Abstract
Antibody diversification by somatic hypermutation, gene conversion, and class switch recombination is completely dependent on activation-induced cytidine deaminase (AID). A recent report showing induction of DNA mutations in Escherichia coli by overexpression of AID, Apobec-1, and related members of the RNA-editing cytidine deaminase family suggested that they may directly modify deoxycytidine in DNA in mammalian cells (DNA-editing model). We therefore examined whether Apobec-1 bona fide RNA-editing enzyme could show somatic hypermutation and class switching activities in murine B lymphocytes and fibroblasts. Unlike AID, Apobec-1 was unable to induce somatic hypermutation or class switching. The results force a reevaluation of the physiological significance of the DNA deaminase activities of AID and Apobec-1 in E. coli and in vitro.
Upon encounter with antigen, B lymphocytes undertake three kinds of genetic alterations in their Ig loci: somatic hypermutation (SHM) and gene conversion in the variable region (V) genes and class switch recombination (CSR) in the heavy-chain constant region (CH) genes (1–4). All of these three phenomena with obviously different molecular mechanisms were found to require a common enzyme, activation-induced cytidine deaminase (AID) (5–8), although its reaction mechanism remains in question.
A hypothesis that AID may be an RNA-editing enzyme has been proposed on the basis of AID's sequence similarity with Apobec-1 and the proximity of the two gene loci on human and mouse chromosomes (9). According to this hypothesis, AID deaminates cytosine bases in putative target mRNA precursors for unknown proteins. The edited mRNAs encode endonucleases as a result of the codon change by C to U conversion. As shown in Apobec-1 (10), AID was suspected to depend on cofactors for recognition of target RNA. In fact, AID was shown to require CSR-specific cofactors and possibly separate cofactors for SHM (11). We have shown that exogenous AID can induce hypermutation in the actively transcribed GFP gene in fibroblasts and in T-cell receptor V and c-myc genes in AID-transgenic mice (12, 13). The results indicate that the target mRNA and editing cofactors should be expressed not only in B cells but also in a wide range of tissues. Recently, the AID activity to induce CSR was shown to depend on additional de novo protein synthesis (14). This result is consistent with the RNA-editing model, because edited mRNA must be translated to execute its biological activity.
However, this hypothesis appears to be contradictory to the finding that AID induces mutations in Escherichia coli (15), because evolutionary conservation of the target RNA and cofactors between mammals and prokaryotes appears to be unlikely. Therefore, the simplest common explanation for mutation induction by AID in fibroblasts, T cells, and E. coli is that AID acts directly on DNA and converts deoxycytidine into deoxyuridine. A fraction of thus-generated deoxyuridine may escape base-excision repair and mismatch repair systems, resulting in elevated mutation frequency. In fact, in the E. coli strain deficient in uracil-DNA glycosylase, which is a key molecule in base-excision repair, AID-induced mutation frequency was greatly augmented (16). This DNA-editing model may also explain CSR and gene conversion, both of which are dependent on DNA strand breaks, because it is known that generation of deoxyuridine/deoxyguanosine mismatches induces DNA strand cleavage by the base-excision repair or mismatch repair systems (16). This model gained additional support from the reports that AID could deaminate deoxycytidine in single-stranded DNA in vitro and in E. coli (17–20).
The recent report that the bona fide RNA-editing enzyme Apobec-1 [a catalytic subunit of the apolipoprotein B (apoB) mRNA-editing complex] can mutate DNA in E. coli raised an important question: whether E. coli mutagenesis by AID represents the events associated with SHM in mammalian cells (21). It is therefore essential to test whether Apobec-1 induces mutations in mammalian cells. In this report, we demonstrated that, unlike AID, Apobec-1 could not induce DNA mutation in mammalian B lymphocytes and fibroblasts, whereas both proteins were mutagenic in E. coli. The results suggest that mutagenesis in E. coli by AID may not represent the physiological reactions in mammalian cells.
Materials and Methods
Assays for SHM, CSR, and ApoB mRNA Editing. FMT cells are transfectants expressing a tetracycline-responsive transactivator and a SHM assay construct, pI, in CH12F3-2 cells (14, 22, 23). AID-FLAG and FLAG-Apobec-1 cDNAs were constructed by PCR with gene-specific primers fused with FLAG-coding sequences (sequences are available on request). These cDNAs were subcloned into a retroviral vector pFB (Stratagene). Production of recombinant retroviruses, infection, and the calculation of infection efficiency were performed as described (24). The frequency of GFP-positive cells was analyzed by FACSCalibur (Becton Dickinson). The efficiency of class switching in AID–/– spleen B cells and circle transcripts were measured as described (14, 25, 26).
For apoB mRNA editing, amplification of the edited site in apoB mRNA extracted from HepG2 cells was done by RT-PCR with Pyrobest (Takara Shuzo, Kyoto) with specific primers as described (27). The obtained fragments were cloned into the T vector (Promega) and sequenced.
Immunoprecipitation and Western Blotting. Cells were lysed in a buffer containing 10 mM Tris·HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and Complete Protein Inhibitor Cocktail (Roche). Immunoprecipitation was performed with Protein G Sepharose (Amersham Biosciences) and anti-FLAG polyclonal antibody (Sigma) and analyzed by Western blotting with anti-FLAG M2 antibody (Sigma).
Mutation Measurements. FLAG-tagged AID and Apobec-1 cDNA (a gift from N. O. Davidson, Washington University, St. Louis) were subcloned into pET16b (Novagen). The mutation assay for E. coli was performed as described (21). Briefly after transformation, 10 independent colonies of E. coli cells were expanded individually overnight with 100 μg/ml ampicillin. Then, 108 cells from each culture were plated on rich agar plates containing 100 μg/ml rifampicin (Wako Pure Chemical, Osaka). The numbers of colonies on 10 agar plates were counted separately, and those medians were calculated.
Immunocytostaining and Cell Fractionation. Cells were fixed in 3% paraformaldehyde for 15 min. Immunostaining was performed by incubating fixed cells with biotinylated anti-FLAG M2 antibody for FLAG-tagged Apobec-1 after incubation with streptavidin-rhodamine. Nuclei were stained with Hoechst 33342 (Wako Pure Chemical). Slides were visualized under a Zeiss fluorescence microscope. The images were processed by using photoshop 7.0 software (Adobe Systems, Mountain View, CA). Nuclear and cytoplasmic fractions were separated with NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce). Nuclear and cytoplasmic fractions were monitored by Western blotting with anti-RNA polymerase II and anti-phosphatidylinositol 3-kinase p85 (Santa Cruz Biotechnology), respectively.
Results and Discussion
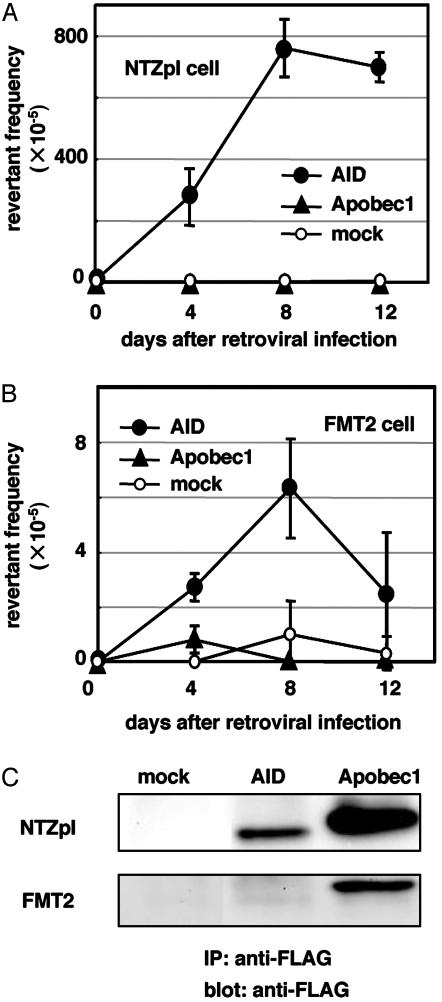
We examined whether Apobec-1 can induce DNA mutations in NIH 3T3 fibroblast lines, in which overexpression of AID can efficiently introduce mutations in actively transcribed GFP genes (13). FLAG-tagged Apobec-1-expressing virus was prepared and used to infect NIH 3T3 fibroblasts harboring the mutant GFP substrate with an artificial stop codon in the coding sequence (NTZpI cells) (28). Induction of SHM was monitored by generation of GFP-expressing cells with flow cytometry. As shown in Fig. 1A, Apobec-1 overexpression did not induce GFP reversion in NTZpI cells. By contrast, smaller amounts of AID expression were sufficient to induce hypermutation (8 × 10–3 bp every 8 days) (Fig. 1 A and C).
Fig. 1.
Appearance of GFP-positive cells by expression of FLAG-tagged AID and Apobec-1 in NTZpI (A) or in FMT2 (B) cells. The frequency of GFP-positive cells was analyzed by FACSCalibur as described in Materials and Methods.(C) FLAG-tagged proteins were immunoprecipitated (IP) from extracts of retrovirus-infected cells and detected by Western blotting.
Next, we wished to study whether Apobec-1 can induce DNA mutations in a B cell line. However, all SHM-prone B cell lines, such as Ramos and BL2, are known to constitutively express AID (29, 30) and are not suitable to assess the effects of Apobec-1 expression on SHM. On the other hand, CH12F3-2 cells can be induced to express AID and undergo CSR by cytokine stimulation (22). However, SHM in Ig V genes was inefficient in stimulated CH12F3-2 cells (T.E. and T.H., unpublished data). To detect low levels of SHM activity, we stably introduced the mutated GFP substrate (28). Twelve of 14 single-copy transfectants showed induction of GFP-positive cells with a frequency of 0.1 × 10–4 to 2.0 × 10–4 on day 3 after cytokine stimulation (data not shown), corresponding to 1 × 10–5 bp per generation at maximum, 1,000-fold higher than the background genome mutation frequency.
FMT2 cells, which expressed GFP most strongly among the 12 transfectants, were used for detailed analysis of mutation induction by cytokine stimulation. Analysis of GFP-coding DNA sequences from 37 GFP-positive FMT2 clones obtained by limiting dilution after FACS sorting confirmed reversion of the stop codon in all cases. There were two patterns of reversion: TAG (stop) to TAC (tyrosine) in 54% (20/37) and TAG to TAT (tyrosine) in 46% (17/37). Both patterns involve the third letter of the triplet and cannot be explained by deamination of deoxycytidine on the opposite strand of DNA followed by deoxyuridine to thymidine conversion by DNA replication (16, 31). Five of 37 clones had additional mutations: four AGC to AAC and one GAC to GGC.
Infection of nonstimulated FMT2 cells with AID virus induced GFP-positive cells at a comparable rate to cytokine-stimulated FMT2 cells (Fig. 1B). In contrast, FMT2 cells infected with Apobec-1 virus did not express GFP. Immunoprecipitation by using anti-FLAG polyclonal antibodies followed by Western blotting with an anti-FLAG monoclonal antibody detected higher levels of Apobec-1 protein than AID in both NIH 3T3 and FMT2 cells (Fig. 1C). These results indicate that exogenous expression of Apobec-1 cannot induce SHM in B cells.
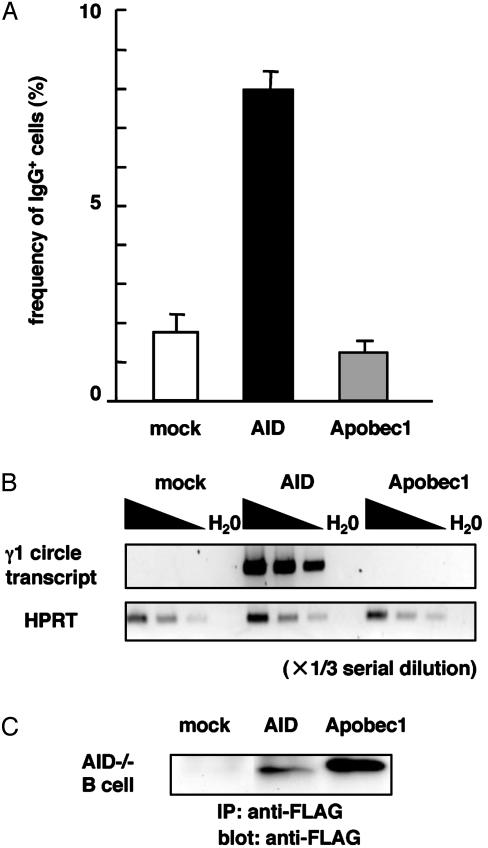
We also tested whether Apobec-1 overexpression can confer CSR in AID-deficient B cells. Spleen cells from AID-deficient mice were cultured in vitro in the presence of lipopolysaccharide and IL-4. After 48 h, Apobec-1- or AID-expressing retrovirus was added to the culture. After 2 days, frequencies of IgG1-expressing cells were measured by flow cytometry. In parallel with the SHM study, expression of AID, but not of Apobec-1, could induce surface expression of IgG1 (Fig. 2A). The absence of IgG1 induction was confirmed by a more sensitive assay to detect γ1 circle transcripts derived from looped-out circular DNA by CSR (26) (Fig. 2B). Immunoprecipitation and Western blotting confirmed protein expression of Apobec-1 in spleen cells, the level of which was higher than that of AID (Fig. 2C). Taken together, these results indicate that Apobec-1 cannot complement AID deficiency with regard to CSR induction.
Fig. 2.
CSR induction in AID-deficient spleen B cells by infection of Apobec-1- or AID-expressing retroviruses. (A) Frequencies of IgG-expressing cells were measured by flow cytometry as described in Materials and Methods. (B) γ1 circle transcripts were measured by RT-PCR. The samples were serially diluted by one-third. HPRT, hypoxanthine–guanine phosphoribosyltransferase. (C) Protein expression of FLAG-tagged proteins in spleen cells was monitored by immunoprecipitation and Western blotting.
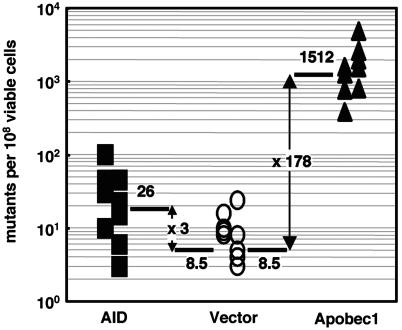
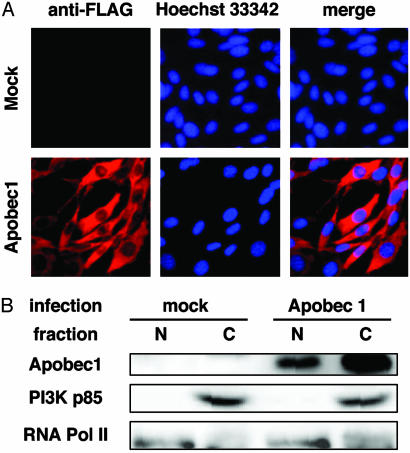
The above conclusion is further supported by the endogenous expression of Apobec-1 in AID–/– spleen B cells and CH12F3-2 cells (T.E. and T.H., unpublished data) although the expression level was less than one-fifth of the transgene. NIH 3T3 cells did not express Apobec-1. Because we constructed a retroviral vector that expresses rat Apobec-1 tagged with the FLAG epitope at its amino terminus, we tested whether the tagged product has intact RNA-editing activity. Apobec-1 tagged with FLAG was introduced in human hepatocellular carcinoma cell line HepG2 cells, which are known to support RNA editing of endogenous apoB 100 mRNA only if Apobec-1 is exogenously provided (27). Sequencing of cloned RT-PCR product amplifying the part of apoB 100 mRNA that surrounds the editing target C at position 6666 revealed conversion of this C to U in 4 of 50 clones sequenced, which was not observed in the mock transfectants (Table 1). This conversion frequency (8%) was comparable to the previous study (27). FLAG tagging of Apobec-1 did not lose induction of rifampicin resistance in E. coli. As shown in Fig. 3, both FLAG-tagged Apobec-1 and AID showed significant increases in rifampicin resistance, which is known to reflect particular mutations in the rpoB gene (21). Apobec-1 has nuclear localization and nuclear exclusion signals and is found in nuclei as well as cytoplasm (32). We examined whether FLAG-tagged Apobec-1 is also found in nuclei by immunocytostaining (Fig. 4A) and biochemical fractionation (Fig. 4B). FLAG-tagged Apobec-1 was expressed in both nucleus and cytoplasm. Thus, it is clear that the absence of SHM and CSR activities in Apobec-1-expressing cells is not caused by the restricted accessibility of FLAG-tagged Apobec-1 to DNA.
Table 1. The apoB mRNA-editing activity of FLAG-tagged Apobec-1.
| Transfection | Clones changed from C to U, % | No. of sequenced clones |
|---|---|---|
| Mock | 0 | 0/50 |
| AID | 0 | 0/50 |
| Apobec-1 | 8 | 4/50 |
Sequencing of cloned RT-PCR product amplifying the part of apoB 100 mRNA surrounding the editing target cytosine at position 6666 was carried out as described in Materials and Methods.
Fig. 3.
Induction of rifampicin resistance in E. coli by FLAG-tagged Apobec-1 and AID. Shown are frequencies of rifampicin-resistant mutants generated after overnight culture of E. coli BL-21 (DE3) carrying the FLAG-tagged Apobec-1 and AID expression plasmid or the vector control. Each point represents the mutation frequency of an independent overnight culture. Mutation frequencies were measured by determining the median number of colony-forming cells that survived rifampicin selection per 108 viable cells. The fold enhancement of the frequency by the Apobec-1 and AID expression is indicated.
Fig. 4.
Subcellular localization of Apobec-1. (A) Hoechst 33342-stained nucleus (blue) and anti-FLAG M2 antibody-stained FLAG-Apobec-1 (red) in infected NIH 3T3 cells. (B) Nuclear (N) and cytoplasmic (C) fractions of NIH 3T3 cells were monitored by the presence of RNA polymerase II and phosphatidylinositol 3-kinase p85, respectively. FLAG-Apobec-1 was immunoprecipitated with anti-FLAG polyclonal antibodies and detected by Western blotting with anti-FLAG M2 monoclonal antibody.
In this paper, we demonstrated that RNA-editing enzyme Apobec-1 could not induce mutagenesis in a B cell line, in which endogenously induced AID can induce mutations in the actively transcribed GFP sequence. The same result was also obtained in the fibroblast line, in which high-level expression of AID induces high-frequency mutations in the GFP substrate (13). These results are in sharp contrast to those from the E. coli system, in which both AID and Apobec-1 induced high-frequency mutations in rpoB and other genes (17, 19, 21). Several possible mechanisms can be discussed. First, the difference may be attributable to the different cell types used (mammalian versus bacterial cells). If this were the case, it would tell us that DNA deamination by Apobec-1 in E. coli does not necessarily represent the real enzymatic activity in mammalian cells. Second, it may be caused by the difference in the target sequence. However, mutations in E. coli were scattered in two genes (rpoB and gyrA) affecting drug sensitivity, indicating that DNA recognition by Apobec-1 does not have strict sequence specificity (21).
In the human genome, there are 10 AID-like cytidine deaminase genes (33), among which at least four gene products (AID, Apobec-1, Apobec3C, and Apobec3G) have DNA deaminase activity in E. coli (21). If all of them are real DNA deaminases in humans, the DNA-editing target should be strictly regulated. Otherwise, uncontrolled mutagenesis would result in frequent tumorigenesis, and the presence of 4–10 redundant DNA deaminases with vague sequence specificity would be dangerous for human survival. Therefore, it appears to be premature to conclude that DNA deamination activity of AID and other cytidine deaminases in E. coli has any physiological relevance in mammalian cells.
In summary, this study pointed out the potential difference in Apobec-1 activity between mammalian cells and E. coli. The same might apply to the activities of AID in E. coli and mammalian cells. More information needs to be accumulated before elucidation of the actual role of AID in vertebrate-specific processes of antibody diversification.
Acknowledgments
We thank Drs. Y. Sakakibara, A. Shimizu, and K. Shimotohno and other members of our lab for discussion; Y. Tabuchi, T. Toyoshima, A. Kawamura, and Y. Sasaki for their technical assistance; and T. Nishikawa, Y. Shiraki, and K. Saito for secretarial help. This research was supported by Center of Excellence Grant 12CE2006 from the Ministry of Education, Science, Sports, and Culture of Japan.
Abbreviations: SHM, somatic hypermutation; CSR, class switch recombination; AID, activation-induced cytidine deaminase; apoB, apolipoprotein B.
References
- 1.Diaz, M. & Casali, P. (2002) Curr. Opin. Immunol. 14 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Honjo, T., Kinoshita, K. & Muramatsu, M. (2002) Annu. Rev. Immunol. 20 165–196. [DOI] [PubMed] [Google Scholar]
- 3.Manis, J. P., Tian, M. & Alt, F. W. (2002) Trends Immunol. 23 31–39. [DOI] [PubMed] [Google Scholar]
- 4.Storb, U. & Stavnezer, J. (2002) Curr. Biol. 12 R725–R727. [DOI] [PubMed] [Google Scholar]
- 5.Muramatsu, M., Kinoshita, K., Fagarasan, S., Yamada, S., Shinkai, Y. & Honjo, T. (2000) Cell 102 553–563. [DOI] [PubMed] [Google Scholar]
- 6.Revy, P., Muto, T., Levy, Y., Geissmann, F., Plebani, A., Sanal, O., Catalan, N., Forveille, M., Dufourcq-Labelouse, R., Gennery, A., et al. (2000) Cell 102 565–575. [DOI] [PubMed] [Google Scholar]
- 7.Arakawa, H., Hauschild, J. & Buerstedde, J. M. (2002) Science 295 1301–1306. [DOI] [PubMed] [Google Scholar]
- 8.Harris, R. S., Sale, J. E., Petersen-Mahrt, S. K. & Neuberger, M. S. (2002) Curr. Biol. 12 435–438. [DOI] [PubMed] [Google Scholar]
- 9.Muramatsu, M., Sankaranand, V. S., Anant, S., Sugai, M., Kinoshita, K., Davidson, N. O. & Honjo, T. (1999) J. Biol. Chem. 274 18470–18476. [DOI] [PubMed] [Google Scholar]
- 10.Chester, A., Somasekaram, A., Tzimina, M., Jarmuz, A., Gisbourne, J., O'Keefe, R., Scott, J. & Navaratnam, N. (2003) EMBO J. 22 3971–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ta, V. T., Nagaoka, H., Catalan, N., Durandy, A., Fischer, A., Imai, K., Nonoyama, S., Tashiro, J., Ikegawa, M., Ito, S., et al. (2003) Nat. Immunol. 4 843–848. [DOI] [PubMed] [Google Scholar]
- 12.Okazaki, I. M., Hiai, H., Kakazu, N., Yamada, S., Muramatsu, M., Kinoshita, K. & Honjo, T. (2003) J. Exp. Med. 197 1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshikawa, K., Okazaki, I. M., Eto, T., Kinoshita, K., Muramatsu, M., Nagaoka, H. & Honjo, T. (2002) Science 296 2033–2036. [DOI] [PubMed] [Google Scholar]
- 14.Doi, T., Kinoshita, K., Ikegawa, M., Muramatsu, M. & Honjo, T. (2003) Proc. Natl. Acad. Sci. USA 100 2634–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin, A. & Scharff, M. D. (2002) Nat. Rev. Immunol. 2 605–614. [DOI] [PubMed] [Google Scholar]
- 16.Petersen-Mahrt, S. K., Harris, R. S. & Neuberger, M. S. (2002) Nature 418 99–103. [DOI] [PubMed] [Google Scholar]
- 17.Pham, P., Bransteitter, R., Petruska, J. & Goodman, M. F. (2003) Nature 424 103–107. [DOI] [PubMed] [Google Scholar]
- 18.Dickerson, S. K., Market, E., Besmer, E. & Papavasiliou, F. N. (2003) J. Exp. Med. 197 1291–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramiro, A. R., Stavropoulos, P., Jankovic, M. & Nussenzweig, M. C. (2003) Nat. Immunol. 4 452–456. [DOI] [PubMed] [Google Scholar]
- 20.Bransteitter, R., Pham, P., Scharff, M. D. & Goodman, M. F. (2003) Proc. Natl. Acad. Sci. USA 100 4102–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris, R. S., Petersen-Mahrt, S. K. & Neuberger, M. S. (2002) Mol. Cell 10 1247–1253. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura, M., Kondo, S., Sugai, M., Nazarea, M., Imamura, S. & Honjo, T. (1996) Int. Immunol. 8 193–201. [DOI] [PubMed] [Google Scholar]
- 23.Shockett, P., Difilippantonio, M., Hellman, N. & Schatz, D. G. (1995) Proc. Natl. Acad. Sci. USA 92 6522–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okazaki, I. M., Kinoshita, K., Muramatsu, M., Yoshikawa, K. & Honjo, T. (2002) Nature 416 340–345. [DOI] [PubMed] [Google Scholar]
- 25.Nagaoka, H., Muramatsu, M., Yamamura, N., Kinoshita, K. & Honjo, T. (2002) J. Exp. Med. 195 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinoshita, K., Harigai, M., Fagarasan, S., Muramatsu, M. & Honjo, T. (2001) Proc. Natl. Acad. Sci. USA 98 12620–12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamuta, M., Oka, K., Krushkal, J., Kobayashi, K., Yamamoto, M., Li, W. H. & Chan, L. (1995) J. Biol. Chem. 270 13042–13056. [DOI] [PubMed] [Google Scholar]
- 28.Bachl, J., Carlson, C., Gray-Schopfer, V., Dessing, M. & Olsson, C. (2001) J. Immunol. 166 5051–5057. [DOI] [PubMed] [Google Scholar]
- 29.Faili, A., Aoufouchi, S., Gueranger, Q., Zober, C., Leon, A., Bertocci, B., Weill, J. C. & Reynaud, C. A. (2002) Nat. Immunol. 3 815–821. [DOI] [PubMed] [Google Scholar]
- 30.Martin, A. & Scharff, M. D. (2002) Proc. Natl. Acad. Sci. USA 99 12304–12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Noia, J. & Neuberger, M. S. (2002) Nature 419 43–48. [DOI] [PubMed] [Google Scholar]
- 32.Yang, Y. & Smith, H. C. (1997) Proc. Natl. Acad. Sci. USA 94 13075–13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarmuz, A., Chester, A., Bayliss, J., Gisbourne, J., Dunham, I., Scott, J. & Navaratnam, N. (2002) Genomics 79 285–296. [DOI] [PubMed] [Google Scholar]