Abstract
The methyl–CpG dinucleotide containing a symmetrical 5-methylcytosine (mC) is involved in gene regulation and genome stability. We report here that methylation-mediated transcriptional repressor methylated DNA-binding domain 1 (MBD1) interacts with methylpurine–DNA glycosylase (MPG), which excises damaged bases from substrate DNA. MPG itself actively represses transcription and has a synergistic effect on gene silencing together with MBD1. Chromatin immunoprecipitation analysis reveals the molecular movement of MBD1 and MPG in vivo: (i) The MBD1–MPG complex normally exists on the methylated gene promoter; (ii) treatment of cells with alkylating agent methylmethanesulfonate (MMS) induces the dissociation of MBD1 from the methylated promoter, and MPG is located on both methylated and unmethylated promoters; and (iii) after completion of the repair, the MBD1–MPG complex is restored on the methylated promoter. Mobility-shift and structural analyses show that the MBD of MBD1 binds a methyl–CpG pair (mCpG × mCpG) but not the methyl–CpG pair containing a single 7-methylguanine (N) (mCpG × mCpN) that is known as one of the major lesions caused by MMS. We further demonstrate that knockdown of MBD1 by specific small interfering RNAs significantly increases cell sensitivity to MMS. These data suggest that MBD1 cooperates with MPG for transcriptional repression and DNA repair. We hypothesize that MBD1 functions as a reservoir for MPG and senses the base damage in chromatin.
DNA methylation at position 5 of cytosine within CpG dinucleotides is the major epigenetic modification of mammalian genomes and is required for gene regulation, chromatin formation, and genome stability (1). The methylation status of DNA is correlated with a wide range of phenomena during mammalian development (2–4). On the other hand, aberrant methylation of tumor-associated gene promoters contributes directly to the progression of some cancer cells (5–7). In the nucleus, cytosine methylation is specifically recognized by transacting factors such as the methyl–CpG-binding domain proteins [methylated DNA-binding domain (MBD) proteins] (1, 8). MBD1, MBD2, MBD3, and MeCP2 act as transcriptional repressors depending on the presence of methyl–CpG pairs in chromatin (9–12). In addition, 5-mC and cytosine have high mutation rates due to spontaneous hydrolytic deamination, giving rise to thymine and uracil, respectively (13). Two DNA glycosylases, MBD4 and thymine DNA glycosylase (TDG), correct the resultant mismatches in the CpG context by excising thymine and uracil (14–16).
DNA glycosylases initiate base excision repair (BER) by severing the glycosylic bonds of numerous damaged bases such as alkylated, deaminated, and oxidized bases due to cellular metabolism and exogenous agents (17). Thus, BER protects the genome against the mutagenic effects of a variety of DNA damage in the cell (18). Among the glycosylase family of proteins, methylpurine–DNA glycosylase (MPG) catalyzes the excision of many kinds of base substrates, including methylpurines, deaminated adenine [hypoxanthine], oxidized guanine [8-oxoguanine], and cyclic etheno adducts on both adenine and guanine (19, 20). The majority of lesions created by alkylating agents such as methylmethanesulfonate (MMS) are 3-methyladenine (3-mA) (10%), 7-methylguanine (7-mG) (65≈80%), and O6-mG (0.3≈7%). Among them, MPG effectively removes 7-mG and 3-mA (21). The presence of these modified bases in the genome leads to cell damage and death, due to inappropriate DNA replication and genome instability with increased mutations (22). In fact, Mpg knockout mice showed high sensitivity to alkylation damage of their DNA (23).
Evidence of multiple connections between transcription and DNA repair has emerged from many studies. In particular, transcription-coupled repair has been documented in nucleotide excision repair that is a main pathway involved in correcting bulky DNA lesions induced by UV light. The rate of repair in actively transcribed genes is significantly faster than in nontranscribed regions of the genome (24). It has also been found that several lesions removed by BER are related to transcriptional status (25, 26), but the relationship between BER and transcription remains to be elucidated. Recently, TDG has been shown to function in transcriptional control, through an interaction with transcription factors and coactivators (27–31). Human endonuclease III, one of the DNA glycosylases, also interacts with nucleotide excision repair–endonuclease XPG and the damage-inducible transcription factor Y box-binding protein 1 (32, 33).
During the investigation of the MBD1-containing complex, we found that MBD1 specifically interacts with MPG. From the observation of molecular movement of MBD1 and MPG in vivo,we found that MBD1 was dissociated from the methylated gene promoter under the conditions of MMS treatment, which may support that repair of specific DNA lesions within the genome relies on local perturbation of chromatin structure (34, 35). We discuss a mechanistic link between DNA repair, gene repression, and chromatin dynamics in response to base damage.
Materials and Methods
Cell Culture, Transfection, and Immunoprecipitation. HeLa, NCL-H1299, and SBC5 cells were cultured and introduced with plasmids by using FuGENE6 (Boehringer Mannheim) or Lipofectamine (Invitrogen) (36, 37). Immunoprecipitation of MBD1 and MPG was done as described in Supporting Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Plasmids and Antibodies. For information on plasmids and antibodies, see Supporting Methods.
In Vitro Binding and GST Pull-Down Assay. The in vitro binding assay was performed as described (ref. 38 and Supporting Methods). For pull-down assay, GST and GST-fusion proteins (200 nM each) were immobilized on glutathione-agarose beads and incubated with 250 μl of the lysates from HeLa cells. The lysates were prepared by using a buffer [0.05% Nonidet P-40/50 mM Tris·HCl (pH 8.0)/100 mM NaCl/protease inhibitors/1 mM sodium orthovanadate].
Luciferase Assay. For information on the luciferase assay, see Supporting Methods.
Growth Inhibition Assay. Cells were seeded at a density of 1 × 105 cells per well on six-well dishes and treated for1hwith MMS. After incubation for 3 days, the cells were quantitatively counted, and the results were shown as a ratio of the number of MMS-treated cells relative to that of untreated cells (percent of control growth) (39).
Chemical Crosslinking and Chromatin Immunoprecipitation. At 48 h after transfection, cells (1 × 106) were treated with 1 mM MMS for 1 h. The cells were treated with dimethyl 3,3′-dithiobispropionimidate-2HCl (5 mM), rinsed, and then crosslinked by addition of 1% formaldehyde for 10 min. The cell lysates were prepared for chromatin immunoprecipitation and PCR amplification of the human p16 gene promoter (refs. 36 and 37 and Supporting Methods).
Electrophoretic Mobility-Shift Assay (EMSA). Oligodeoxynucleotides containing 7-mG were prepared by a primer extension method (refs. 40 and 41 and Supporting Methods). For the EMSA, binding reactions were carried out in 10 μl of a buffer [10 mM Tris·HCl (pH 8.0)/5 mM MgCl2/5 mM DTT/5% glycerol] by using 0.8 μg of GST-fused MBD of MBD1 and 200 nM of the 30-mer oligonucleotide duplex containing cytosine or mC in combination with G or mG, for 30 min at room temperature. The reaction mixtures were loaded on a 6% polyacrylamide gel, and the gel was stained with SYBR green I (Molecular Probes).
Small Interfering RNA (siRNA) Knockdown of MBD1. siRNA duplexes were designed for targeting mRNA encoding human MBD1 (Japan Bioservice, Saitama, Japan): 5′-GGCAUCUUGUGCUAUCCAGTT-3′ and 5′-CUGGAUAGCACAAGAUGCCTT-3′. The siRNAs for lamin A/C and GL3 were previously reported (42). The siRNAs were transfected into the cells by using Oligofectamine (Invitrogen).
Results
MBD1 Interacts with MPG. To identify factors that interact with MBD1, we performed a yeast two-hybrid screen by using the C-terminal TRD as a bait (Fig. 1A). From a screening of ≈7 × 106 independent transformants of a human HeLa cDNA library, we isolated two independent cDNA clones encoding the MPG.
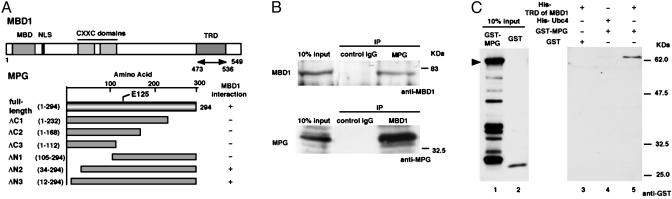
Fig. 1.
Interaction of MBD1 with MPG. (A) Structure of MBD1 and MPG. MBD1 (isoform v3) contains the MBD, nuclear localization signal (NLS), cysteine-rich CXXC domains, and TRD (9, 37). TRD (amino acids 473–536) was used in a yeast two-hybrid screen as a bait. MPG has an enzymatically active site at glutamic acid (E) 125. In full-length and deletion mutants of MPG, the presence and absence of MBD1 interaction in Fig. 2 A are indicated by plus and minus, respectively. (B) Association between endogenous MBD1 and MPG in HeLa cells. (C) Direct binding of the TRD of MBD1 to MPG in vitro. Bacterially expressed (His)6-TRD of MBD1 and GST-MPG were used for nickel-NTA resin affinity chromatography (38). The arrowhead indicates the full-length MPG fused to GST.
To demonstrate an interaction between MBD1 and MPG in vivo, we investigated an association of these endogenous proteins in HeLa cells. The cells were lysed and then subjected to immunoprecipitation with anti-MPG or anti-MBD1 antibodies. Western blot analysis showed that MBD1 was present in the immunoprecipitates with MPG but not in the control lane (Fig. 1B Upper). Endogenous MPG was coprecipitated with MBD1, but not with the control antibodies (Fig. 1B Lower).
To demonstrate whether the TRD of MBD1 directly binds MPG, (His)6-TRD of MBD1 and GST-fused MPG were bacterially prepared for affinity chromatography as described (38). The purified (His)6-TRD of MBD1 was incubated with GST or GST-MPG in the presence of the nickel-NTA resin (Fig. 1C). The GST-MPG was released specifically from the resin bound by TRD of MBD1 (lane 5), indicating the association of MPG with the TRD of MBD1. As a control, GST was not released from the same resin (lane 3), and GST-MPG did not associate with the resin bound by unrelated (His)6-Ubc4, one of the ubiquitin-conjugating enzymes (lane 4). In addition, there were several degradation products of GST-MPG in the input (lane 1), because this protein was reported to have many protease hypersensitive sites (43). These degradation products did not bind the TRD of MBD1, suggesting that almost full-length of MPG is required for this interaction (as shown in Fig. 2).
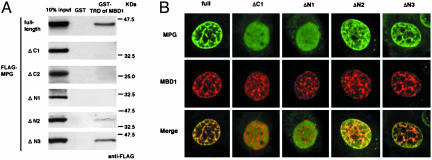
Fig. 2.
Regions of MPG for interacting with MBD1. (A) Association of MPG with the TRD of MBD1. GST-fused TRD of MBD1 or GST was immobilized on glutathione agarose beads and incubated with the lysates from HeLa cells expressing full-length or deletion mutants of MPG. (B) Localization of MPG and MBD1 in the nucleus. The full-length or deletion mutants of FLAG-MPG were coexpressed with DsRed-fused MBD1.
Regions of MPG for Interacting with MBD1. To determine the regions of MPG that are crucial to the interaction with TRD of MBD1, six deletion mutants of FLAG-MPG (shown in Fig. 1A) were expressed in HeLa cells. GST-fused TRD of MBD1 or GST was immobilized on glutathione-agarose beads and incubated with the lysates from HeLa cells expressing the full-length or deletion mutants of MPG. After repeated washings, the proteins bound on the beads were detected by anti-FLAG antibodies (Fig. 2A). The full-length MPG bound the TRD of MBD1 but not GST alone. The deletions of C-terminal regions of MPG did not associate with the TRD of MBD1 [ΔC1, -2, and -3 (data not shown)]. Further, the N-terminal deletions ΔN2 and -3 bound the TRD of MBD1, but ΔN1 mutant lost the ability to bind it. Collectively, these results suggested that MPG directly interacts with TRD of MBD1 through both terminal regions of MPG.
To investigate the localization of MPG and MBD1 in the nucleus, full-length or deletion mutants of MPG fused to FLAG and fluorescent protein-tagged MBD1 were coexpressed and visualized by using a confocal laser-scanning microscope (Fig. 2B). Previous reports clarified that the punctate localization of MBD1 depends on the distribution of methyl–CpG sequences on the genome (9, 10, 36, 37). Full-length ΔN2 and ΔN3 of MPG colocalized with MBD1 at multiple foci in the nuclei. In contrast, both ΔC1 and ΔN1 mutants diffusely distributed throughout the nuclei and significantly weakened the specific colocalization with MBD1. These results represented >90% of the nuclei in the cells studied, suggesting that both terminal regions of MPG are important for the association with MBD1.
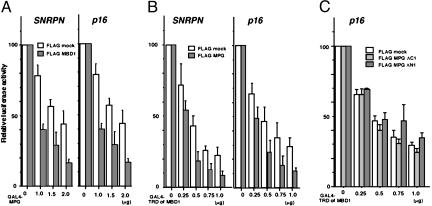
MBD1 and MPG Cooperate for Transcriptional Repression. Molecules in the DNA repair system, like TDG, are actively involved in transcriptional regulation (27, 28, 30, 31). To check whether MPG modulates gene promoter activities, we expressed GAL4-MPG and FLAG-MBD1 (Fig. 3A), GAL4-TRD of MBD1 and FLAG-MPG (Fig. 3B), and GAL4-TRD of MBD1 and FLAG-MPG mutants (ΔC1 and ΔN1) (Fig. 3C) in HeLa cells. The effects of these combinations were examined by using a Photinus pyralis luciferase reporter that contains five GAL4-binding elements just upstream of human SNRPN and p16 gene promoters. Both gene activities are known to be affected by the methylation status in cells (44). GAL4-MPG alone repressed transcription from both promoters in a dose-dependent manner (Fig. 3A; FLAG mock). To assess the functional implication of the MBD1-MPG association, we examined the effect of FLAG-MBD1 on the repression by GAL4-MPG. Expression of MBD1 enhanced the MPG-mediated repression of both promoter activities (Fig. 3A; FLAG-MBD1). Next, we elucidated the effect of MPG on repression by GAL4-fused TRD of MBD1 (Fig. 3B). TRD efficiently repressed the luciferase activities of both promoters in agreement with previous reports (9, 10). The coexistence of GAL4-TRD of MBD1 and FLAG-MPG produced a synergistically repressive effect on these promoters. The level of these effects was very similar to that in the case of TDG and thyroid transcription factor-1 (30). However, FLAG-MPG mutants (ΔC1 and ΔN1) did not affect the repression by TRD of MBD1 (Fig. 3C), emphasizing the importance of the interaction between MBD1 and MPG. These results suggested that MPG itself can actively repress transcription, and that MBD1 and MPG cooperate to enhance their repressive activities.
Fig. 3.
Cooperation of MBD1 and MPG for transcriptional repression. GAL4-MPG and FLAG-MBD1 (A), GAL4-TRD of MBD1 and FLAG-MPG (B), and GAL4-TRD of MBD1 and FLAG-MPG mutants (C) were expressed in HeLa cells to examine their effects on a luciferase reporter that contains five GAL4-binding elements just upstream of human SNRPN or the p16 gene promoter. The cells were transfected with plasmids (1 μg) to express FLAG-tagged proteins, together with indicated amounts of plasmids for GAL4-fused proteins. FLAG-mock was used as a control. The luciferase activities from insertless GAL4-mock were normalized to 100. Values are given as means and standard deviations of results from more than three independent experiments.
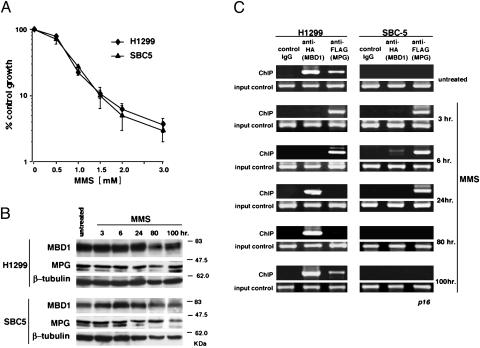
Molecular Dynamics of MBD1 and MPG Under MMS-Induced DNA Damage. The alkylating agent MMS is known to induce genome-wide base damage, including 7-mG, which is the major substrate for MPG in vivo. To investigate the association of MBD1 and MPG with chromosomal gene promoters under conditions of MMS treatment, we chose the p16 tumor suppressor gene in which hypermethylation of the promoter-associated CpG island causes transcriptional repression in many cancers (5, 7). NCI-H1299 cells possess methylated p16 promoter, whereas the same DNA region is unmethylated in SBC-5 cells (36, 44, 45). We first tested a dose-dependent effect of MMS on the treated cells by using a cell growth inhibition assay (Fig. 4A). The growth inhibition usually corresponds to the level of genome-wide base damage (39). Both cell lines were treated with MMS (0≈3 mM) for 1 h and grown for 3 days, resulting in a similar growth inhibition in proportion to the MMS concentrations. Repeated experiments showed that MMS (1 mM)-treated cells resumed their proliferation after temporary growth arrest (data not shown), indicating that this treatment is not excessively toxic for the cells studied.
Fig. 4.
Molecular dynamics of MBD1 and MPG under MMS-induced DNA damage. (A) MMS-dependent growth inhibition of NCI-H1299 and SBC-5 cells. Cell numbers were examined after MMS treatment for 1 h and after incubation for 3 days. (B) Expression of MBD1, MPG, and β-tubulin in untreated and MMS-treated cells. At 48 h after transfection, both cell lines expressing HA-MBD1 and FLAG-MPG were treated with 1 mM MMS for 1 h. Western blot analysis was done with anti-MBD1, anti-FLAG, and anti-β-tubulin antibodies. (C) Association of MBD1 and MPG with chromosomal p16 gene promoters under MMS treatment. The p16 gene promoter is highly methylated in NCI-H1299 cells and unmethylated in SBC-5 cells (36, 37). For chromatin immunoprecipitation, the cells expressing HA-MBD1 and FLAG-MPG were treated with 1 mM MMS for 1 h and incubated for the indicated times. The coprecipitated DNAs with the indicated antibodies were PCR-amplified by using a set of primers for p16 promoter sequences. Genomic DNAs in the input cell lysates before the immunoprecipitation were used as a control. All data are from more than three independent experiments.
We next investigated the dynamics of MBD1 and MPG on chromosomal genes in vivo (Fig. 4 B and C). For chromatin immunoprecipitation, HA-MBD1 and FLAG-MPG were equally expressed in NCI-H1299 and SBC-5 cells, and these cells were then treated with 1 mM MMS for 1 h. Western blot analysis revealed that exogenously expressed MBD1 and MPG as well as endogenous β-tubulin were present at the indicated time points after MMS treatment (Fig. 4B). These cells were crosslinked with dimethyl 3,3′-dithiobispropionimidate-2HCl and then with formaldehyde. Coprecipitated DNAs with the indicated antibodies were subjected to PCR amplification by using a set of primers for p16 promoter sequences. The amplified sequence is 217 bp long, containing 23 CpG dinucleotides (C+G content 75.1%; CpG/GpC = 0.68). These CpG sites are highly methylated in NCI-H1299 cells (36, 37, 44, 45). The MBD1–MPG complex was normally present on the methylated, but not unmethylated, p16 gene promoter (Fig. 4C; untreated). At 3 h after treatment (referred to as the active stage of BER (refs. 46 and 47, and Fig. 7A, which is published as supporting information on the PNAS web site), MBD1 was unexpectedly dissociated from the methylated promoter (Fig. 4C; 3 h). This dissociation of MBD1 from the methylated promoter was also found at6hafterMMStreatment. In contrast, MPG existed on the both methylated and unmethylated promoters, probably for excision of damaged bases. At 24 h after treatment, the presumed terminal stage of the repair process (refs. 46 and 47, and Fig. 7A), MBD1 was restored on the methylated promoter (Fig. 4C; 24 h). MPG was associated with the unmethylated promoter until 24 h after treatment and was little found on the promoter regardless of the methylation status at 80 h after treatment (Fig. 4C;80h). MPG may leave this promoter and target other damaged sites on the genome at this stage. At 100 h after MMS treatment, the MBD1–MPG complex was restored on the methylated, but not unmethylated, p16 gene promoter (Fig. 4C; 100 h). Additional analyses at the other chromosomal region, such as the imprinted SNRPN gene promoter, gave consistent results (data not shown). These observations suggested that MBD1 and MPG change their localization on the genome in response to MMS-induced base damage (see Discussion).
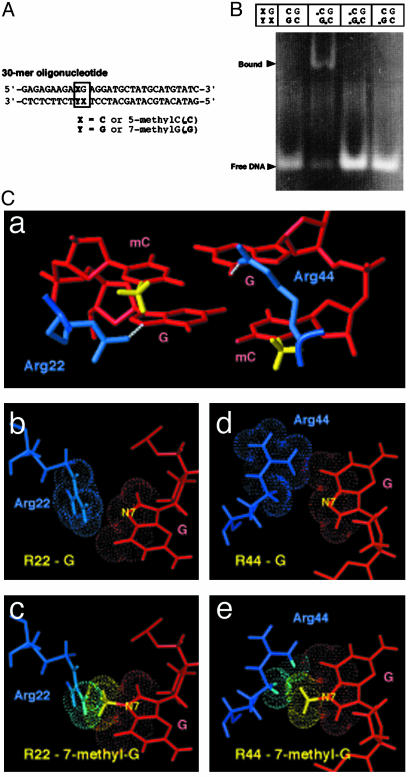
Presence of 7-mG in a Methyl–CpG Pair Abolishes Methylated DNA-Binding Ability of MBD1. We focused on understanding our observation that MBD1 was dissociated from the methylated region under the damage caused by MMS. MBD, which is highly conserved among the MBD family of proteins, is reported to be essential for their localization on methylated regions (10, 37, 48–50). Our previous structural analyses by using NMR revealed that MBD residues not only distinguish the two methyl groups of cytosines but also recognize the two guanine bases at the methyl–CpG site (51). Therefore, it is of great interest to investigate whether the ability of MBD to bind a methyl–CpG pair is affected by base damage. The major lesion created by methylating agents such as MMS is a 7-mG (21). Further, recent studies have indicated that cytosine methylation at CpG* sequences enhances mitomycin-induced alkylation of the guanine (*), compared with unmethylated CpG sites (52). Because there is the possibility that chemical guanine methylation by MMS occurs at a methyl–CpG site, we analyzed whether MBD of MBD1 is able to bind the CpG pair containing a mG (Fig. 5). We first prepared four 30-base paired oligodeoxynucleotides containing CG × CG, mCG × mCG, mCG × mCmG, or CG × CmG in a unique position by using a primer extension reaction as described (Fig. 5A) (40, 41). An electrophoretic mobility-shift assay was performed by using these double-stranded DNAs and bacterially expressed MBD of MBD1 (Fig. 5B) (10, 51). MBD specifically bound the oligodeoxynucleotide-containing mCG × mCG, whereas the DNA containing mCG × mCmG did not associate with MBD. The oligodeoxynucleotide containing CG × CG or CG × CmG also did not bind MBD. This result suggested that the occurrence of a single mG in the methyl–CpG pair abolished the ability of MBD1 to bind the methylated DNA.
Fig. 5.
Abrogation of methylated DNA binding of MBD by the presence of 7-mG in a methyl–CpG pair. (A) Construction of 30 base-paired oligodeoxynucleotides containing 5-mC and/or 7-mG. The unique dinucleotides, CG × CG, mCG × mCG, mCG × mCmG, and CG × CmG, were introduced into position (XG × XY). mG or G was incorporated into position Y of the synthesized strand. (B) Electrophoretic mobility-shift assay by using the oligodeoxynucleotides and MBD from MBD1. (C) Structural model of the interaction between MBD and methylated DNA. A proposed interaction between the methyl–CpG site and two arginine side chains at the interface, on the basis of NMR structure determination, is shown in A. The interactions between the guanine bases and the arginine side chains (b and d) can be perturbed by alkylations at N7 positions of the guanine bases (c and e). Arginine 22 and 44, blue; DNA, red; hydrogen bond between arginine and guanine, white; methyl groups in methyl–CpG sequence, yellow.
Structure analysis by NMR further supported the important participation of the modified guanine bases in the association between MBD of MBD1 and the methyl–CpG dinucleotide (Fig. 5C). Two methyl groups of cytosines in the methyl–CpG site are distinguished by major groove contacts made by five residues of the MBD: Val-20, Arg-22, Trp-34, Arg-44, and Ser-45. Among them, Arg-22 and -44 also recognize the two guanine bases within the dinucleotide (a). The guanidiums of these residues are poised to donate a hydrogen bond to the guanine bases in the methyl–CpG site (b and d). Conversion of either of these guanine bases to 7-mG by alkylation could significantly interfere with guanidium–G interactions. The methyl group introduced to one of the guanine bases would cause a steric clash with the guanidium group of Arg-22 (c); the methyl group of the guanine base also can interfere with δ-methylene of Arg-44 (e). These arginine residues play integral roles in DNA recognition, because substitution of either these residues totally abolished DNA binding (10, 51). So far, these findings suggest that the incidence of damaged guanine in a methyl–CpG pair leads to dissociation of MBD from methylated DNAs.
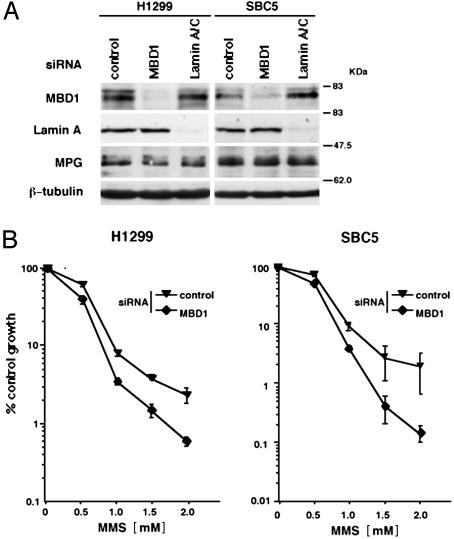
Increased Sensitivity to MMS in MBD1 Knockdown Cells. To demonstrate the role of MBD1 in MPG-mediated DNA repair, we finally used specific siRNAs capable of degrading target transcripts in a highly selective manner in vivo. Western blot analysis demonstrated the effectiveness of siRNA knockdown of MBD1 and lamin A/C (Fig. 6A). Both siRNAs did not change the levels of MPG and β-tubulin. Under such conditions, we examined whether MBD1 can affect cell growth inhibition with MMS treatment. At 24 h after the transfection of siRNAs, these two cell lines were treated with indicated concentrations of MMS for 1 h and then incubated for 3 days. The growth inhibition by MMS moderately but significantly increased in the MBD1 knockdown cells, in contrast to the control cells (Fig. 6B). This result was comparable to the higher sensitivity to alkylation damage in MPG knockout cells (23). Finally, we quantified abasic sites in MMS treated control and MBD1-knockdown cells by using aldehyde reaction probes (40). This result suggested that MBD1 knockdown disturbed the efficient repair of base lesions induced by MMS (Fig. 7B). Taken together, these data suggest that MBD1 in cooperation with MPG contributes to DNA repair response.
Fig. 6.
MBD1 knockdown increases cell sensitivity to MMS. (A) Effectiveness of siRNA knockdown of MBD1. The cells were transfected with siRNAs targeting mRNAs encoding MBD1 and lamin A/C. MPG and β-tubulin are also detected. (B) Effect of MBD1 knockdown on cell growth inhibition by MMS. Growth inhibition assay was performed as in Fig. 4A. GL3 siRNA was used as a control (41). Values are given as means and standard deviations from five independent experiments.
Discussion
In the present study, we reported that MBD1 specifically interacts with MPG. Both terminal regions of MPG were potentially critical for the association with the TRD of MBD1. The 3D structure of MPG complexed with the substrate DNA showed that MPG binds a DNA adduct without any requirements of additional proteins in vitro (53). It was, however, pointed out that MPG protein used for the structural analysis is an enzymatically active fragment that lacked the N terminus (residues 1–79) of the protein. In living cells, interaction with MBD1 and other molecules is likely to contribute to the functional regulation of MPG. In addition, in vitro gel-shift assays demonstrated that MPG can bind DNAs without sequence specificity and regardless of the presence of modified bases (Fig. 8A, which is published as supporting information on the PNAS web site). MPG seemed to interact with fully methylated DNAs and with MBD1 on the methylated DNAs (Fig. 8B).
As in the cases of TDG and endonuclease III, MPG functions as a transcriptional repressor. By using a luciferase reporter analysis, we found that MPG-dependent repression is not alleviated by histone deacetylase (HDAC) inhibitors (data not shown). TDG acts as a repressor of the transcriptional factors, and TDG-mediated repression was also resistant to the HDAC inhibitors (30). Our recent investigations revealed that MPG physically binds transcriptional factor Sp1 and some components of TFIIH (data not shown). These associations may play a role in MPG-mediated gene repression. The TRD of MBD1 produces strong transcriptional repression activities via interaction with MPG as well as recently identified MBD1-containing chromatin associating factor (MCAF) (36). Interestingly, MPG stably interacts with TRD of MBD1 in the presence of MCAF (data not shown).
On the basis of our data in Fig. 4, we propose a model of dynamics of both proteins for the repair process. MBD1 and MPG normally exist on the methylated promoter, probably for repression and in readiness for DNA damage. On DNA damage by MMS that mostly creates 7-mG, MBD1 is dissociated from the damaged methyl–CpG sites just like a sensor for the damaged bases. Because MBD1 lies in the bottom of the chromatin on methylated DNA regions, the dissociation of MBD1 would stimulate chromatin remodeling and further release the molecules packed in the chromatin. At this stage, MPG distributes widely on the genome to remove the damaged bases and inhibit transcription from these sites. After completion of the repair, MBD1 returned on the repaired methyl–CpG sites together with MPG to reconstruct the repressive chromatin. Recently, Martin et al. (54) proposed a model in which the telomeric heterochromatin serves as a reservoir for many chromatin factors such as Ku and the nucleosome-binding SIR proteins in yeast that are involved in DNA damage response. Similarly, MBD1-based heterochromatin may serve as a reservoir for MPG that responds to numerous base damage.
Compared with 3-methyladenine, it has been thought that 7-mG is relatively innocuous to cells because it appears not to directly interfere with DNA replication (55). Our study, however, suggested that 7-mG in a methyl–CpG pair can alter the chromatin structure due to the inability of MBD1 to bind the damaged methyl–CpG dinucleotide. Alternatively, the dissociation of MBD1 may result from unidentified chromatin change after DNA damage. In transcription-coupled repair, actively transcribed genes are repaired significantly earlier than nontranscribed regions of genome (34). However, it is also important that base damage in nontranscribed heterochromatin regions is properly repaired, because genome-wide DNA damage directly induces chromosome abnormalities and genomic instability (56). Thymine glycosylase MBD4 binds preferentially to methyl–CpG × TpG for the mismatch repair that originates from a methyl–CpG pair (16). On the other hand, MPG coexists with MBD1 in the methylated DNA regions and repairs base damage in both the methylated and unmethylated regions. As was the case of MBD4, the interaction of MBD1 with MPG may function as one of the DNA repair system associated with methyl–CpG dinucleotides. Our study sheds light on the close link between BER, transcription, and chromatin dynamics.
Supplementary Material
Acknowledgments
We thank K. Kubo (Osaka Prefecture University, Osaka) for MPG-expressing Escherichia coli, H. Ide and N. Saitoh for helpful suggestions, and H. Saya for much appreciated support. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas; by a Grantin-Aid for 21st Century Center of Excellence Research from the Ministry of Education, Culture, Sports, Science and Technology; and by a grant from the Uehara Memorial Foundation (to M.N.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: siRNA, small interfering RNA; MBD, methylated DNA-binding domain; TDG, thymine DNA glycosylase; MPG, methylpurine–DNA glycosylase; MMS, methylmethanesulfonate; TRD, transcriptional repression domain; BER, base excision repair; mG, methylguanine; mC, methylcytosine.
References
- 1.Bird, A. (2002) Genes Dev. 16 6–21. [DOI] [PubMed] [Google Scholar]
- 2.Panning, B. & Jaenisch, R. (1998) Cell 93 305–308. [DOI] [PubMed] [Google Scholar]
- 3.Okano, M., Bell, D. W., Haber, D. A. & Li, E. (1999) Cell 99 247–257. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Porath, I. & Cedar, H. (2000) Curr. Opin. Genet. Dev. 10 550–554. [DOI] [PubMed] [Google Scholar]
- 5.Baylin, S. B., Herman, J. G., Graff, J. R., Vertino, P. M. & Issa, J. P. (1998) Adv. Cancer Res. 72 141–196. [PubMed] [Google Scholar]
- 6.Jones, P. A. & Baylin, S. B. (2002) Nat. Rev. Genet. 3 415–428. [DOI] [PubMed] [Google Scholar]
- 7.Baylin, S. & Bestor, T. H. (2002) Cancer Cell 1 299–305. [DOI] [PubMed] [Google Scholar]
- 8.Wade, P. A. & Wolffe, A. P. (2001) Nat. Struct. Biol. 8 575–577. [DOI] [PubMed] [Google Scholar]
- 9.Ng, H. H., Jeppesen, P. & Bird, A. (2000) Mol. Cell. Biol. 20 1394–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujita, N., Shimotake, N., Ohki, I., Chiba, T., Saya, H., Shirakawa, M. & Nakao, M. (2000) Mol. Cell. Biol. 20 5107–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, Y., Ng, H. H., Erdjument-Bromage, H., Tempst, P., Bird, A. & Reinberg, D. (1999) Genes Dev. 13 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wade, P. A., Gegonne, A., Jones, P. L., Ballestar, E., Aubry, F. & Wolffe, A. P. (1999) Nat. Genet. 23 62–66. [DOI] [PubMed] [Google Scholar]
- 13.Lindahl, T. (1993) Nature 362 709–715. [DOI] [PubMed] [Google Scholar]
- 14.Neddermann, P., Gallinari, P., Lettieri, T., Schmid, D., Truong, O., Hsuan, J. J., Wiebauer, K. & Jiricny, J. (1996) J. Biol. Chem. 271 12767–12774. [DOI] [PubMed] [Google Scholar]
- 15.Bellacosa, A., Cicchillitti, L., Schepis, F., Riccio, A., Yeung, A. T., Matsumoto, Y., Golemis, E. A., Genuardi, M. & Neri, G. (1999) Proc. Natl. Acad. Sci. USA 96 3969–3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendrich, B., Hardeland, U., Ng, H. H., Jiricny, J. & Bird, A. (1999) Nature 401 301–304. [DOI] [PubMed] [Google Scholar]
- 17.Scharer, O. D. & Jiricny, J. (2001) BioEssays 23 270–281. [DOI] [PubMed] [Google Scholar]
- 18.Hoeijmakers, J. H. J. (2001) Nature 411 366–374. [DOI] [PubMed] [Google Scholar]
- 19.Wyatt, M. D., Allan, J. M., Lau, A. Y., Ellenberger, T. E. & Samson, L. D. (1999) BioEssays 21 668–676. [DOI] [PubMed] [Google Scholar]
- 20.Hurley, L. H. (2002) Nat. Rev. Cancer 2 188–200. [DOI] [PubMed] [Google Scholar]
- 21.Beranek, D. T. (1990) Mutat. Res. 231 11–30. [DOI] [PubMed] [Google Scholar]
- 22.Elder, R. H., Jansen, J. G., Weeks, R. J., Willington, M. A., Deans, B., Watson, A. J., Mynett, K. J., Bailey, J. A., Cooper, D. P., Rafferty, J. A., et al. (1998) Mol. Cell. Biol. 18 5828–5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelward, B. P., Weeda, G., Wyatt, M. D., Broekhof, J. L., de Wit, J., Donker, I., Allan, J. M., Gold, B., Hoeijmakers, J. H. & Samson, L. D. (1997) Proc. Natl. Acad. Sci. USA 94 13087–13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bohr, V. A., Smith, C. A., Okumoto, D. S. & Hanawalt, P. C. (1985) Cell 40 359–369. [DOI] [PubMed] [Google Scholar]
- 25.Cooper, P. K., Nouspikel, T., Clarkson, S. G. & Leadon, S. A. (1997) Science 275 990–993. [DOI] [PubMed] [Google Scholar]
- 26.Le Page, F., Klungland, A., Barnes, D. E., Sarasin, A. & Boiteux, S. (2000) Proc. Natl. Acad. Sci. USA 97 8397–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chevray, P. M. & Nathans, D. (1992) Proc. Natl. Acad. Sci. USA 89 5789–5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Um, S., Harbers, M., Benecke, A., Pierrat, B., Losson, R. & Chambon, P. (1998) J. Biol. Chem. 273 20728–20736. [DOI] [PubMed] [Google Scholar]
- 29.Minty, A., Dumont, X., Kaghad, M. & Caput, D. (2000) J. Biol. Chem. 275 36316–36323. [DOI] [PubMed] [Google Scholar]
- 30.Missero, C., Pirro, M. T., Simeone, S., Pischetola, M. & Di Lauro, R. (2001) J. Biol. Chem. 276 33569–33575. [DOI] [PubMed] [Google Scholar]
- 31.Tini, M., Benecke, A., Um, S. J., Torchia, J., Evans, R. M. & Chambon, P. (2002) Mol. Cell 9 265–277. [DOI] [PubMed] [Google Scholar]
- 32.Bessho, T. (1999) Nucleic Acids Res. 27 979–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marenstein, D. R., Ocampo, M. T. A., Chan, M. K., Altamirano, A., Basu, A. K., Boorstein, R. J., Cunningham, R. P. & Teebor, G. W. (2001) J. Biol. Chem. 276 21242–21249. [DOI] [PubMed] [Google Scholar]
- 34.Meijer, M. & Smerdon, M. J. (1999) BioEssays 21 596–603. [DOI] [PubMed] [Google Scholar]
- 35.Green, C. M. & Almouzni, G. (2002) EMBO Rep. 3 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujita, N., Watanabe, S., Ichimura, T., Ohkuma, Y., Chiba, T., Saya, H. & Nakao, M. (2003) Mol. Cell. Biol. 23 2834–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujita, N., Takebayashi, S.-i., Okumura, K., Kudo, S., Chiba, T., Saya, H. & Nakao, M. (1999) Mol. Cell. Biol. 19 6415–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miao, F., Bouziane, M., Dammann, R., Masutani, C., Hanaoka, F., Pfeifer, G. & O'Connor, T. R. (2000) J. Biol. Chem. 275 28433–28438. [DOI] [PubMed] [Google Scholar]
- 39.Horton, J. K., Prasad, R., Hou, E. & Wilson, S. H. (2000) J. Biol. Chem. 275 2211–2218. [DOI] [PubMed] [Google Scholar]
- 40.Asaeda, A., Ide, H., Asagoshi, K., Matsuyama, S., Tano, K., Murakami, A., Takamori, Y. & Kubo, K. (2000) Biochemistry 39 1959–1965. [DOI] [PubMed] [Google Scholar]
- 41.Asagoshi, K., Terato, H., Ohyama, Y. & Ide, H. (2002) J. Biol. Chem. 277 14589–14597. [DOI] [PubMed] [Google Scholar]
- 42.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411 494–498. [DOI] [PubMed] [Google Scholar]
- 43.Roy, R., Kumar, A., Lee, J. C. & Mitra, S. (1996) J. Biol. Chem. 271 23690–23697. [DOI] [PubMed] [Google Scholar]
- 44.Merlo, A., Herman, J. G., Mao, L., Lee, D. J., Gabrielson, E., Burger, P. C., Baylin, S. B. & Sidransky, D. (1995) Nat. Med. 1 686–692. [DOI] [PubMed] [Google Scholar]
- 45.Otterson, G. A., Khleif, S. N., Chen, W., Coxon, A. B. & Kaye, F. J. (1995) Oncogene 11 1211–1216. [PubMed] [Google Scholar]
- 46.Wang, W., Sitaram, A. & Scicchitano, D. A. (1995) Biochemistry 34 1798–1804. [DOI] [PubMed] [Google Scholar]
- 47.Plosky, B., Samson, L., Engelward, B. P., Gold, B., Schlaen, B., Millas, T., Magnotti, M., Schor, J. & Scicchitano, D. A. (2002) DNA Rep. 1 683–696. [DOI] [PubMed] [Google Scholar]
- 48.Lewis, J. D., Meehan, R. R., Henzel, W. J., Maurer-Fogy, I., Jeppesen, P., Klein, F. & Bird, A. (1992) Cell 69 905–914. [DOI] [PubMed] [Google Scholar]
- 49.Nan, X., Campoy, F. J. & Bird, A. (1997) Cell 88 471–481. [DOI] [PubMed] [Google Scholar]
- 50.Hendrich, B. & Bird, A. (1998) Mol. Cell. Biol. 18 6538–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohki, I., Shimotake, N., Fujita, N., Jee, J., Ikegami, T., Nakao, M. & Shirakawa, M. (2001) Cell 105 487–497. [DOI] [PubMed] [Google Scholar]
- 52.Li, V. S., Reed, M., Zheng, Y., Kohn, H. & Tang, M. (2000) Biochemistry 39 2612–2618. [DOI] [PubMed] [Google Scholar]
- 53.Lau, A. Y., Scharer, O. D., Samson, L., Verdine, G. L. & Ellenberger, T. (1998) Cell 95 249–258. [DOI] [PubMed] [Google Scholar]
- 54.Martin, S. G., Laroche, T., Suka, N., Grunstein, M. & Gasser, S. M. (1999) Cell 97 621–633. [DOI] [PubMed] [Google Scholar]
- 55.Ezaz-Nikpay, K. & Verdine, G. L. (1994) Chem. Biol. 1 235–240. [DOI] [PubMed] [Google Scholar]
- 56.Engelward, B. P., Allan, J. M., Dreslin, A. J., Kelly, J. D., Wu, M. M., Gold, B. & Samson, L. D. (1998) J. Biol. Chem. 273 5412–5418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.