1 therefore correlates with promoter binding by the transcription factor and with high levels of LTR promoter activation specifically in the LoVo cell line (see Figs. 6 and 7). This finding supports the conclusion that HNF-1 is at least partially responsible for tissue-specific activation of the β3Gal-T5 LTR promoter.
Fig. 8.
Detection of HNF-1α transcripts in human tissues and colorectal cancer cell lines. Total cDNAs derived from normal human tissues and the DLD-1 and LoVo cell lines were used in RT-PCR, with primers specific for GAPDH (Upper) or HNF-1α (Lower). Approximate molecular weights are indicated on the left.
Discussion
In this study, we show that an ERV-L LTR serves as one of at least two alternative promoters for the β3Gal-T5 gene, with highest activity in the gastrointestinal tract and mammary gland. Our demonstration that LTR promoter activity in colorectal cancer cell lines correlates with expression and promoter binding by HNF-1 identifies a tissue-specific transcription factor involved in gene promotion by an ERV LTR. The correlation between HNF-1 expression and LTR promoter activity is not perfect, however. For example, HNF-1 is expressed in the kidney and liver (Fig. 8; see also ref. 27). Whereas the LTR promoter was active at low levels in these tissues, overall levels of β3Gal-T5 expression were extremely low (Fig. 4). It is possible that mechanisms such as LTR methylation prevent promoter activation by HNF-1 in these tissues, although this seems unlikely because of the low G/C content of the LTR in this case. Alternatively, a tissue-specific silencer or repressor element may also be involved in regulation of the β3Gal-T5 LTR promoter. More work is required to determine whether any such element is located within the LTR or surrounding genomic DNA.
HNF-1 expression was not detected in the mammary gland, thymus, or trachea, although the LTR promoter was active in these tissues (Figs. 4 and 8). This may represent a shortcoming of the RT-PCR assay used, as HNF-1 expression has been detected in the thymus (27). However, it is more likely that transcription factors other than HNF-1 activate the LTR promoter in these tissues. As shown in Fig. 7, various non-HNF-1 protein complexes specifically bound WT HNF-1 site 2. Because these complexes were present in both DLD-1 and LoVo cells, any transcriptional effect of protein binding may be nonspecific. However, these proteins may alter the ability of HNF-1 or other proteins to activate the LTR promoter in certain tissues. In addition, whereas putative binding site 1 did not bind HNF-1, mutation of this site decreased promoter activity (Figs. 6 and 7). This finding suggests that proteins not yet identified may bind this site, although no other predicted transcription factor binding site was identified in this region.
Although further work remains to be done to identify the DNA elements and transcription factors responsible for regulation of the β3Gal-T5 LTR promoter, the correlation between HNF-1 expression and promoter activity is robust in tissues and cell lines of gastrointestinal origin. This finding is consistent with a recent report, published after completion of this study, which confirmed that HNF-1 site 2 and an overlapping Cdx binding site are responsible for β3Gal-T5 expression in colorectal cancer cell lines (30). This report also demonstrated that decreased HNF-1 and Cdx expression correlated with the decreased level of β3Gal-T5 expression in colorectal tumors.
The mechanism of LTR promoter regulation is of particular interest in the colon, where the majority (74%) of β3Gal-T5 transcripts are driven by the LTR. This is highly unusual compared with other reported LTR-promoted genes such as apolipoprotein C-I, EDNRB, and Mid1, where the LTR contributes a maximum of 15%, 30%, and 38% of total transcripts, respectively, in selected tissues (5, 9). As an increasingly large number of LTR gene promoters are being identified (7, 31), it seems clear that LTR elements are not always deleterious to the organism and, in fact, may enhance the range of transcriptional regulatory signals available to a nearby gene. In the case of β3Gal-T5, the ERV-L LTR is deeply fixed in the primate genome as it is found in higher apes and Old World monkeys (unpublished observations). Thus, this element has been retained over millions of years and now plays a major role in expression of β3Gal-T5, particularly in the large intestine. This is an intriguing example of adoption of an ancient retro-element for usage by the host.
Acknowledgments
We thank Dr. Josette Landry for helpful advice and discussions. This work was supported by a grant from the Canadian Institutes of Health Research (to D.L.M.) with core support provided by the BC Cancer Agency. P.M. was supported by a fellowship from the Knut and Alice Wallenberg Foundation and the Nilsson-Ehle Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: β3Gal-T5, β1,3-galactosyltransferase 5; HERV, human endogenous retrovirus; HNF-1, hepatocyte nuclear factor 1.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY372061).
References
- 1.International Human Genome Sequencing Consortium (2001) Nature 409 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Bock, M. & Stoye, J. P. (2000) Curr. Opin. Genet. Dev. 10 651–655. [DOI] [PubMed] [Google Scholar]
- 3.Stoye, J. P. (2001) Curr. Biol. 11 R914–R916. [DOI] [PubMed] [Google Scholar]
- 4.Landry, J. R. & Mager, D. L. (2002) Genomics 80 499–508. [PubMed] [Google Scholar]
- 5.Medstrand, P., Landry, J. R. & Mager, D. L. (2001) J. Biol. Chem. 276 1896–1903. [DOI] [PubMed] [Google Scholar]
- 6.Schulte, A. M., Lai, S., Kurtz, A., Czubayko, F., Riegel, A. T. & Wellstein, A. (1996) Proc. Natl. Acad. Sci. USA 93 14759–14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van de Lagemaat, L. N., Landry, J. R., Mager, D. L. & Medstrand, P. (2003) Trends Genet. 19 530–536. [DOI] [PubMed] [Google Scholar]
- 8.Bieche, I., Laurent, A., Laurendeau, I., Duret, L., Giovangrandi, Y., Frendo, J. L., Olivi, M., Fausser, J. L., Evain-Brion, D. & Vidaud, M. (2003) Biol. Reprod. 68 1422–1429. [DOI] [PubMed] [Google Scholar]
- 9.Landry, J. R., Rouhi, A., Medstrand, P. & Mager, D. L. (2002) Mol. Biol. Evol. 19 1934–1942. [DOI] [PubMed] [Google Scholar]
- 10.Chen, H. J., Carr, K., Jerome, R. E. & Edenberg, H. J. (2002) DNA Cell Biol. 21 793–801. [DOI] [PubMed] [Google Scholar]
- 11.Landry, J. R. & Mager, D. L. (2003) J. Virol. 77 7459–7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulte, A. M., Malerczyk, C., Cabal-Manzano, R., Gajarsa, J. J., List, H. J., Riegel, A. T. & Wellstein, A. (2000) Oncogene 19 3988–3998. [DOI] [PubMed] [Google Scholar]
- 13.Amado, M., Almeida, R., Carneiro, F., Levery, S. B., Holmes, E. H., Nomoto, M., Hollingsworth, M. A., Hassan, H., Schwientek, T., Nielsen, P. A., et al. (1998) J. Biol. Chem. 273 12770–12778. [DOI] [PubMed] [Google Scholar]
- 14.Cole, S. E., Mao, M. S., Johnston, S. H. & Vogt, T. F. (2001) Mamm. Genome 12 177–179. [DOI] [PubMed] [Google Scholar]
- 15.Isshiki, S., Togayachi, A., Kudo, T., Nishihara, S., Watanabe, M., Kubota, T., Kitajima, M., Shiraishi, N., Sasaki, K., Andoh, T. & Narimatsu, H. (1999) J. Biol. Chem. 274 12499–12507. [DOI] [PubMed] [Google Scholar]
- 16.Salvini, R., Bardoni, A., Valli, M. & Trinchera, M. (2001) J. Biol. Chem. 276 3564–3573. [DOI] [PubMed] [Google Scholar]
- 17.Zhou, D., Henion, T. R., Jungalwala, F. B., Berger, E. G. & Hennet, T. (2000) J. Biol. Chem. 275 22631–22634. [DOI] [PubMed] [Google Scholar]
- 18.Zhou, D., Berger, E. G. & Hennet, T. (1999) Eur. J. Biochem. 263 571–576. [DOI] [PubMed] [Google Scholar]
- 19.Thurin, M. & Kieber-Emmons, T. (2002) Hybrid Hybridomics 21 111–116. [DOI] [PubMed] [Google Scholar]
- 20.Kent, W. J., Sugnet, C. W., Furey, T. S., Roskin, K. M., Pringle, T. H., Zahler, A. M. & Haussler, D. (2002) Genome Res. 12 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wheelan, S. J., Church, D. M. & Ostell, J. M. (2001) Genome Res. 11 1952–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wingender, E., Chen, X., Hehl, R., Karas, H., Liebich, I., Matys, V., Meinhardt, T., Pruss, M., Reuter, I. & Schacherer, F. (2000) Nucleic Acids Res. 28 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medstrand, P., Lindeskog, M. & Blomberg, J. (1992) J. Gen. Virol. 73 2463–2466. [DOI] [PubMed] [Google Scholar]
- 24.Shao, J., Fujiwara, T., Kadowaki, Y., Fukazawa, T., Waku, T., Itoshima, T., Yamatsuji, T., Nishizaki, M., Roth, J. A. & Tanaka, N. (2000) Oncogene 19 726–736. [DOI] [PubMed] [Google Scholar]
- 25.Benit, L., Lallemand, J. B., Casella, J. F., Philippe, H. & Heidmann, T. (1999) J. Virol. 73 3301–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cordonnier, A., Casella, J. F. & Heidmann, T. (1995) J. Virol. 69 5890–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendel, D. B. & Crabtree, G. R. (1991) J. Biol. Chem. 266 677–680. [PubMed] [Google Scholar]
- 28.Tronche, F., Ringeisen, F., Blumenfeld, M., Yaniv, M. & Pontoglio, M. (1997) J. Mol. Biol. 266 231–245. [DOI] [PubMed] [Google Scholar]
- 29.Shah, R. N., Ibbitt, J. C., Alitalo, K. & Hurst, H. C. (2002) Oncogene 21 8251–8261. [DOI] [PubMed] [Google Scholar]
- 30.Isshiki, S., Kudo, T., Nishihara, S., Ikehara, Y., Togayachi, A., Furuya, A., Shitara, K., Kubota, T., Watanabe, M., Kitajima, M. & Narimatsu, H. (2003) J. Biol. Chem. 278 36611–36620. [DOI] [PubMed] [Google Scholar]
- 31.Jordan, I. K., Rogozin, I. B., Glazko, G. V. & Koonin, E. V. (2003) Trends Genet. 19 68–72. [DOI] [PubMed] [Google Scholar]