Abstract
p53, the most commonly mutated gene in human tumors, is believed to play a crucial role in the prevention of cancer by protecting cells from mutation, a theory commonly known as the “Guardian of the Genome” hypothesis. There are two hypotheses as to how this can occur. In the first, p53 protects the genome by retarding the cell cycle, thus allowing more time for DNA repair. In the second, p53 reduces cancer by initiating apoptosis in damaged cells, thus making it impossible for these cells to become carcinogenic. This study directly tested these two theories in primary murine embryonic fibroblasts on a common genetic background with and without p53, using a lacI transgene as a mutational target. The data demonstrate that, as a direct consequence of cell cycle delay, p53 slowed the induction of mutations and decreased their frequency but had little effect on the frequency of apoptosis. This indicates that the function of p53 in cell cycle control is more important than the role of p53 in apoptosis, for mutation prevention, in any uniform cell population. Moreover, p53-mediated protection is further improved in slowly dividing cells, suggesting that p53 may be particularly important in protecting stem cells from mutation. The role of apoptosis in vivo, however, may be to remove whole tissue subpopulations that can be renewed by less sensitive stem cells.
Keywords: cell cycle, carcinogenesis, lacI
Functional mutations in the p53 tumor suppressor gene have been documented in diverse tumor types (1–3), establishing p53 as the most frequent gene mutation found in cancer (4). The fundamental role of p53 in controlling functions such as G1/S transition, DNA damage repair, and apoptosis is well established. In models that describe tumorigenesis as a multistep process, p53 is thought to suppress the onset of carcinogenesis by decreasing the somatic mutation rate. This reduction in mutation is likely to be achieved by one or both of the following mechanisms. The first involves the activation of apoptosis by means of p53 after DNA damage (5), selectively removing severely damaged cells from the cell population (6). In the second mechanism, p53 prevents genomic instability by initiating cell cycle arrest, thus allowing the repair of damaged DNA before DNA synthesis (7, 8). Although many researchers have been able to document an elevation in mutation frequency in p53-deficient cells (9–13), others have not (14, 15), and the relationships among p53, apoptosis, cell proliferation, and mutagenesis have remained uncertain.
To investigate the role of p53 in mutagenesis, murine cell strains were established from mice with a p53-null mutation, which was introduced via homologous recombination (16). Cells with this knockout lack both p53 mRNA and P53 protein (16). p53-nullizygous mice were bred with Big Blue mice (Stratagene). Subsequently, a cross between the resultant F1 animals generated the embryos from which the cell strains were established. Big Blue mice harbor a λ shuttle vector that is 45 kb in size and contains the lacI and αlacZ genes (LIZ). Each cell contains ≈80 copies of the λ shuttle vector integrated into the genome on chromosome 4 (17). The utility of this vector is its ability to act as a target for mutations that can subsequently be “shuttled” to an Escherichia coli cell for analysis. The vector is recovered from the genomic DNA by using Transpack in vitro λ packaging extracts (Stratagene). The mutation assay is a simple blue–white plaque color screening analysis. Each plaque generated on an E. coli bacterial lawn supplemented with 5-bromo-4-chloro-3-indolyl β-galactopyranoside (X-Gal) is representative of an individual target gene. The ratio of mutant blue plaques to the total number of generated plaques gives the mutation frequency.
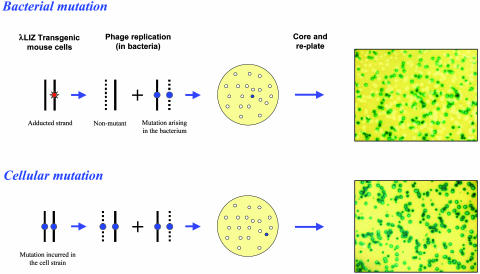
The Big Blue system has been adapted to measure rates of DNA repair (18), because it became evident that mutant plaques recovered by in vitro packaging of mouse DNA can arise from mutations sustained either in mouse cells or in the bacteria (18–20). The proportion of mutant phage contained within a mutant plaque distinguishes these two types of mutations (Fig. 1) (18). This was shown experimentally; mutations formed in mouse cells yield >90% mutant phage (when a plaque is cored and replated) because both DNA strands are mutant. On the other hand, mutations formed in the bacteria from adducted DNA yield only ≤50% mutant phage per plaque because one of the DNA strands is WT (18). Any decline in the frequency of these mosaic mutant plaques signifies the removal of adducts from the mouse DNA. Adduct reduction by repair would decrease the total number of mutant plaques recovered, whereas mutation fixation would increase the frequency of those plaques that contain >90% mutant phage (18). Because the frequency of both DNA adducts and mutations can be measured concurrently, this system makes it possible to study the kinetics of both repair and mutation fixation in cells that harbor the Big Blue λ LIZ shuttle vector, including the p53 cell strains that we have established.
Fig. 1.
The generation of mosaic and homogenous mutant plaques. In the first example (Upper), a lesion created within the cell strain forms a mutation in the bacterium during phage replication. This lesion is fixed into a mutation during the first round of phage replication, producing a mosaic plaque that is composed of a 1:1 ratio of mutant to WT phage, evident after replating. In the second example (Lower), a mutation sustained within the mouse cells directs the formation of a nearly homogenous mutant plaque, again verified by replating.
The objective of the present study was to use this system to elucidate the relationship between p53 and mutation. More specifically, because it is possible to monitor DNA adducts, mutation fixation, and apoptosis in cell strains on the same genetic background, our intention was to determine whether p53 influences the relationships among these endpoints.
Materials and Methods
Primary Cell Strain Establishment. Nullizygous C57BL/6 TSG-p53 N12 knockout mice (Taconic), developed by Donehower and Bradley (16) at Baylor College of Medicine, were crossed with Big Blue C57BL/6 homozygous animals (Stratagene). Embryos were obtained from matings between p53+/–, λ LIZ+/– mice (F1 generation). Detection of a vaginal plug was denoted as day 0 (conception), and the development of the embryos was timed from this date. Embryos were isolated for 12 days postconception and used to establish fibroblasts in culture, as described in ref. 21. Embryonic fibroblasts were grown for three passages, at which time an aliquot of cells was harvested for DNA isolation and genotyping; the remaining cells were cryostored. All experimental trials were performed in two separately derived cell strains for each genotype in four independently conducted experiments.
Genotyping. Initially, all cell strains were screened for the presence of the Big Blue λ LIZ shuttle vector with the following primers: forward, 5′-AATTAAACCACACCTATGGTG-3′; and reverse, 5′-CCTCTGCCGAAGTTGAGTATTT-3′. These primers flank the bacteriophage λ cII gene and generate a 400-bp fragment that is verified by standard agarose gel electrophoresis. Cell strains positive for the λ LIZ shuttle vector are either hemizygous or homozygous for this construct. The status of this, however, does not influence the mutational response of the transgene to spontaneous or N-ethyl-N-nitrosourea (ENU)-induced mutation (R. N. Winn and G. Douglas, personal communication). Multiplex PCR was used to ascertain the p53 genotype of those primary cell strains, which proved to be transgenic. The multiplex primers used were as follows: WT forward, 5′-GTGTTTCATTAGTTCCCCACCTTGAC-3′; WT reverse, 5′-ATGGGAGGCTGCCAGTCCTAACCC-3′; neo forward, 5′-GGGAATTCTGGGACAGCCAAGTCTGT-3′; and neo reverse, 5′-TTTACGGAGCCCTGGCGCTCGATGT-3′. A 320-bp fragment is generated with the WT primers, whereas a 150-bp fragment is generated with the neo primers. Strains WT for p53 were confirmed by the amplification of a single 320-bp fragment, nullizygous p53 knockout strains were verified by the generation of a 150-bp fragment, and cells heterozygous for p53 were identified by the amplification of both the 150- and 320-bp products.
Cell Culture. Primary embryonic fibroblast cells were cultured in DMEM (GIBCO/BRL) containing 10% (vol/vol) FBS (GIBCO), 1% l-glutamine, and 1% penicillin/streptomycin (GIBCO). A culture atmosphere of 5–7% CO2 was maintained in a humidified incubator at 37°C. Stock cultures were grown in 150-mm tissue culture dishes (Sarstedt).
To induce quiescence, cultures were seeded in 100-mm dishes (Sarstedt) with 1.2 × 106 cells in the presence of serum and incubated overnight. After a 24-h growth period, the cultures were washed three times with PBS and reincubated with serum-free medium (DMEM/1% l-glutamine/1% penicillin/streptomycin). The serum-free medium was renewed at 4-day intervals. Quiescent cells were transferred from 100-mm culture dishes to 150-mm dishes and subsequently cultured under standard conditions to induce cell proliferation.
Acute Mutagen Treatment. All cultures were prepared by seeding 1.2 × 106 cells in 100-mm culture dishes and incubating under standard growth conditions. Proliferating (24 h after plating) and quiescent (14 days after serum removal) cells were treated with ENU (200 μg/ml, Sigma) for 30 min in serum-free media at standard cell culture conditions. Cultures were then washed three times with PBS. Both quiescent and proliferating cells were later incubated in standard growth media (see above). Cells were trypsinized 0, 1, 2, 3, and 4 days after treatment, pelleted by centrifugation (1,000 × g), quick-frozen with liquid nitrogen, and stored at –80°C until further analysis.
Subacute Mutagen Treatment. Proliferating cells were treated with an initial ENU dose (200 μg/ml) and incubated for 4 days to allow for complete mutation fixation. The culture was then split; half of the cells were quick-frozen for DNA isolation and mutation measurement, and the remaining cells were seeded and treated again subsequent to an overnight incubation. This procedure was repeated to yield the compound dose values 400, 600, 800, and 1,000 μg/ml ENU.
After 14 days of serum deprivation, quiescent cells were treated with ENU (200 μg/ml) and then induced to proliferate. Passage of the cells into new tissue culture plates containing standard growth media (see above) induced proliferation. Four days after cellular restart, the culture was split again; half the cells were quick-frozen and stored for analysis, and the remaining cells were challenged with subsequent mutagen treatments, as in the experiments with acute treatments.
DNA Isolation. Cells were thawed and then resuspended in proteinase K solution (2 mg/ml, Sigma). Genomic DNA was purified from the cell suspension after an incubation time of 2 h at 55°C, followed by phenol/chloroform (1:1) extraction and precipitation with ethanol as described in ref. 22. The precipitated DNA was spooled onto a hooked glass Pasteur pipette, air-dried, and dissolved in 50 μl of Tris-EDTA buffer. The DNA was allowed to dissolve overnight at room temperature. The concentration of DNA was then determined spectrophotometrically after diluting the DNA solution 1:100 in Tris-EDTA (Beckman, DU 520 General Purpose UV-Vis) at 260 nm.
lacI Mutagenesis Assay. Transpack (Stratagene) packaging extracts were used to package and screen [on a bacterial lawn (strain SCS-8) supplemented with 5-bromo-4-chloro-3-indolyl β-galactopyranoside (X-Gal)] isolated mouse DNA for lacI mutations according to the manufacturer's recommendations, with the following enhancements (advantages outlined in ref. 23): (i) LB broth was substituted for NZY broth in all steps of the standard Big Blue protocol, (ii) the SCS-8 same-day culture was diluted to an OD600 of 2.0 in equal volumes of 1 M MgSO4 and LB broth (standard dilution = OD600 of 0.5 in 10 mM MgSO4), and (iii) on each assay tray the top X-Gal-enriched layer of medium was supplemented with 10 g/liter agarose as opposed to 7.5 g/liter.
Determination of Mutant Plaque Mosaicism. Well isolated (>4 mm apart) mutant plaques were cored (24 h after initial plating) with a 200-μl pipette tip and resuspended in 500 μl of phage buffer with 50 μl of chloroform and stored at 4°C. The next day, the phage was adsorbed in a bacterial suspension for 30 min at 37°C and screened for mosaicism after overnight plate incubation. Plaques containing ≥90% mutant phage represent mutations sustained in the murine cells, whereas plaques that contained ≤50% mutant phage result when an adduct suffered in the mouse cell is fixed into a mutation within the bacterium (18). No ambiguous plaques (50–90% mutant) were recovered. The mutation frequency is calculated by the ratio of blue mutant plaques, which contain ≥90% mutant phage, to total number of plaques screened, whereas the ratio of plaques that contain ≤50% mutant phage to the total number of plaques denotes the adduct frequency.
Quantification of Apoptosis by Cell Morphology. To identify apoptotic cells, cell monolayers were fixed with 70% ethanol at room temperature for 10 min. The cells were then rinsed with PBS and stained with acridine orange (10 μg/ml in buffer 1:2 PBS/ ddH2O; 10 min). Cellular morphology was assayed by fluorescence microscopy. Cells were classified as apoptotic if the nucleus was fragmented and the fragments contained condensed chromatin. Typically, such cells also showed cytoplasm condensation and surface blebbing (24). One thousand cells were counted for each determination. Data were analyzed by using spss 10.0 (Statistical Package for the Social Sciences, SPSS, Chicago) for Windows. F tests to determine homogeneity of variance were conducted to ensure that the data were normally distributed. Student's t test was performed to determine statistical significance.
Cell Cycle Analysis. Discrimination of cells in G0/G1 versus S versus G2/M phases of the cell cycle was determined by cellular DNA content. Cells (1 × 106) were collected and suspended in 5 ml of PBS and then centrifuged (6 min at 200 × g). The cells were triturated with a Pasteur pipette to ensure a single cell suspension in 0.5 ml of PBS and transferred to tubes containing 4.5 ml of 70% ice-cold ethanol for storage at –20°C. Immediately before flow cytometric analysis, the ethanol-suspended cells were centrifuged (5 min at 200 × g), resuspended in 5 ml of PBS, incubated at room temperature for 1 min, centrifuged (5 min at 200 × g), resuspended in 1 ml of propidium iodide/Triton X-100 staining solution with RNase A, and incubated at room temperature for 30 min. The propidium iodide fluorescence was collected with the FACSCalibur system (BD Immunocytometry Systems) by using the FL3 channel. The doublet-discrimination mode was applied to eliminate aggregates by using cellquest v3.3 software (BD Immunocytometry Systems). Chick erythrocyte nuclei (singlets) provided an internal standard.
Results
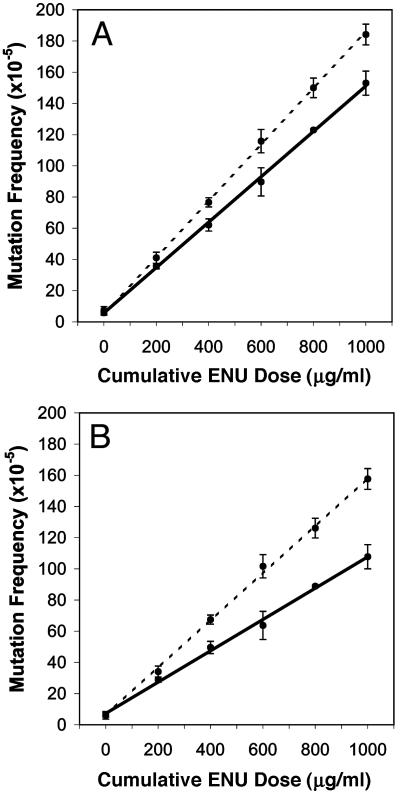
Subacute Mutagen Treatment. To investigate the role of p53 on mutation, homozygous, heterozygous, and nullizygous p53 embryonic fibroblasts were established harboring a lacI mutational target. Proliferating and quiescent cultures were treated with ENU to yield the compound dose values 400, 600, 800, and 1,000 μg/ml. Fig. 2A demonstrates a clear difference in mutation accumulation between proliferating nullizygous and WT cell strains.
Fig. 2.
Subacute mutagen treatment induced numerous mutations in both rapidly dividing (A) and “slowly dividing” (B) mouse cells. In both treatment groups, p53-nullizygous cells (broken line) accumulated more ENU-induced mutations (±SD) compared with cells that possess functional p53 (solid line).
Stem cells, which are persistent in self-renewing tissue populations, are the cells most at risk for the accumulation of multiple mutations (25–27), particularly in the proliferating cell populations where most cancers arise, because the differentiated cells only exist for short periods of time and only divide a few times. Thus, because cancer arises from single cells with several mutations, the importance of stem cells in carcinogenesis is clear. In an attempt to mimic a stem cell's cycle, embryonic cells were induced into quiescence by serum deprivation and contact inhibition (in all cell strains, DNA synthesis and cell division fell below 1% of the normal rate after 14 days of serum deprivation) and were later stimulated to proliferate after mutagen treatment. This procedure was repeated four times. The rate of mutation accumulation is greater in p53-nullizygous cells compared with p53 proficient cell strains (Fig. 2B); furthermore, this divergence is enhanced in relation to similarly treated proliferating cells (Fig. 2 A). It should be noted that the heterozygous cell strains demonstrated the same rate of mutation accumulation as WT cells to both treatment regimes, because experimental observations gathered from the heterozygous cell strains in respect to DNA repair, proliferation, and apoptotic rates also paralleled those data obtained from the WT p53 cell strains (data not shown).
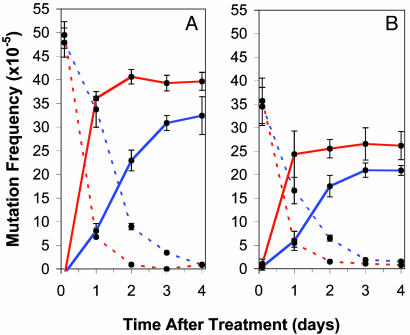
DNA Repair. To elucidate whether the reduction of mutation observed in WT cell strains is a result of heightened DNA repair, ENU adduct stability was monitored in p53-proficient and -deficient strains after mutagen treatment. Fig. 3 compares the repair and mutation fixation rates in p53-nullizygous and WT cell strains in both proliferating (Fig. 3A) and arrested cells induced to proliferate, subsequent to mutagen treatment (Fig. 3B). The ENU treatment resulted in a similar measure of DNA damage (adducts) in both p53-nullizygous and WT cell strains within treatment groups (proliferating and quiescent). Independent of genotype, however, the number of ENU-induced adducts was ≈28% lower in quiescent cultures compared with their WT counterparts. Subsequent to mutagen treatment for both quiescent and proliferating treated cultures, p53-nullizygous cell strains fixed and repaired ENU-induced adducts more rapidly than WT cells. Fig. 3A shows that proliferating nullizygous cell strains require ≈2 days after ENU treatment to eliminate all induced adducts from the transgene, whereas WT cells require ≈4 days. The same trend holds true for the quiescent treatment regime (Fig. 3B); about one additional day (after proliferating induction) is required for WT cells to remove all ENU-induced adducts from the transgene compared with nullizygous strains (≈3 versus 2 days, respectively). However, in both treatment groups, more ENU-induced adducts were repaired rather than fixed into stable mutations in WT cells compared with nullizygous cells (Fig. 3). In proliferating cultures, the ENU treatment induced an adduct frequency of 49.5 ± 2.8 × 10–5 in nullizygous cells and 47.9 ± 3.1 × 10–5 in WT cells; 4 days later, the resulting mutation frequency was 40.2 ± 2.0 × 10–5 and 33.1 ± 4.5 × 10–5, respectively (Fig. 2 A). This represents the repair of only 20% of the adducts in p53-nullizygous cells in contrast to 31% in WT cells. This trend holds true for the quiescent treatment group; correspondingly, an initial adduct frequency of 35.7 ± 4.8 × 10–5 and 34.5 ± 4.1 × 10–5 ensued after treatment in nullizygous and WT cells. At 4 days posttreatment, the resulting mutant frequency was lower in WT cells, 20.9 ± 1.0 × 10–5 versus 26.1 ± 3.9 × 10–5 in nullizygous cells (Fig. 3B). Therefore, 27% of ENU-induced adducts in nullizygous cell strains did not produce a mutation, in contrast to the 40% of ENU-induced adducts in WT cells, which did not produce a mutation in p53-proficient cells (Fig. 2B).
Fig. 3.
The kinetics of repair and mutation in p53-proficient (blue) and -deficient (red) mouse cells. The frequency (±SD) of mosaic (broken line) and homogenous (solid line) mutant plaques generated from DNA isolated from proliferating cells (A) and arrested cells induced to proliferate after mutagen treatment (B). All trials were performed in two separately derived cell strains for each genotype. The mean mutation and adduct frequencies were generated from data accumulated from four independently conducted experiments in each cell strain.
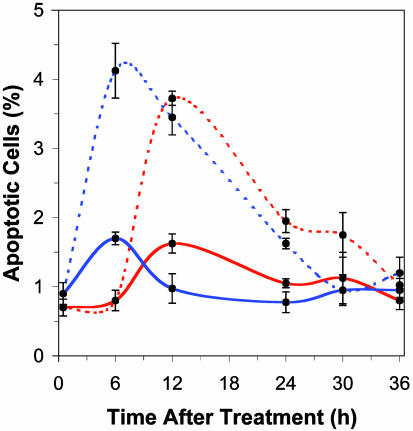
Apoptosis. To investigate whether apoptosis had a role in the disparity of mutation in p53-proficient and -deficient cells, the level of apoptosis was monitored subsequent to the mutagen treatments (Fig. 4). ENU increased apoptosis in both nullizygous and WT cells, yet the induction of apoptosis occurs more rapidly in WT cells. The maximal percentage of apoptotic cells in culture was observed at 6 h posttreatment in p53-proficient cultures versus at 12 h posttreatment in p53-deficient cultures after an acute treatment of either 200 or 1,000 μg/ml ENU (Fig. 4). These results are summarized in Fig. 5, which also illustrates that the subacute treatment regime did not increase the percentage of apoptotic cells over that induced by a single acute dose (i.e., that sensitization did not occur). After the last 200 μg/ml ENU treatment in the subacute treatment regime (a total cumulative dose of 1,000 μg/ml ENU), the level of apoptosis was equivalent to that experienced after a single acute ENU dose in both nullizygous (P = 0.38) and WT cultures (P = 0.41). The kinetics of ENU-induced apoptosis observed in proliferating cultures (Fig. 4) is similar to apoptotic induction in quiescently treated cultures (data not shown), albeit at a reduced frequency. The maximum percentage of apoptotic cell death induced by 200 μg/ml ENU in quiescent cultures was 1.08 ± 0.09% in WT cultures and 1.00 ± 0.09% in nullizygous cultures versus 1.70 ± 0.15% and 1.63 ± 0.18%, respectively, in proliferating cultures (Fig. 5).
Fig. 4.
Induction of apoptosis (percent ± SE) in proliferating WT (blue) and nullizygous (red) p53 cell strains after either an acute treatment of 200 (solid line) or 1,000 (broken line) μg/ml ENU.
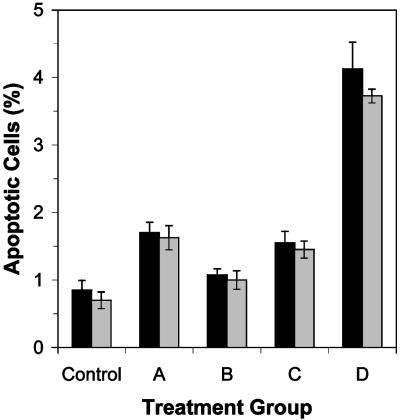
Fig. 5.
A summary of the maximum level of apoptosis (percent ± SE) in p53-proficient (black) and -deficient (gray) cultures induced in proliferating cells treated with 200 μg/ml ENU (A), quiescent cells treated with 200 μg/ml ENU (subsequent to proliferation induction) (B), proliferating cells treated with a subacute cumulative dose of 1,000 μg/ml ENU (see Materials and Methods) after the final treatment (C), and proliferating cells treated with an acute dose of 1,000 μg/ml ENU (D) is illustrated.
Cell Proliferation. Because proliferation is directly linked to mutation fixation, and because p53 is believed to aid the cell by delaying cell proliferation before DNA synthesis to aid DNA repair, the rate of cell division was constantly monitored. Under standard cell culture conditions (see Materials and Methods), p53-deficient cells doubled every 24.5 h compared with 30.5 h for WT cells. Flow cytometric analysis of proliferating cells showed that G1 was reduced from ≈22 h in WT cells to <1 h in p53-nullizygous cells, S is unchanged, and G2/M was increased >5-fold. During quiescence, WT cells are mostly in G1 (87%), whereas nullizygous cells are mostly in G2/M (81%), a pattern that was also found in cells 24 h after their treatment and release from quiescence (data not shown). After mutagen treatment (200 μg/ml) of proliferating cells, the generation time was dramatically increased in WT cells to 51.2 h, whereas this treatment had little effect on rate of proliferation in p53-deficient cells, marginally increasing the doubling time to 28.9 h. After ENU treatment of quiescent cultures, WT cells required 61.5 h to double after stimulated to proliferate, whereas nullizygous cells doubled after 33.9 h subsequent to release from quiescence. Clearly, after mutagen treatment, the p53-nullizygous cells had much less time for DNA repair before DNA synthesis.
Discussion
Elucidation of the role that p53 plays in carcinogenesis is obviously of crucial importance, given that half of all human cancers contain a mutation in the p53 gene (28). Unfortunately, the mechanism by which p53 exerts its protective effect remains uncertain, particularly as biochemical and molecular studies of p53 show it to be involved in a large number of cellular pathways. Nonetheless, it is believed that p53 reduces the frequency of mutation and, as a consequence, carcinogenesis, by either increasing DNA repair or removing cells prone to be carcinogenic via p53-induced apoptosis. We have presented data that clearly demonstrate that p53 reduces the mutant frequency after exposure to a mutagen by increasing DNA repair rather than by increasing apoptosis.
The mechanism by which p53 acts to decrease mutation seems to be via cell cycle delay, slowing cells before S phase, thus providing additional time for DNA repair or the induction of DNA repair enzymes before DNA replication and mutation fixation. This can be seen as DNA damage greatly delays cell turnover and thus mutation fixation in p53-proficient cells but not in p53-deficient cells. This is important, because DNA adducts at the lacI mutational target are only converted into mutations after DNA replication (18). This p53-dependent mechanism, which leads to a decrease in mutation, is equally evident in cells treated during both quiescence and active proliferation. Actively proliferating cells, however, suffer both more DNA damage and more mutation than quiescently treated cells when an equivalent mutagen treatment is administered. Nevertheless, it is clear that p53 is more protective in quiescent cells after proliferation induction than in actively proliferating cells. This is important, because stem cells in which most cancerous mutations must arise (25–27) are also quiescent much of the time.
The results obtained are also informative with respect to the induction of apoptosis and its role in reducing the risk of a cancerous mutation, because they demonstrate that it must be DNA adducts, and not newly induced mutations, that induce apoptosis: This is because the level of apoptosis returns to normal after mutation fixation, regardless of treatment.
In the uniform cell populations that we investigated, p53-dependent apoptosis played little role in preventing mutation. Because virtually all adducts are either repaired or fixed into persistent mutations in one cell cycle, apoptosis could only be effective in preventing mutation if it acted before replication. The overall level of apoptosis, however, as measured over the first cell cycle, was similar in WT and p53-nullizygous cells, even though apoptotic induction occurred later in p53-deficient cells. Furthermore, because the p53-deficient cells were not as delayed as WT cells after DNA damage, apoptosis occurred at a later point in their cell cycle than in WT cells. It seems likely, then, that the mechanism by which apoptosis arises in p53-deficient cells is different than that in WT cells and that the longer time required to induce apoptosis reflects a more indirect induction mechanism.
It is remarkable that only a small proportion of the established primary cultures became apoptotic, given that the cells were genetically identical, arrested in a uniform physiological state, and treated homogeneously. One would expect an all-or-none response under these conditions. It may be that apoptosis occurs only in a minor cell population or a very short period in the cell cycle. We speculate that apoptosis is not triggered by all adducts but rather by adducts on specific DNA sequences. If these significant sequences were genes playing critical roles in preventing cancer, apoptosis would reduce cancer and the mutant frequency at these genes but leave the mutation frequency at most genes, including our mutational target, lacI, unaltered.
Although apoptosis does not reduce the mutation frequency of most genes in a uniform population of cells, it is possible for it to reduce the mutation frequency in a tissue, because tissues have several populations of cells. According to the standard model of tissue renewal, a small number of slowly dividing stem cells give rise to a much larger number of relatively short-lived proliferating cells (transit cells), and these produce the nondividing differentiated cell population (27). In the small intestine of the mouse, there are at least 100 differentiated cells and 100 transit cells per stem cell. The differentiated cells are immutable, at least by the mutagen we have used, because they do not divide (18), and thus they cannot become cancerous. The transit cells, on the other hand, are much more numerous than the stem cells, and are more mutable, so they will have a large number of mutations. Because they are transitory, they are unlikely to acquire more than one of the several mutations required to become cancerous at this stage, but they could acquire the last one needed. Apoptosis in this population would eliminate a large number of mutated and potentially cancerous cells in the tissue, even though the mutation frequency in the surviving transit cells was unaltered. The missing transit cells would be replaced by the progeny of the less mutated stem cells, with the consequence that apoptosis would in this way lower the overall mutant frequency in the tissue and the risk of cancer.
Our data clearly demonstrate that in a uniform cell population, WT p53 protects cells by delaying the cell cycle, thereby facilitating DNA repair and decreasing mutation. Apoptosis does not reduce the formation of mutations but is induced by DNA damage. Nevertheless, in vivo apoptosis may play an important role in the prevention of cancer by removing damaged cells from within a tissue. These results should be typical of normal cells as the cell strains are not transformed and are on a common genetic background. Moreover, the system permitted us to measure, with considerable precision, all of the relevant endpoints: adducts, mutations, proliferation, and apoptosis.
Acknowledgments
We thank Larry Donehower for providing a multiplex strategy, developed by Terry Timme and Tim Thompson at Baylor College of Medicine, for the genotypic analysis of our murine cell strains. We also thank Stephen Dertinger of Litron Laboratories for performing the flow cytometric analysis and Lorien Newell for her patience while editing numerous versions of this manuscript. This work was supported by the Cancer Research Society, the National Cancer Institute of Canada, and the Natural Sciences and Engineering Research Council of Canada.
Abbreviation: ENU, N-ethyl-N-nitrosourea.
References
- 1.Greenblatt, M. S., Bennett, W. P., Hollstein, M. & Harris, C. C. (1994) Cancer Res. 54 4855–4878. [PubMed] [Google Scholar]
- 2.Nigro, J. M., Baker, S. J., Preisinger, A. C., Jessup, J. M., Hostetter, R., Cleary, K., Bigner, S. H., Davidson, N., Baylin, S., Devilee, P., et al. (1989) Nature 342 705–708. [DOI] [PubMed] [Google Scholar]
- 3.Hollstein, M., Sidransky, D., Vogelstein, B. & Harris, C. C. (1991) Science 253 49–53. [DOI] [PubMed] [Google Scholar]
- 4.Vogelstein, B. (1990) Nature 348 681–682. [DOI] [PubMed] [Google Scholar]
- 5.Gottlieb, T. M. & Oren, M. (1998) Semin. Cancer Biol. 8 359–368. [DOI] [PubMed] [Google Scholar]
- 6.Lozano, G. & Elledge, S. J. (2000) Nature 404 24–25. [DOI] [PubMed] [Google Scholar]
- 7.Lakin, N. D. & Jackson, S. P. (1999) Oncogene 18 7644–7655. [DOI] [PubMed] [Google Scholar]
- 8.Ko, L. J. & Prives, C. (1996) Genes Dev. 10 1054–1072. [DOI] [PubMed] [Google Scholar]
- 9.Amundson, S. A., Xia, F., Wolfson, K. & Liber, H. L. (1993) Mutat. Res. 286 233–241. [DOI] [PubMed] [Google Scholar]
- 10.Honma, M., Hayashi, M. & Sofuni, T. (1997) Mutat. Res. 374 89–98. [DOI] [PubMed] [Google Scholar]
- 11.Honma, M., Zhang, L. S., Hayashi, M., Takeshita, K., Nakagawa, Y., Tanaka, N. & Sofuni, T. (1997) Mol. Cell. Biol. 17 4774–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honma, M., Momose, M., Tanabe, H., Sakamoto, H., Yu, Y., Little, J. B., Sofuni, T. & Hayashi, M. (2000) Mol. Carcinog. 28 203–214. [DOI] [PubMed] [Google Scholar]
- 13.Yu, Y., Li, C. Y. & Little, J. B. (1997) Oncogene 14 1661–1667. [DOI] [PubMed] [Google Scholar]
- 14.Nishino, H., Knoll, A., Buettner, V. L., Frisk, C. S., Maruta, Y., Haavik, J. & Sommer, S. S. (1995) Oncogene 11 263–270. [PubMed] [Google Scholar]
- 15.Sands, A. T., Suraokar, M. B., Sanchez, A., Marth, J. E., Donehower, L. A. & Bradley, A. (1995) Proc. Natl. Acad. Sci. USA 92 8517–8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donehower, L. A., Harvey, M., Slagle, B. L., McArthur, M. J., Montgomery, C. A., Jr., Butel, J. S. & Bradley, A. (1992) Nature 356 215–221. [DOI] [PubMed] [Google Scholar]
- 17.Dycaico, M. J., Provost, G. S., Kretz, P. L., Ransom, S. L., Moores, J. C. & Short, J. M. (1994) Mutat. Res. 307 461–478. [DOI] [PubMed] [Google Scholar]
- 18.Bielas, J. H. & Heddle, J. A. (2000) Proc. Natl. Acad. Sci. USA 97 11391–11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishino, H., Buettner, V. L. & Sommer, S. S. (1996) Mutat. Res. 372 97–105. [DOI] [PubMed] [Google Scholar]
- 20.Paashuis-Lew, Y., Zhang, X. B. & Heddle, J. A. (1997) Mutat. Res. 373 277–284. [DOI] [PubMed] [Google Scholar]
- 21.Freshney, R. I. (1994) Culture of Animal Cells: A Manual of Basic Technique (Wiley, New York).
- 22.Tinwell, H., Lefevre, P. A. & Ashby, J. (1994) Mutat. Res. 307 169–173. [DOI] [PubMed] [Google Scholar]
- 23.Bielas, J. H. (2002) Mutat. Res. 518 107–112. [DOI] [PubMed] [Google Scholar]
- 24.Gregory, C. D., Dive, C., Henderson, S., Smith, C. A., Williams, G. T., Gordon, J. & Rickinson, A. B. (1991) Nature 349 612–614. [DOI] [PubMed] [Google Scholar]
- 25.Cairns, J. (1975) Nature 255 197–200. [DOI] [PubMed] [Google Scholar]
- 26.Cairns, J. (2002) Proc. Natl. Acad. Sci. USA 99 10567–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heddle, J. A., Cosentino, L., Dawod, G., Swiger, R. R. & Paashuis-Lew, Y. (1996) Environ. Mol. Mutagen. 28 334–341. [DOI] [PubMed] [Google Scholar]
- 28.Harris, C. C. & Hollstein, M. (1993) N. Engl. J. Med. 329 1318–1327. [DOI] [PubMed] [Google Scholar]