Abstract
This article extends our previous quantitative analysis of the relationship between the dynamics of the primary structure of DNA and mutagenesis associated with single-strand lesions to an analysis of the production and processing of endogenous double-strand breaks (EDSBs) and to their implications for oncogenesis. We estimate that in normal human cells ≈1% of single-strand lesions are converted to ≈50 EDSBs per cell per cell cycle. This number is similar to that for EDSBs produced by 1.5–2.0 Gy of sparsely ionizing radiation. Although EDSBs are usually repaired with high fidelity, errors in their repair contribute significantly to the rate of cancer in humans. The doubling dose for induced DSBs is similar to doubling doses for mutation and for the induction of carcinomas by ionizing radiation. We conclude that rates of production of EDSBs and of ensuing spontaneous mitotic recombination events can account for a substantial fraction of the earliest oncogenic events in human carcinomas.
The somatic mutations that are so characteristic of cancer may be induced by environmental agents but also occur spontaneously. In a previous communication, we analyzed ionizing radiation (IR)-induced mutations, both germinal and somatic, as a function of dose rate, and identified a narrow range of rates at which induced mutations per unit of dose rate reached a minimum (1). In those analyses, we compared the rates of production of various kinds of radiation damage to DNA with those that occur spontaneously at background levels. However, we were able to consider only single-strand DNA lesions (SSL), because of insufficient published data on the rates of production and background levels of double-strand breaks (DSBs) (2),¶ which play a major role in carcinogenesis, e.g., in chromosomal rearrangements and deletions, and in mitotic recombination (MR) in somatic cells. Now, with recently published data at hand, we present a quantitative analysis of the production of endogenous DSBs (EDSBs) from different kinds of abundant SSLs, and discuss plausible mechanisms of EDSB signaling and repair in normal cells and their relationship to the genetic phenomenology of cancer cells. Functional consequences of the production of DSBs include activation of oncogenes, as in many leukemias, lymphomas, and sarcomas, and loss or inactivation of tumor suppressor (ts) genes, as in many solid tumors, including most carcinomas.
Background Occurrence of DSBs
Rates of Production of DSBs in Human Cells. Normal cells grown in vitro display very little evidence of phenomena commonly associated with DSBs, namely, chromosome and chromatid breaks, large deletions, and rearrangements. Is this because DSBs are rare or because they are so precisely repaired that they leave little microscopic evidence of their occurrence? One indirect manifestation of DSBs is sister chromatid exchange (SCE), which is a signature of homologous recombination (HR), in which crossing-over occurs between sister chromatids. In Bloom syndrome (BS), an inherited condition that predisposes strongly to cancer, including carcinomas, this assay has revealed a rate of SCE ≈10 times more frequent per cell cycle than is observed in control cells (3), whose rate is ≈5 SCEs per cell cycle. This latter value establishes a minimal rate for the spontaneous occurrence of DSBs. If, as is thought, a major function of Blm protein is to process aberrant DNA structures at sites of stalled and broken replication forks and prevent HR events, including SCEs (4), then the 50 or so SCEs observed per cell cycle in BS cells may reveal the number of EDSBs produced per cell cycle. To determine whether this is the case, we turn to data generated during the study of DSBs produced by IR and of cellular responses to them.
One of the earliest responses to an IR-induced DSB is the phosphorylation of a histone, H2AX, at serine 139, yielding a focal product (γ-H2AX) that can be detected by a fluorescent antibody (5–7). Other proteins then colocalize at these sites of DSBs and so become candidate signatures for processing and repair of this damage. These nuclear foci are strikingly apparent after their induction by IR and can be counted. If DSBs occur at a significant rate, without IR exposure, would the same foci appear in normal cells? The answer is, impressively, yes, at least for normal human fibroblasts. Knowing the background rate of focus formation of seven per cell cycle, and comparing it with the number of foci, 52, found in a cell exposed to a given dose (12 Gy) of IR (7), one can calculate a doubling dose (DD) of IR, i.e., the dose of IR that would double the number of foci. From the n-fold increase in the magnitude of the signal after exposure to a dose D of IR, the DD can be calculated as the dose that induces signal with a magnitude equal to the background level, i.e., when n = 2. Thus, if the magnitude of the background signal is one relative unit, and a dose D increases this level n-fold (and therefore, the induced magnitude of the signal is n–1 relative units), the DD = D/(n–1). A summary of these and other published results on foci with γ-H2AX and another protein (7) is presented in Table 1.
Table 1. Protein foci formed in response to DSBs.
| Number |
|||
|---|---|---|---|
| Type | Control | Rad 12 Gy | Induced |
| γH2AX | 7 | 52 | 45 |
| γH2AX/Rad50 | 6 | 32 | 26 |
| γH2AX/Brca1 | 2 | 25 | 23 |
| γH2AX/Rad51 | 2 | 19 | 17 |
| All types | 17 | 128 | 111 |
From these data, we estimate that the overall DD for γ-H2AX foci is 1.5 Gy (= ≈1.5 Sv for γ-rays). Inasmuch as the number of IR-induced DSBs is linearly dependent on the dose of IR, with a slope of ≈30 DSBs per cell per Gy (8, 9), and because the number of foci is probably a constant fraction of induced DSBs (7), we can calculate that a DD of 1.5 Gy would induce ≈45 DSBs above a background of 45 EDSBs. Consequently, the estimated rate of γ-H2AX focus formation for normal human cells corresponds to about the same number of DSBs as that estimated from SCEs observed in BS cells. We note, incidentally, that individual therapeutic doses of IR are usually in the range of this DD for DSBs (≈2 Gy). As we show elsewhere (1), the mutational response of mammalian cells to the genotoxic action of IR depends on the rate of production of spontaneous damage. So, the rates of production of EDSBs are relevant for both spontaneous and environmental mutagenesis, as well as for radiotherapy and some forms of chemotherapy. From a consideration of these data and of data from BS cells, we conclude that the spontaneous rate of production of EDSBs is ≈50 per cell per cell cycle.
Production in Non-Human Cells. DNA replication-associated production of DSBs has also been measured in demembranated sperm nuclei by using extracts from eggs of the African clawed frog, X. laevis, which were immunodepleted of the Mre11-containing complex of proteins to minimize the repair of EDSBs that arise during normal DNA replication. Each chromosome accumulated 5–10 DSBs (10). These haploid nuclei revealed 90 DSBs per sperm DNA equivalent (or 180 per diploid equivalent), a rate that is three to four times higher than that calculated for mammalian somatic cells when the genomic DNA content in Xenopus is taken into account (11).
The production of EDSBs has also been studied in the yeast, S. cerevisiae, whose genome consists of 12 × 106 bp (12). Here, the protein Rad52, which is essential for homologous recombination repair (HRR) and for repair of radiation-induced DSBs in other eukaryotes, relocalizes to form foci after exposure to IR, as it also does in 22% of unexposed S-phase haploid cells (12). Thus, the spontaneous rate per base pair would be at least 0.22 per 12 × 106 bp, or 2 × 10–8 per base pair per cell cycle.
This rate of occurrence of EDSBs seems to apply even to prokaryotes (13, 14). For example, at least 10% of E. coli cells sustain one spontaneously produced DSB per cell cycle (reviewed in ref. 14). Because the E. coli genome comprises 4 × 106 bp, this translates to at least 2.5 × 10–8 DSBs per base pair per cell cycle.
We see for a wide range of species that spontaneous DSBs occur in both germinal and somatic cells at rates, in vitro and, perhaps, in vivo, of 50–180 per 6 × 109 bp, or about one DSB in 108 bp (approximate DNA content of average human chromosome) per cell cycle (Table 2).
Table 2. Rate of production of EDSBs in different species.
| BPs per genome |
EDSBs |
||
|---|---|---|---|
| Species | Cell cycle | 108 bp | |
| Homo sapiens | 6 × 109 | 50 | 0.8 |
| Xenopus laevis | 6 × 109 | 160 | 2.7 |
| Saccharomyces cerevisiae | 1.2 × 107 | One per 4-5 cells | ≈2 |
| Escherichia coli | 4 × 106 | One per 1-10 cells | ≥2 |
Calculations and references can be found in the text.
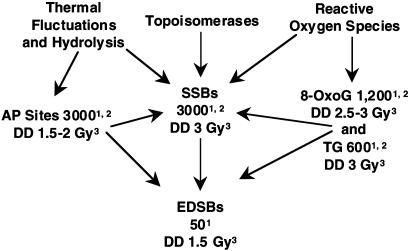
Origin of EDSBs from SSLs. From these considerations it appears that EDSBs are produced at a rate of approximately one per 108 bp per cell cycle and repaired at a probable rate of at least 95% over a broad range of organisms. The production of such breaks is a dynamic process that occurs in two steps. The first step is the abundant production of SSLs of different types during a normal cell cycle, in amounts equivalent to that induced by radiation delivered at a dose rate of ≈0.5 cGy/min (1), and the second step is the conversion of some SSLs into DSBs during the S phase of a cell cycle (Fig. 1).
Fig. 1.
Flow chart of the rates of production of EDSBs from SS lesions in mammalian cells under normal conditions. 1, Rate of production per S phase of cell cycle; 2, see ref. 1; 3, Number in Gy equivalent of IR (= DD).
The most abundant spontaneous SSLs are single-strand breaks (SSBs), apurinic/apyrimidinic (AP) sites, the oxidation products 8-oxoguanine (8-oxoG) and thymine glycol (TG), and some endogenous alkylation products, including 3-methyladenine (3-MA) (1, 15–20). We estimated the rate of production of SSBs and AP sites in vivo at ≈600 per h and of 8-oxoG and TG at a minimum of ≈200 and 100 per cell per h, respectively (ref. 1 and references therein). We do not have an estimate for the rate of production of 3-MA, but it seems to be an order of magnitude below that for SSBs and oxidation base products. The total rate of production of these SSLs is probably ≈1,000 per cell per h and, therefore, on the order of 5 × 103 per S phase, or 10–6 per base pair of DNA per S phase. This rate is ≈100 times the rate calculated for production of EDSBs per cell during DNA replication.
The vast majority of SSLs are repaired by error-free mechanisms, but at least 1% escape repair and are not bypassed. These lesions cause collapse or stalling of replication forks from SSBs or base damage, respectively. Some of these lesions are then converted to DSBs. For example, as shown in bacterial cells, an unrepaired SSB causes collapse of a replication fork and formation of a DSB (14), whereas many TGs, 3-MAs, apurinic sites, and, to a lesser extent, 8-oxoGs, in the template strand cause a block of DNA replication (21–25) and formation of nicks and gaps in DNA that can lead to DSBs and activation of HR. Precise estimates of the rates of these events are not available, but as concluded above, the 5 × 103 SSLs that occur in a normal cell cycle evidently lead to 50 or so EDSBs per cell per cell cycle, a conversion rate of ≈1%. Virtually all of these conversions occur during S phase; therefore, nondividing cells are expected to produce EDSBs at a very low rate. Data for rates of production are not available for such cells (26, 27), but their ambient level is very low (estimated at ≈0.05 per cell) in resting human diploid fibroblasts (27).
We note, too, that this analysis provides a basis for understanding the role of reactive oxygen species (ROS) in the origin of EDSBs under normal conditions. These ROS do not produce DSBs directly at a biologically relevant rate but, rather, indirectly through the formation of SSLs (Fig. 1), which can be converted to DSBs, just as happens for EDSBs in replicating cells under normal conditions.
Repair of EDSBs
HRR. What, then, are the mechanisms for such effective EDSB repair? One such mechanism is revealed by the occurrence of endogenous SCEs and of their large increase in cells from individuals with BS (3), in which there is a defect in the gene that encodes a helicase that is critical in regulating HRR. Apparently in BS cells, the 90% of DSBs that are normally repaired with no detectable effect, as discussed (28–30), are instead repaired by SCEs. To a far lesser extent, there is recombination between chromosomes (MR), which is increased 50- to 100-fold in BS cells (3), as discussed later. An observable effect of this second phenomenon in somatic cells can be loss of heterozygosity (LOH) for markers distal to the site of recombination, one daughter cell becoming homozygous for one set of alleles and the other for the remaining set. Analysis of BS cells reveals that HRR is an important mode of repair/processing of EDSBs and that errors in this repair may contribute substantially to the origin of common human “spontaneous” cancers.
The product of the gene that is mutated in BS, Blm, is one of multiple proteins that are involved in the repair/processing of DSBs by HR (2, 28–36), the key component of which is Rad51. In both undamaged and IR-exposed cells, Brca1 and Brca2 proteins associate with Rad51 in nuclear foci (34, 35). A still larger Brca1-associated genome surveillance complex (BASC) is presumed to be active in the repair of abnormal DNA structures (36), including DSBs. The initial step in HRR is resection of broken DNA ends, catalyzed by a complex of Mre11, Rad50, and Nbs1 proteins, which interacts with Brca1 to enable DNA strand invasion and displacement by Rad51 for “search homology.” The critical role of Brca1 and Brca2 in the HRR of EDSBs is further shown by the extensive chromosomal breakage found in cells nullizygous for both BRCA1 and BRCA2. The fact that germline mutations in BRCA1 and BRCA2 predispose to breast cancer is probably due to the mutagenic effects of estrogen in all tissues, but especially in a tissue expressing estrogen receptors, and suggests that estrogen can be an important factor in the production of EDSBs, whose repair requires Brca1 and Brca2.
Nonhomologous End-Joining (NHEJ). The other major pathway is NHEJ, which operates in all phases of the cell cycle but is particularly active during G1. The proteins that mediate NHEJ include DNA-dependent protein kinase (DNA-PK), which is composed of a catalytic subunit, DNA-PKcs, and a Ku70/Ku80 heterodimer that recruits DNA-PKcs, which in turn reacts with the XRCC4-ligase IV heterodimer to rejoin the broken DNA ends.
The balance between the utilization of the HRR and NHEJ pathways within cells can change. Uniquely HRR proteins (e.g., Rad51 and Rad52) and uniquely NHEJ proteins (Ku70, Ku80, and DNA-PKcs) both bind to DNA ends at DSBs; loss of one can be partially compensated for by the other (37). NHEJ is the chief mode for repair of IR-induced DSBs (2, 8). We have emphasized IR induction of DSBs because of excellent quantitation of dose, but we also note the importance of NHEJ for response to viral infection. For example, HIV infection is thought to kill cells at least partly by activation of an apoptotic cell death pathway. Therefore, the discovery of Daniel et al. (38) that infection of cells that are mutationally defective in NHEJ results in increased apoptosis suggests that the NHEJ pathway can be involved in non-error prone repair of not only IR-induced DSBs but also other induced DSBs.
Single-Strand Annealing (SSA). The third identified major mechanism for the repair of DSBs is SSA, which operates principally in repair of lesions in repetitive DNA sequences, such as Alu elements, and can create chromosomal deletions, insertions, and translocations. Because there is so much DNA that is repetitive, SSA could be a common mechanism of repair. Indeed, translocations between nonhomologous chromosomes are common occurrences in yeast (39). However, this mechanism seems to be suppressed in normal mammalian cells but perhaps not in cancer cells. A possible mechanism in cancer is suggested by the observed stimulation of SSA by the loss of wild-type BRCA2, which in turn is associated with an increased lethality from mitomycin C-induced DNA cross-links. Such a mechanism could explain much of the chromosomal structural instability seen in cancer cells and points to SSA as a possible therapeutic target, because cancer cells would be much more vulnerable than normal cells (40).
Fidelity of Repair of DSBs as a Function of the Rate of Their Production. The fidelity of repair of EDSBs by HRR or NHEJ should be high in normal cells. However, DSBs induced by IR delivered at a high dose rate are repaired with a lower fidelity, in good agreement with a predominant role for NHEJ in rejoining radiation-induced, but not endogenous, DSBs. This seems to imply that HRR, a relatively error-free mechanism, is the principal pathway for repair of EDSBs. But there is evidence that the fidelity of NHEJ in human diploid fibroblasts is strongly dependent on the dose rate of IR (41), being error-prone at high dose rates but not measurably so at low dose rates. Such dependence has been suggested also from the analysis of dose-rate effects on somatic and germ-line mutations (1). IR is usually used experimentally at dose rates of ≈1 Gy/min or more. At this dose rate, the production of mutations is considerably higher per unit of radiation delivered and the fidelity of repair is clearly less than at a dose rate of 0.5 cGy/min. Therefore, it is important to estimate the DSB DD rate, i.e., the dose rate of IR that produces DSBs at a rate similar to that with which EDSBs are produced. Because DNA replication occurs over a period of ≈6 h, EDSBs are produced at a rate of ≈50 every 360 min, or 0.14 per min. IR induces DSBs with a yield of 30 per cell per Gy, or 0.30 per cell per cGy (9). Consequently, at a dose rate of 0.47 cGy/min, IR would induce DSBs at a rate equivalent to that at which EDSBs are produced in replicating cells such as cultured normal human fibroblasts, i.e., 0.47 cGy/min would be a DD rate. Such comparison indicates that the fidelity of NHEJ in the repair of radiation-induced DSBs may be no less than that of HR in the repair of EDSBs. This conclusion is in good agreement with data on the dependence of the fidelity of repair of DSBs on the dose rate in normal human fibroblasts (41); DSBs induced by IR delivered at a dose rate of ≈0.4 cGy/min are repaired with a very low error rate, primarily by NHEJ. Thus, NHEJ can be an effective mode of repair of EDSBs when the usual mechanism of their repair by HRR is overloaded or the rate of their formation is increased by failure of restoration of stalled replication forks, as discussed earlier.
Signaling from DSBs for Their Repair
Local and Global Signaling. A well known response to DNA damage, including DSBs, is the activation of cell cycle checkpoints to allow time for repair of induced DNA damage before its conversion into irreparable genetic alterations or induction of apoptotic cell death. Among the most proximal identified participants in these responses in mammalian cells are three proteins of the lipid kinase phosphatidylinositol 3-kinase family (PI3K) [also called PI3K-like protein kinases (PIKKs)]: Atm (ataxia telangiectasia mutated), Atr (Atm- and Rad3-related), and the DNA-dependent protein kinase complex DNA-PK. After exposure to IR, the activities of Atm, Atr, and DNA-PK are up-regulated and constitute the initiating signal for cellular responses to DSBs. Atr can also be activated by UV light-induced damage and especially by hydroxyurea and other agents that induce replicative stress.
However, activation of DNA-PK in response to DSBs results immediately in activation of NHEJ, which rapidly rejoins DSBs (2, 8, 9, 41). Therefore, signaling from DNA-PK requires a shorter time and is more localized than signaling from Atm and Atr, which also play roles in checkpoint activation. Accordingly, two kinds of DSB signaling can be envisioned, local and global. As noted previously, a key step in HRR is the formation by Rad51 (with the assistance of Brca2) of a nucleoprotein filament on the strand exposed after the initial resection. The message from Atm to Rad51 is mediated by Abl, which thus plays a role in HRR (42, 43).
The DD for the Intensity of DSB Signaling. We estimated earlier the DD for radiation-induced phosphorylation of histone H2AX and its conversion into γ-H2AX protein. Recent evidence indicates that any of the three PIKKs, Atm, Atr, and DNA-PK, can be responsible for this phosphorylation (7, 44). What, then, is the DD for the activation of Atm and DNA-PK (both good candidates for the initiation of signaling from IR-induced DSBs)? If DSBs initiate a signaling pathway for repair, their DDs might be expected to be rate-limiting and therefore similar to the DDs for important steps on the repair pathways. These latter DDs can be calculated from published data (45).
In the normal human lymphoblast cell line GM0536, a clearly detectable increase in Atm kinase activity was observed as early as 5 min postirradiation (5 Gy of γ-irradiation), and the activity was maximally up-regulated (≈4-fold) at 60 min postirradiation (45). If the response in the range of 1–5 Gy were linear, the DD for activation of Atm would be ≈1.7 Gy.
The activation of DNA-PK in response to radiation-induced DSBs is determined by the recruitment of DNA-PKcs to a DSB through binding of the Ku70/Ku80 complex to the DNA ends. A basal level of activated DNA-PK was also measured in normal human lymphoblastoid cells before and after their exposure to 5 Gy of γ-irradiation. DNA-PKcs kinase activity reached a maximum, ≈3-fold, at 120 min postirradiation (45), thereby indicating a DD of ≈2.5 Gy for the activation of this key protein in signaling from, and processing/repair of, DSBs.
Abl is up-regulated after exposure to IR or certain anticancer drugs, such as cisplatin or mitomycin C, that can produce DSBs (43, 45–48). In normal human lymphoblast cell lines, upregulation of Abl kinase activity was detected within 15 min, with maximal activation of 4- to 5-fold at 60 min, after irradiation by 5Gyof γ-irradiation (45). Thus, the presumed DD for activation of Abl kinase in these cells would be in the range of 1.2–1.7 Gy. In normal mouse embryonic fibroblasts (MEFs), activity of the enzyme rises ≈3.5-fold in cells exposed to 5 Gy of γ-irradiation, suggesting a DD of 2 Gy.
Putative Sensors of EDSBs. The estimates of DDs for production of DSBs and for signaling from them all fall in a range of 1.2–2.5 Gy, suggesting that either these kinases themselves or proteins that directly interact with both DSBs and the kinases represent the sensor(s) of the breaks, and that the initial event, the DSB, is rate-limiting for the chain of events that follows. For DNA-PK, there is convincing evidence that the Ku heterodimer is a sensor of DSBs, which, after interaction with the broken ends, attracts and activates the DNA-PKcs.
However, activation of Atm-initiated, and probably also Atr-initiated, signaling depends not only on the presence of DSBs but on a number of other factors whose amounts/activities could be cell-type-specific and dependent on conditions of culture and, of course, the cell's mitotic activity. For example, phosphorylated H2AX recruits a mediator of DNA damage checkpoint protein 1 (MDC1) to DSB sites, thereby apparently enhancing phosphorylation of H2AX and amplifying the response of ATM to DSBs synergistically (49). Therefore, we do not expect that all mammalian cells, cultured under different conditions, would show identical DDs for activation of Atm and proteins that are activated by Atm.
An important publication from Kastan's laboratory (50) reports the activation of Atm by IR-induced damage, presumably by DSBs, based on autophosphorylation. Observations concerning the mechanisms of initiation of DSB signaling by Atm (ref. 50 and references therein), combined with the previously considered findings of almost immediate phosphorylation of histone H2AX in response to DSBs in a dose-dependent manner, strongly suggest a role for chromatin dynamics in the response of mammalian cells to both spontaneous and IR-induced DSBs. However, the DD for activation of autophosphorylation of Atm by IR and of phosphorylation of different substrates by Atm could be different.
The concept of monitoring EDSBs immediately anticipates, in agreement with previous data (34, 35, 51, 52), the presence of at least some DSB signaling proteins in chromatin during S phase. In this context, EDSBs may be referred to as physiological DSBs (with regards to signaling). These EDSBs provide a “noisy” background against which induced DSBs can be recognized by signaling pathways. For the sensing of stimuli in biological systems, recognition of a signal is most effective in the presence of a relatively low, but not too low, background, as discussed (1). Thus, a general principle is emerging: the detection of, and response to, exogenously induced DNA damage of different forms is a comparative process (1). Physiological signaling from EDSBs is certainly quite different from that induced by acute, high-dose-rate IR, which is usually used in both radiobiological and “cell signaling” studies. However, as estimated above, if IR were delivered at a dose rate of ≈0.5 cGy/min, the magnitude of signaling would be comparable with that for endogenous DSB produced during DNA replication. There should be a clear dependence of DSB signaling on the rate of their induction.
Relationships Between Spontaneous and IR-Induced Cancer and DSBs
Oncogenic Events and Errors of DSB Repair. The first events in carcinogenesis are most commonly either translocations that activate or form an oncogene, or mutations or losses of the two alleles of a ts gene. The former characterize many leukemias, lymphomas, and sarcomas, whereas the latter are usual for the common carcinomas. Translocations can result from errors in NHEJ, SSA, or, to a lesser extent, HRR (28–30, 53, 54); however, in most lymphomas, translocations are associated with mechanisms unique to the lymphoid lineage of cells. Mutation and loss of ts alleles by MR constitute major mechanisms in the production of premalignant lesions on the path to carcinomas. The latter show extensive chromosomal instability, including interchromosomal rearrangements, which can obscure early events, and are not discussed here.
HRR and MR. HRR can occur between identical sister chromatids, with or without resulting SCEs, without oncogenic consequence (28, 30, 55, 56). With very low frequency, however, HRR can lead to recombination between chromatids of homologous chromosomes, i.e., MR for alleles beyond the site of recombination. The chief oncogenic consequence of such an event is the conversion of a cell that is heterozygous for a mutation in a ts gene into a cell that is homozygous for the mutation. Judging from data available for several of the best-studied tumors, including retinoblastoma and colorectal adenomas, LOH for a ts gene is a common second event (57–60), and MR is a common cause of LOH (59, 60). Although MR is a common second event during spontaneous oncogenesis, deletion is an important second event during IR-induced oncogenesis. Obviously, MR cannot be a first event because this reaction would be silent in the absence of a mutation.
LOH events due to MR are frequent second events in the inactivation of ts genes, but their frequency does not directly measure the frequency of HRR itself. Although MR relies on the rare somatic pairing of two homologous chromosomes followed by HRR, it affects many genes. Of course, the segment of a chromosome affected by MR varies considerably in length, according to the position of the site of recombination. An estimate of the mean length of an affected segment can be made by considering that the 22 autosomes have a total of 39 arms (5 are acrocentric) and that, if the average event resulted in MR for one-half of the genes on a chromosomal arm, each MR event would affect ≈1% (1/2 × 1/39) of the genome. What, then, is the frequency of MR? In an interesting study of BS cells, Langlois et al. (3) found that the M and N alleles at the glycophorin A locus were recombined with a frequency of 10–3 per gene per cell, whereas these alleles recombined in control cells at a frequency of ≈10–5. Although it is not possible to calculate the frequency of MR per mitosis directly, it seems that it is at least as high as the rate of point mutations (≈2 × 10–7), because a frequency of 10–5 reflects a rate of MR per mitosis that is greater than specific locus mutation rates. However, one recombination event would affect ≈1% of the genome, so a frequency of 10–5 per gene would correspond more closely with a frequency of 10–7 for a MR event. For comparison, suppose, in agreement with available information, that one per million EDSBs is repaired by MR and that 1% of all genes would be affected, as estimated above. Then 50 EDSBs per S phase would yield 5 × 10–7 recombinants per gene per cell per cell cycle. This probably explains why a point mutation is the most common first event in the inactivation of a ts gene but may be even less common than MR as the second event; MR occurs less frequently than does intragenic mutation, but the resultant LOH for a particular locus may be more frequent. For example, in a study of second events at the APC locus in adenomatous polyps, Tomlinson and colleagues (59) reported that 60% of second events were due to MR, the remainder being due to point mutations or small (1–13 bp) intragenic deletions. This study supports the idea that MR occurs in vivo at a rate that is of the same order of magnitude as the rate for intragenic mutations.
As reviewed recently (61), data on the Min mouse model for polyposis (62) also “pointed to MR as the cause of the loss of the normal Apc allele” (61). Furthermore, an increase in LOH rates due to MR was observed in intestinal lesions from Min mice (heterozygous for mutation of the Apc gene) bred on a BS (BLM–/–) background and was accompanied by a great increase in the number of adenomatous polyps (63).
DD for Mutation and Cancer Incidence in Humans. If DSBs contribute substantially to both mutation and the incidence of solid tumors, we may expect to find that the DDs for all three phenomena are similar. Indeed, they are all in the range of 1.2–1.5 Sv. (The biological effectiveness of 1 Sv of IR is the same as that of 1 Gy for sparsely IR such as x-rays and γ-rays.) Mutation data were collected by assays of somatic glycophorin A (GPA) variants, namely, by the occurrence, in a population of erythrocytes from patients of genotype MN, of MM or M_ cells, reflecting the mechanisms of MR (MM) or deletion (M_), respectively (64). The response to IR was linear, with a DD of 1.20 Sv. Data for cancer incidence, obtained from the Life Span Study of atomic bomb survivors in Japan, revealed a DD of 1.59 Sv (64–66). We estimate a DD of 1.52 Sv by confining the analysis to the 12 major solid tumors that meet the criteria set by the United Nations Scientific Committee on Atomic Radiation that “adequate epidemiological data are available from the Life Span Study” (Table 3, which is published as supporting information on the PNAS web site, www.pnas.org, and references therein).
Conclusions
We conclude from our analysis of published data that EDSBs are produced at sites of common SSLs during S phase at a rate of ≈50 or so per cell cycle in mammalian cells. This analysis reconciles an apparent discrepancy that on one hand EDSBs were until recently regarded as too infrequent to be biologically relevant, and that on the other hand, HR is important during normal DNA replication. The rate of EDSB production is estimated as approximately equal to that produced by 1.5 Gy of IR, i.e., 1.5 Gy is a DD for DSBs. Because the length of S phase for cells such as human diploid fibroblasts is ≈6 h, the rate of EDSB production during cell replication is nearly identical to that of DSB induction by IR at a dose rate of 0.5 cGy/min. At this dose rate, mammalian cells show a minimal yield, per unit dose, of IR-induced mutations (1); i.e., repair of induced DSBs is most efficient and precise when they are produced at the above-estimated rates. Evidently, the mechanisms of repair of DSBs have evolved to deal with the dynamic instability of the primary structure of DNA during its replication.
Although the rate and fidelity of repair of EDSBs are optimized, the ensuing low level of errors can account for an important fraction of oncogenic events in humans, notably to the inactivation of ts genes, which are the usual targets in premalignant lesions in the genesis of carcinomas. The first event in the inactivation of a ts gene allele is usually a SSL, and the second event is frequently the result of EDSB-initiated MR. In contrast, IR-induced second events often result from deletions due to errors in repair by NHEJ. Although MR is a rare event, we calculate, taking into account the rate of production of EDSBs, that its rate of targeting the second allele of a ts gene is of the same order of magnitude as the rates of deletion or point mutation and deduce that its consequence is selectable when a cell that is heterozygous for a ts mutation is converted to a homozygous cell (LOH phenomenon).
We have considered, for the sake of simplicity, that DSBs arise uniformly across the genome, although we realize that the well known phenomena of mutational “hot spots” and chromosomal “fragile sites” may be indicative of heterogeneity for vulnerability to EDSBs as well. We also realize that there are significant differences in the repair of DSBs between rodents and humans, so we have relied on human data wherever possible for quantitative conclusions. Further research, using new knowledge of DNA repair, may help to explain differences in oncogenesis between rodents and humans.
Finally, we conclude that many cancers are dysregulated for repair of EDSBs, especially those with loss of function of TP53, and might be targeted by new therapeutic agents. For example, inhibitors of topoisomerases I and II induce DSBs predominantly in S phase, as with EDSBs, and HRR is the predominant mechanism of the repair of both induced and endogenous DSBs produced in this manner. We also note that SSA, a mechanism seldom used for repair of DSBs in human cells, is recruited for repair of EDSBs in cancer cells and also could be a therapeutic target for induction of lethal genomic instability.
Supplementary Material
Acknowledgments
M.M.V. is very grateful to Professor Arthur K. Balin and Dr. Loretta Pratt of The Sally Balin Medical Center for their support. We thank Drs. Anna Marie Skalka and David Livingston for their constructive comments on the manuscript. This research was supported by an appropriation from the Commonwealth of Pennsylvania and a core grant from the National Cancer Institute (CA06927) to the Fox Chase Cancer Center.
Abbreviations: Atm, ataxia telangiectasia mutated; Atr, Atm- and Rad3-related; BS, Bloom syndrome; DD, doubling dose; DNA-PK, DNA-dependent protein kinase; DNA-PKcs, catalytic subunit of DNA-PK; DSB, double-strand break; EDSB, endogenous DSB; HRR, homologous recombination repair; IR, ionizing radiation; LOH, loss of heterozygosity; MR, mitotic recombination; NHEJ, nonhomologous end-joining; SCE, sister chromatid exchange; SSA, single-strand annealing; SSB, single-strand break; SSL, single-strand lesion; ts, tumor suppressor.
Footnotes
Endogenous DNA Damage Symposium, American Association for Cancer Research, Nov. 11–16, 1998, Fort Myers, FL.
References
- 1.Vilenchik, M. M. & Knudson, A. G., Jr. (2000) Proc. Natl. Acad. Sci. USA 97 5381–5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeggo, P. A. (1998) Adv. Genet. 38 185–218. [DOI] [PubMed] [Google Scholar]
- 3.Langlois, R. G., Bigbee, W. L., Jensen, R. H. & German, J. (1989) Proc. Natl. Acad. Sci. USA 86 670–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Constantinou, A., Davies, A. A. & West, S. C. (2001) Cell 104 259–268. [DOI] [PubMed] [Google Scholar]
- 5.Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. (1998) J. Biol. Chem. 273 5858–5868. [DOI] [PubMed] [Google Scholar]
- 6.Rogakou, E. P., Boon, C., Redon, C. & Bonner, W. M. (1999) J. Cell Biol. 146 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paull, T. T., Rogakou, E. P., Yamazaki, V., Kirchgessner, C. U., Gellert, M. & Bonner, W. M. (2000) Curr. Biol. 10 886–895. [DOI] [PubMed] [Google Scholar]
- 8.Iliakis, G. (1991) BioEssays 13 641–648. [DOI] [PubMed] [Google Scholar]
- 9.Stenerlow, B., Karlsson, K. H., Cooper, B. & Rydberg, B. (2003) Radiat. Res. 159 502–510. [DOI] [PubMed] [Google Scholar]
- 10.Costanzo, V., Robertson, K., Bibikova, M., Kim, E., Grieco, D., Gottesman, M., Carroll, D. & Gautier, J. (2001) Mol. Cell 8 137–147. [DOI] [PubMed] [Google Scholar]
- 11.Tymowska, J. (1977) Cytogenet. Cell Genet. 18 165–182. [DOI] [PubMed] [Google Scholar]
- 12.Lisby, M., Rothstein, R. & Mortensen, U. H. (2001) Proc. Natl. Acad. Sci. USA 98 8276–8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blaisdell, J. O. & Wallace, S. S. (2001) Proc. Natl. Acad. Sci. USA 98 7426–7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuzminov, A. (2001) Proc. Natl. Acad. Sci. USA 98 8461–8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vilenchik, M. M. (1971) in Mechanisms of DNA Damage and Repair (Biological Center of the Academy of Sciences, Puschino, Russia), pp. 266–267.
- 16.Lindahl, T. (1993) Nature 362 709–715. [DOI] [PubMed] [Google Scholar]
- 17.Ames, B. N. (1989) Mutat. Res. 214 41–46. [DOI] [PubMed] [Google Scholar]
- 18.Adelman, R., Saul, R. L. & Ames, B. N. (1988) Proc. Natl. Acad. Sci. USA 85 2706–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vilenchik, M. M. (1987) DNA Instability and Late Effects of Ionizing Radiation (in Russian) (Energoatomizdat, T-7-3, P-2b-79, Moscow).
- 20.Ichikawa, N., Watanabe, K., Tomikawa, S., Nagao, T. & Uchida, H. (1996) Transplant Proc. 28 1765–1766. [PubMed] [Google Scholar]
- 21.Loeb, L. A. & Preston, B. D. (1986) Annu. Rev. Genet. 20 201–230. [DOI] [PubMed] [Google Scholar]
- 22.McNulty, J. M., Jerkovic, B., Bolton, P. H. & Basu, A. K. (1998) Chem. Res. Toxicol. 11 666–673. [DOI] [PubMed] [Google Scholar]
- 23.Rouet, P. & Essigmann, J. M. (1985) Cancer Res. 45 6113–6118. [PubMed] [Google Scholar]
- 24.Maccabee, M., Evans, J. S., Glackin, M. P., Hatahet, Z. & Wallace, S. S. (1994) J. Mol. Biol. 236 514–530. [DOI] [PubMed] [Google Scholar]
- 25.Engelward, B. P., Allan, J. M., Dreslin, A. J., Kelly, J. D., Wu, M. M., Gold, B. & Samson, L. D. (1998) J. Biol. Chem. 273 5412–5418. [DOI] [PubMed] [Google Scholar]
- 26.Bonner, W. M. (2003) Proc. Natl. Acad. Sci. USA 100 4973–4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothkamm, K. & Lobrich, M. (2003) Proc. Natl. Acad. Sci. USA 100 5057–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, R. D. & Jasin, M. (2001) Biochem. Soc. Trans. 29 196–201. [DOI] [PubMed] [Google Scholar]
- 29.Jasin, M. (2001) in DNA Damage and Repair, eds. Nicholoff, J. A. & Hoekstra, M. F. (Humana, Totowa, NJ), Vol. 3, pp. 207–231. [Google Scholar]
- 30.Pierce, A. J., Stark, J. M., Araujo, F. D., Moynahan, M. E., Berwick, M. & Jasin, M. (2001) Trends Cell Biol. 11 S52–S59. [DOI] [PubMed] [Google Scholar]
- 31.Cox, M. M. (2001) Proc. Natl. Acad. Sci. USA 98 8173–8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfeiffer, P. (1998) Toxicol. Lett. 96–97 119–129. [DOI] [PubMed] [Google Scholar]
- 33.van den Bosch, M., Lohman, P. H. & Pastink, A. (2002) Biol. Chem. 383 873–892. [DOI] [PubMed] [Google Scholar]
- 34.Scully, R., Chen, J., Plug, A., Xiao, Y., Weaver, D., Feunteun, J., Ashley, T. & Livingston, D. M. (1997) Cell 88 265–275. [DOI] [PubMed] [Google Scholar]
- 35.Sharan, S. K., Morimatsu, M., Albrecht, U., Lim, D. S., Regel, E., Dinh, C., Sands, A., Eichele, G., Hasty, P. & Bradley, A. (1997) Nature 386 804–810. [DOI] [PubMed] [Google Scholar]
- 36.Wang, Y., Cortez, D., Yazdi, P., Neff, N., Elledge, S. J. & Qin, J. (2000) Genes Dev. 14 927–939. [PMC free article] [PubMed] [Google Scholar]
- 37.Allen, C., Kurimasa, A., Brenneman, M. A., Chen, D. J. & Nickoloff, J. A. (2002) Proc. Natl. Acad. Sci. USA 99 3758–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daniel, R., Katz, R. A. & Skalka, A. M. (1999) Science 284 644–647. [DOI] [PubMed] [Google Scholar]
- 39.Haber, J. E. & Leung, W. Y. (1996) Proc. Natl. Acad. Sci. USA 93 13949–13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tutt, A., Bertwistle, D., Valentine, J., Gabriel, A., Swift, S., Ross, G., Griffin, C., Thacker, J. & Ashworth, A. (2001) EMBO J. 20 4704–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rothkamm, K., Kuhne, M., Jeggo, P. A. & Lobrich, M. (2001) Cancer Res. 61 3886–3893. [PubMed] [Google Scholar]
- 42.Chen, G., Yuan, S. S., Liu, W., Xu, Y., Trujillo, K., Song, B., Cong, F., Goff, S. P., Wu, Y., Arlinghaus, R., et al. (1999) J. Biol. Chem. 274 12748–12752. [DOI] [PubMed] [Google Scholar]
- 43.Slupianek, A., Schmutte, C., Tombline, G., Nieborowska-Skorska, M., Hoser, G., Nowicki, M. O., Pierce, A. J., Fishel, R. & Skorski, T. (2001) Mol. Cell 8 795–806. [DOI] [PubMed] [Google Scholar]
- 44.Burma, S., Chen, B. P., Murphy, M., Kurimasa, A. & Chen, D. J. (2001) J. Biol. Chem. 276 42462–42467. [DOI] [PubMed] [Google Scholar]
- 45.Shangary, S., Brown, K. D., Adamson, A. W., Edmonson, S., Ng, B., Pandita, T. K., Yalowich, J., Taccioli, G. E. & Baskaran, R. (2000) J. Biol. Chem. 275 30163–30168. [DOI] [PubMed] [Google Scholar]
- 46.Foray, N., Marot, D., Randrianarison, V., Venezia, N. D., Picard, D., Perricaudet, M., Favaudon, V. & Jeggo, P. (2002) Mol. Cell. Biol. 22 4020–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kharbanda, S., Pandey, P., Jin, S., Inoue, S., Bharti, A., Yuan, Z. M., Weichselbaum, R., Weaver, D. & Kufe, D. (1997) Nature 386 732–735. [DOI] [PubMed] [Google Scholar]
- 48.Wang, J. Y. (2000) Oncogene 19 5643–5650. [DOI] [PubMed] [Google Scholar]
- 49.Lou, Z., Chini, C. C., Minter-Dykhouse, K. & Chen, J. (2003) J. Biol. Chem. 278 13599–13602. [DOI] [PubMed] [Google Scholar]
- 50.Bakkenist, C. J. & Kastan, M. B. (2003) Nature 421 499–506. [DOI] [PubMed] [Google Scholar]
- 51.Andegeko, Y., Moyal, L., Mittelman, L., Tsarfaty, I., Shiloh, Y. & Rotman, G. (2001) J. Biol. Chem. 276 38224–38230. [DOI] [PubMed] [Google Scholar]
- 52.Smith, G. C., Cary, R. B., Lakin, N. D., Hann, B. C., Teo, S. H., Chen, D. J. & Jackson, S. P. (1999) Proc. Natl. Acad. Sci. USA 96 11134–11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gellert, M., Hesse, J. E., Hiom, K., Melek, M., Modesti, M., Paull, T. T., Ramsden, D. A. & van Gent, D. C. (1999) Cold Spring Harbor Symp. Quant. Biol. 64 161–167. [DOI] [PubMed] [Google Scholar]
- 54.Richardson, C. & Jasin, M. (2000) Nature 405 697–700. [DOI] [PubMed] [Google Scholar]
- 55.Haber, J. E. (1999) Trends Biochem. Sci. 24 271–275. [DOI] [PubMed] [Google Scholar]
- 56.Kanaar, R., Hoeijmakers, J. H. & van Gent, D. C. (1998) Trends Cell. Biol. 8 483–489. [DOI] [PubMed] [Google Scholar]
- 57.Zhu, X., Dunn, J. M., Goddard, A. D., Squire, J. A., Becker, A., Phillips, R. A. & Gallie, B. L. (1992) Cytogenet. Cell Genet. 59 248–252. [DOI] [PubMed] [Google Scholar]
- 58.Thiagalingam, S., Laken, S., Willson, J. K., Markowitz, S. D., Kinzler, K. W., Vogelstein, B. & Lengauer, C. (2001) Proc. Natl. Acad. Sci. USA 98 2698–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sieber, O. M., Heinimann, K., Gorman, P., Lamlum, H., Crabtree, M., Simpson, C. A., Davies, D., Neale, K., Hodgson, S. V., Roylance, R. R., et al. (2002) Proc. Natl. Acad. Sci. USA 99 16910–16915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hagstrom, S. A. & Dryja, T. P. (1999) Proc. Natl. Acad. Sci. USA 96 2952–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tischfield, J. A. & Shao, C. (2003) Nat. Genet. 33 5–6. [DOI] [PubMed] [Google Scholar]
- 62.Haigis, K. M. & Dove, W. F. (2003) Nat. Genet. 33 33–39. [DOI] [PubMed] [Google Scholar]
- 63.Luo, G., Santoro, I. M., McDaniel, L. D., Nishijima, I., Mills, M., Youssoufian, H., Vogel, H., Schultz, R. A. & Bradley, A. (2000) Nat. Genet. 26 424–429. [DOI] [PubMed] [Google Scholar]
- 64.Mendelsohn, M. L. (1996) Jpn. J. Cancer Res. 87 inside front cover. [PubMed]
- 65.Thompson, D. E., Mabuchi, K., Ron, E., Soda, M., Tokunaga, M., Ochikubo, S., Sugimoto, S., Ikeda, T., Terasaki, M., Izumi, S., et al. (1994) Radiat. Res. 137 S17–S67. [PubMed] [Google Scholar]
- 66.Pierce, D. A. & Preston, D. L. (2000) Radiat. Res. 154 178–186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.