Abstract
The study of immunodominance within microbe-specific CD8 T cell responses has been challenging. We used a previously undescribed approach to create unbiased panels of CD8 cytotoxic T lymphocyte clones specific for herpes simplex virus type 2, a pathogen with a complex genome encoding at least 85 polypeptides. Circulating herpes simplex virus type 2-specific cells were enriched and cloned after sorting for expression of the skin homing-associated receptor, cutaneous lymphocyte-associated antigen, bypassing restimulation with antigen. The specificity of the resultant cytotoxic clones was determined. Clonal frequencies were compared with each other and with the total number of cytotoxic clones. For each subject within the homing receptor-positive compartment, the CD8 cytotoxic response was dominated by T cells specific for only a few peptides. Previously undescribed antigens and epitopes in viral tegument, capsid, or scaffold proteins were immunodominant in some subjects. Clone enumeration analyses were confirmed in some subjects with dominance studies by using herpes simplex mutants, vaccinia recombinants, and/or enzyme-linked immune spots. We conclude that among circulating cells expressing a homing-associated receptor, during chronic herpes type 2 infection, the CD8 T cell response becomes quite focused despite the presence of many potential antigenic peptides.
Keywords: cytotoxic T lymphocytes, cutaneous lymphocyte-associated antigen, lymphocyte homing receptors
Virus-specific T cell receptor αβ+ CD8+ lymphocytes recognize short peptides derived from viral proteins. Large herpes virus genomes encode many potentially antigenic fragments. These agents cause chronic infections with periodic episodes of lytic replication. The immune response contributes to an equilibrium in which periodic viral shedding provides opportunities for transmission, but in which dissemination, organ system dysfunction, and malignant transformation are normally limited.
Herpes simplex virus type 2 (HSV-2) infects ≈22% of U.S. adults (1) and causes mucocutaneous genital ulcerations. CD8 T cells are a critical component of the response to HSV-2. The local infiltration of CD8 cells and HSV-2-specific cytotoxic T lymphocyte (CTL) correlates temporally with viral clearance (2). Cross-sectional studies of HSV-2/HIV-1-coinfected persons show the precursor frequency of virus-specific cytotoxic CD8 T cells is inversely related to the severity of HSV-2 disease (3) and HSV-2 shedding.j Therefore, there is interest in defining “immunodominant” CD8 T cell antigens in HSV-2 for natural history and vaccine research.
Sercarz et al. (4) have defined immunodominant epitopes as those that account for the bulk of the global virus-specific CD8 T cell response on a cell number basis. We use this definition to study CTL responses to HSV-2. To isolate HSV-2-specific CD8 CTL without secondary restimulation with antigen, which could bias the relative abundances of epitope-specific lymphocytes, we used a strategy based on the expression of a tissue-specific homing-associated molecule. HSV-2 infects skin and genital epithelia. A determinant linked to cutaneous lymphocyte-associated antigen (CLA) (5) binds to E-selectin to assist lymphocyte trafficking to skin. Circulating HSV-2-specific CD8 T cells express CLA, bind E-selectin (6), and also express CD28 (6). We used these facts to enrich HSV-2-specific CD8 T cells from peripheral blood mononuclear cells (PBMC) and detailed the fine specificity of resultant CTL.
Materials and Methods
Subjects and Specimens. Subjects were HIV-1 seronegative, HSV-2 seropositive (7) for >1 yr, and were not taking anti-HSV therapy or experiencing symptomatic HSV at blood collection. Protocols were approved by the Institutional Review Board. PBMC were cryopreserved. Some subjects had daily sampling to detect HSV shedding (8). HLA typing used genotypic or serologic methods (9).
Cells, Viruses, and Antigens. Epstein–Barr virus-transformed B cells were cultured from PBMC (10). Vero and BSC40 used DMEM-α with 10% heat-inactivated FCS. CD4 clone 1A.B.25.1 is reactive with HSV-2 virion protein (VP)16 (11). To enrich CTL, PBMC (5 × 106) were stained with FITC-anti-CLA and phycoerythrin (PE)-anti-CD28 (Pharmingen) and PE-Cy5-anti-CD8α (Caltag, Burlingame, CA) and sorted (Vantage II, Becton Dickinson) gating on CD8high CD28+ lymphocytes with >101.6 or <101.0 fluorescence for CLA. Cells were rested overnight in T cell medium with 50 units/ml human IL-2 (Immunex), cloned, and expanded with anti-CD3 (12). For bulk expansion, sorted CD8high CD28+ CLAbright or CD8high CD28+ CLAnegative cells were stimulated with phytohemagglutinin and IL-2 (12) and tested after 14–20 d. To compare CLA-containing and -depleted responders in enzyme-linked immunospot (ELISPOT), PBMC were labeled with biotinylated anti-CLA (Pharmingen) and depleted with magnetic antibiotin beads (Miltenyi Biotec, Auburn, CA); flow-through cells were analyzed with FITC-anti-CLA and phycoerythrin–anti-CD8. Myeloid dendritic cells for ELISPOT are described in Supporting Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Recombinant vaccinia expressing HSV-2 protein-infected cell protein (ICP)27 (13) of HSV-2 have been described. Vaccinia expressing full-length HSV-2 strain 333 (14) unique long part (UL, HSV genome with ORF number) 47 (VP13/14) and UL49 (VP22) were created (15) with vaccinia WR and pSC11 (16) after cloning full-length HSV-2 genes by PCR. Stocks were grown and titered on BSC-40 cells. For proliferation assays, viruses were UV irradiated (11) and used at 1:100.
CTL assays used HSV-2 333 (14) and HSV-1 E115 (17). HSV-2 strains with disrupted or repaired gene UL47, which encodes VP13/14, are detailed in Supporting Methods.
T Cell Epitope Discovery. An expression cloning system using genomic HSV-2 DNA libraries (6, 12) was used as detailed in Supporting Methods and a recent paper (18).
Lymphocyte Functional Assays. Cyotoxicity was tested in 51Cr assays (12). Clone screening used autologous Epstein–Barr virus-transformed B cells targets with or without HSV-2 infection. Bulk T cell lines and established clones were tested in triplicate. HSV and vaccinia infections and peptide loading were done as described (6, 11). Proliferation assays used 3H thymidine incorporation with 104 cloned T cells, 105 autologous irradiated PBMC as antigen-presenting cells per well (11), and UV-irradiated virus with labeling at 72–86 h. ELISPOT is described in Supporting Methods.
Flow Cytometry. Cells were stained with tetramers of HLA B*0702 and VP22 49–57 (B7-RPR) followed by anti-CLA and anti-CD8 (Caltag) (6, 12).
Results
Circulating HSV-2-Specific CTL Express CLA. To evaluate the hypothesis that HSV-2-specific CTL were preferentially present in the CLA+ fraction of CD8+ PBMC, we evaluated sorted and polyclonally expanded CD8+, CD28+ cells expressing either high or low levels of CLA and subsequently tested them for HSV-2-specific cytotoxicity. Virus-specific killing was observed only in CLAhigh cells (Fig. 2, which is published as supporting information on the PNAS web site). In contrast, CLAlow cells did not show detectable cytotoxicity at effector-to-target ratios of up to 50:1. The association of CLA expression and HSV-2-specific cytotoxicity was then examined by cloning sorted CD8+, CD28+, CLAhigh cells with a nonspecific mitogen. Among 10 consecutively studied HSV-2-infected adults (Table 4, which is published as supporting information on the PNAS web site), HSV-2-specific CD8 CTL clones were derived from nine adults. Overall, 14.8% of clones from the CLAhigh fractions were HSV-2-specific. Within individual subjects, the range was 0–31.5% with a mean of 10.5% (Table 1).
Table 1. Clones derived from CD8+ CD28+ CLAhigh PBMC with HSV-2-specific CTL activity.
| Subject | Clones screened | CTL clones* | Percent CTL clones |
|---|---|---|---|
| 1 | 241 | 76 | 31.5 |
| 2 | 319 | 95 | 29.8 |
| 3 | 86 | 14 | 16.3 |
| 4 | 64 | 4 | 6.2 |
| 5 | 96 | 6 | 6.2 |
| 6 | 84 | 5 | 5.9 |
| 7 | 288 | 10 | 3.5 |
| 8 | 194 | 6 | 3.1 |
| 9 | 93 | 2 | 2.1 |
| 10 | 12 | 0 | 0 |
| 11† | 58 | 0 | 0 |
Clones with >25% specific release for autologous HSV-2-infected B cell targets and <10% specific release for autologous uninfected targets.
Seronegative for HSV-1 and -2, control subject.
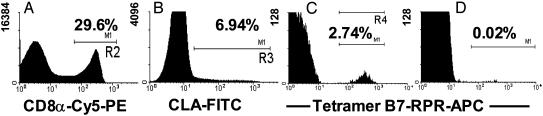
Data consistent with enrichment of HSV-2-specific CD8 T cells in the CLA-positive fraction of PBMC were also obtained by direct staining of PBMC. PBMC from HLA B*0702-positive HSV-2-infected persons, gated for CD8 and CLA expression, were analyzed for binding of tetramer B7-RPR (6). In one example (subject 5), among CD8αhigh CLA+ cells, 2.74% stained with tetramer B7-RPR (Fig. 1). CD8αhigh CLA– cells were 0.02% tetramer-positive. Among six HLA B7+/HSV-2-infected persons, between 0.42% and 2.74% of CLAhigh CD8high lymphocytes stained with tetramer B7-RPR. Staining among CD8high, CLAlow lymphocytes was uniformly <0.05%. Taken together, these data indicate that HSV-2-specific CD8 CTL express CLA in persons with chronic HSV-2 infection.
Fig. 1.
Enrichment of HSV-2-specific cells in the circulating CLAhigh compartment, presented as histograms of cell number vs. fluorescence intensity after sequential multireagent staining. (A) Binding of anti-CD8α antibody to lymphocytes (gated on forward and side scatter, not shown) from subject 6376. (B) CD8α+ cells from A were analyzed for expression of CLA. (C) CLA+ cells from B were analyzed for binding of allophycocyanin-conjugated tetramer B7-RPR (6). (D) CLA– cells from B were analyzed for binding of tetramer B7-RPR. The y axes differ for A–D. The percentages of positive cells are shown.
Definition of HSV-2 CD8 T Cell Antigens and Epitopes. To examine patterns of dominance in the CD8 CTL response to HSV-2, we determined the fine specificity of panels of HSV-2-specific clones. For subjects 1 and 2 (Table 1), well growing CTL screen-positive clones were randomly selected for expansion (19) and detailed studies due to resource limitations. Clones restricted by HLA A*0201 or B0702 were checked with known epitopes in VP13/14, VP22, ICP0, or glycoprotein D or B (6, 10, 12, 20, 21). Some reacted with previously known epitopes (Table 2), but these accounted for a minority of the CTL clones.
Table 2. Fine specificity of HSV-2-specific CD8 CTL clones made from directly sorted CLAhigh PBMC.
| Subject identification, HLA | Clones tested | No. of clones (%) | Restricting allele† | HSV-2 ORF‡ | HSV-2 protein‡ | Amino acid‡ | HSV reactivity§ |
|---|---|---|---|---|---|---|---|
| 1 | 34 | 18 (53) | A*0201 | UL47 | VP13/14 | 551-559 | TS |
| A*02, *23 B*58, *35 | 7 (20) | B*5801 | UL54 | ICP27 | Unknown | TS | |
| 5 (15) | A*0201 | UL47 | VP13/14 | 289-298 | TS | ||
| 2 (6) | A*0201 | UL54 | ICP27 | Unknown | TS | ||
| 2 (6) | Unknown | ||||||
| 2 | 11 | 6 (55) | B*1402 | UL25 | UL25 | 405-413 | TS |
| A*01, *32 B*14, *27 | 3 (27) | B*1402 | UL7 | UL7 | 174-186 | TS | |
| 1 (9) | B*27052 | US6 | gD2 | 365-373 | TS | ||
| 1 (9) | DR*01¶ | Unknown | |||||
| 3 | 14 | 9 (64) | B*1402 | UL25 | UL25 | 405-413 | TS |
| A 2,3 B 14 | 5 (36) | B*1402 | UL7 | UL7 | 174-186 | TS | |
| 4 | 4 | 4 (100) | A*0201 | UL47 | VP13/14 | 551-559 | TS |
| A*02, *24 B*44 | |||||||
| 5 | 6 | 4 (67) | B*0702 | UL49 | VP22 | 49-57 | TS |
| A*02, *03 B*07, *44 | 2 (34) | B*0702 | UL26 | UL26| | 475-483 | TS | |
| 6 | 5 | 2 (40) | B*0702 | RL2 | ICP0 | 743-751 | TS |
| A1, 26 B7, 8 | 2 (40) | A*0101 | UL46 | VP11/12 | 354-362 | TC | |
| 1 (20) | B*0702 | UL49 | VP22 | 49-57 | TS | ||
| 7 | 7 | 7 (100) | B*37 | Unknown** | TS/TC** | ||
| A*02, *24 B*07, *37 | |||||||
| 8 | 5 | 2 (40) | B*5701 | US8 | gE2 | 518-526 | TS |
| A1, 2 B8, 57 | 2 (40) | A*0201 | UL47 | VP13/14 | 551-559 | TS | |
| 1 (20) | A*0201 | UL47 | VP13/14 | 289-298 | TS |
Data from cDNA positive in Cos-7 cotransfection assay except for partially matched APC (subject 7) or mAb inhibition (subject 2, HLA DR-restricted clone).
From ref. 22. Not all HSV proteins have names separate from gene designations.
Type-specific (TS) clones lyse only HSV-2-infected cells. Type-common (TC) clones lyse both HSV-1- and -2-infected cells.
A single DR*01-restricted CD4+ CD8smear+ TCRαβ+ CD3+ CTL clone was detected (data not shown).
Nomenclature for proteins encoded by overlapping UL26/UL26.5 genes is complex (see text).
Cell culture failure prevented workup. One clone is HSV TC, and six are HSV-2 TS.
Epitope discovery for the remaining clones began with determining their HLA-restricting alleles. Clear evidence of HLA class I restriction was obtained (not shown), with a few exceptions (below). One or two restricting HLA class I alleles were detected per subject (Table 2). As HSV-2 DNA fragments encoding previously undescribed antigens were uncovered (Table 3) by expression cloning (12, 18), the remaining HLA-appropriate clones were assayed with these fragments. As peptide epitopes were defined (below), clones with suitable HLA restriction were retested with relevant peptides.
Table 3. HSV-2 DNA fragments stimulating IFN-γ secretion by CD8 CTL clones with specificities derived by CLA-based sorting of PBMC.
| Subject | CD8 clone | Cotransfected HLA cDNA | HSV-2 genomic DNA fragment | HSV-2 gene(s)† | HSV-2 protein fragment(s): amino acids |
|---|---|---|---|---|---|
| 3 | F8 | B*1402 | 17,406-17,824 | UL7 | UL7: 50-192 |
| 2 | 1F3 | B*1402 | 49,999-50,287 | UL25 | UL25: 322-417 |
| 5 | 1E4 | B*0702 | 52,594-52,910 | UL26/UL26.5 | UL26: 404-627 |
| 6 | E2 | A*0101 | 99,085-100,838 | UL46 | VP11/12: 254-722 |
| 2 | 2B9 | B*27052 | 142,038-142,393 | US6 | gD2: 342-393 |
| 8 | 2H1 | B*5701 | 145,347-146,693 | US8, US9‡ | gE2: 503-545, US9: 1-89 |
HSV-2 genes predicted to be forward and in-frame with vector translational start or to follow their endogenous promoter (22).
A portion of US8.5 is forward but out of frame. A portion of US10 is present but backward.
Six previously undescribed CTL epitopes were discovered during the study of 36 clones from five subjects. In four cases, the initial positive HSV-2 genomic DNA library fragments contained portions of a single known HSV-2 ORF (Table 3): UL7, UL25, UL46, or unique short part (HSV genome with ORF number) (US)6. In contrast, clone 8.2H1 (Table 3) reacted with a 1.35-kB genomic fragment containing part of gene US8, all of US9 and its promoter, and parts of US8.5 and US10 of frame or backward. Only US8, which encodes glycoprotein E, was active in cotransfection assays with HLA B*5701 (not shown). CD8 clone 5.1E4 (Table 3) reacted with 316-bp fragment in-frame both the UL26 and internally overlapping UL26.5 ORFs (22). Each previously undescribed antigen was confirmed with a peptide (Table 2 and Fig. 3, which is published as supporting information on the PNAS web site). For CD8 clone 3.F8 (Table 3), truncation analysis reduced the antigenic region to UL7 amino acids 150–192 (not shown), followed by evaluation of overlapping 13-mer peptides in CTL assays. For the other clones (Table 3), sequence data, HSV-1 and -2 reactivity, and HLA peptide-binding motifs allowed targeted syntheses of candidate peptides. Each CTL clone recognized a synthetic peptide at low concentrations, with 50% responses near 1 nM (Fig. 3).
Two of the previously undescribed antigens were HSV-2 virion tegument proteins. Clone 6.E2 (Table 3) recognized VP11/12 (23), the product of gene UL46. Clone 3.F8 (Table 3) and similar clones recognized the product of gene UL7 (24). Two of the antigens were capsid or capsid-associated proteins. VP26, encoded by UL25 (25), was recognized by CTL from two subjects. A protein in the capsid scaffold, encoded by UL26 or UL26.5, was also recognized. The scaffold is a framework for capsid assembly (25). Other previously undescribed CD8 epitopes were in glycoproteins. Clone 2.2B9 reacted with glycoprotein D (22, 26). Clone 8.2H1 recognized a peptide in glycoprotein E. CD8 responses to gE, UL46, UL7, scaffold (UL25), or capsid (UL26) proteins of HSV-2 are previously undescribed. Peptide sequences are in GenBank (accession no. NC_001798).
Patterns of Antigen and Epitope Immunodominance. CD8 CTL responses were detected to a maximum of three antigens per person (Table 2). For subject 4, each clone reacted with a single epitope in VP13/14; this expansion was detected in this subject with direct tetramer staining of PBMC (6). For subject 1, all 34 clones reacted with either VP13/14 or ICP27 (see below). Subjects 2 and 3 both had the HLA B*1402 genotype and predominantly recognized UL7 and UL25 in the context of HLA B*1402. HLA B*1402-restricted responses were dominant: subject 3 had a single B*2705-restricted clone recognizing gD2. Two-thirds of the clones from subject 5 recognized VP22 49–57; this expansion was detected with direct tetramer staining of PBMC [0.6% of CD8hi cells (6)]. Although the specificity of clones from subject 7 was not determined, all seven HSV-2-reactive clones were HLA B*37-restricted, consistent with an oligoclonal response, although it is possible that up to seven different peptides were recognized.
Immunodominance can also be studied within populations. HLA A*0201 is the most common HLA allele in most ethnic groups (27). Among six HLA A2-positive persons, we detected reactivity with VP13/14 amino acids 551–559 in three persons and clonal reactivity with 289–298 in two. Among three HLA B7-positive subjects, two had clones reactive with VP22 49–57 and one had an ICP0 743–751-reactive clone. Some subjects with the appropriate HLA alleles had no detectable responses to peptides restricted by these alleles. For example, neither subject 3 nor 7, both HLA A2-positive, had A2-restricted responses among 23 clones studied, and subject 7 had no HLA B7-restricted clones detected. Among other subjects, HLA A2 or HLA B7 dominated the response.
The relative immunogenicity of different structural and kinetic classes of HSV-2 proteins was compared within the population. Among 86 independent HSV-2-specific CD8 CTL clones derived by CLA-sorting, 45 (52.3%) recognized tegument, 15 (17%) recognized capsid, 3 (3%) recognized envelope glycoproteins, and 2 (2%) recognized scaffold proteins (Table 2). An additional 11 (13%) recognized nonstructural immediate early proteins, either ICP0 or ICP27 (below). The specificities of 10 clones (12%) were not determined. All seven subjects analyzed had tegument-specific clones.
Immunodominance Studies with Bulk CTL Cultures. In selected subjects, we investigated the specificity of CLA+ HSV-2-specific CTL by using alternate assay formats. Bulk lymphocyte cultures derived from sorted PBMC were tested with target cells expressing, or failing to express, defined HSV-2 genes (Fig. 2 and Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org). To isolate responses to VP13/14, previously undescribed reagents were validated. VP13/14-specific clones failed to lyse del47, a deletion mutant not expressing UL47, but recognized 47rev, a rescue virus (see Supporting Methods). The disruption strategy for UL47 risked disturbing proteins VP11/12 and VP16, encoded by adjacent genes UL46 and UL48. VP11/12- and VP16-specific T cell clones recognized the del47 and 47rev. Southern blots of restriction endonuclease-digested DNA (28) from cells infected with HG52, 47del, and rev47 gave the expected patterns with probes from UL46 and UL47 and eGFP (data not shown). Recombinant vaccinia expressing VP13/14 (gene UL47) were specifically lysed (Fig. 4) by relevant CD8 CTL (12).
For subjects 1 and 4, deletion of UL47 (VP13/14) was associated with significantly decreased lysis of HSV-2-infected cells (Fig. 2). Restoration of UL47 restored lysis to wild-type levels. Bulk effectors also lysed target cells infected with vaccinia-VP13/14. Target cells pulsed with a single peptide, VP13/14 551–559, were recognized by bulk CTL from subject 4, in agreement with clonal analysis (Table 2). Subject 1 showed significant lysis of cells infected with vaccinia-ICP27, consistent with clonal analysis.
Comparison of CLA-Expressing and -Depleted Responder Populations. In previous work, bulk cultures and clones suitable for CTL assays required preliminary bulk expansion with HSV-2-infected cells as antigen-presenting cells (20, 29–31). Our present approach, based on CLA expression, avoids this step. We compared whole and CLA-depleted PBMC responder populations directly ex vivo by using an IFN-γ secretion readout. Flow cytometry documented >90% depletion of CLA+ cells among CD8+ lymphocytes from each subject. Subjects were evaluated for responses to known HLA-appropriate epitopes by using ELISPOT with synthetic peptides. Reduction in spot-forming units/106 responders, usually >50%, was observed after depletion of CLA+ cells for most subjects and epitopes (Table 5, which is published as supporting information on the PNAS web site). Subdominant responses, which were not detected in the clonal enumeration of clones with CTL activity, were also detected.
Type Specificity of HSV-Reactive CD8 CTL. Among the previously undescribed CD8 T cell epitopes (Table 3), one, HSV-2 UL46 amino acids 354–362, is identical in the analogous HSV-1 protein (32, 33). Clones recognizing this epitope lysed both HSV-1- and –2-infected cells (data not shown). Every clone tested recognizing the other five previously undescribed epitopes (Table 3), previously described epitopes (6, 12), and ICP27-specific clones from subject 1 was HSV-2 type-specific. Among the seven HLA B*37-restricted CTL clones from subject 7, one was type-common (not shown). Overall, one of nine precisely known HSV-2 CD8 epitopes and three of 86 HSV-2-reactive CD8 CTL clones described in this report were type-common.
Discussion
In this work, we use a previously undescribed one-step purification method, based on the expression of a skin homing-associated receptor, CLA, to enrich HSV-2-specific CD8 T cells. HSV-2 is in the subfamily Alphaherpesvirinae, a group with tropism for the skin. By using this strategy, we find a striking concentration, within the CLA-expressing subset of PBMC, of virus-specific CD8 CTL responses to just a few antigens and epitopes per subject.
The CTL we recovered by CLA sorting recognized diverse viral antigens. Responses to tegument proteins VP13/14 and VP22 and the immediate early protein ICP0, observed in HSV-2-infected tissue (9, 12), were confirmed in blood. Clonal recognition of ICP27 confirms work with bulk CTL (30). Responses to structural envelope, capsid, and scaffold proteins were also observed. The UL25 gene product, VP26, is present at low levels (≈40 copies) in mature capsids (25). The protein products of the UL26 and UL26.5 genes are a complex polypeptide family (34) in the capsid scaffold. Analyses of HSV CD8 T cell antigens should therefore span the whole proteome.
By producing panels of independently derived CTL clones from several subjects and determining their fine specificity, we could study, within the CLA-expressing compartment, the frequency distribution of CTL precursors recognizing specific viral epitopes. In most subjects, only a few HSV peptides accounted for most of the circulating CTL precursors. For subjects 1 and 7, identification of peptide epitopes was not completed for each CTL clone, and it is possible that responses were diverse in these individuals. For subject 1, the observed response was spread over a minimum of four epitopes, but these occurred within two proteins (VP13/14 and ICP27). Strikingly, two persons with the B*1402 allele, subjects 2 and 3, both displayed immunodominant B*1402-restricted responses to the same two epitopes among their CLA-positive blood lymphocytes. In this regard, HSV-2 may be similar to HIV-1, in which specific epitopes tend to be immunodominant in HLA B*1402-positive persons (35).
CLA expression appears to be a common property of HSV-specific CTL: aggregate published (6) and current peptide, HLA restriction, and vaccinia data indicate that at least 13 HLA class I-restricted HSV-2 CD8 epitopes are recognized by circulating CLA+ cells. We cannot exclude that CLA-negative CD8 cells may recognize additional, unknown, and possibly immunodominant HSV-2 epitopes. Experiments with peptides or whole-ORF reagents covering the entire proteome will be required to address this question. CD28 expression was used as an adjunctive criterion for cell sorting (6). The expression of CD28 by virus-specific CD8 T cells appears to vary among viruses (36) and can vary among epitopes and individuals (37, 38). Further research is required to determine whether significant levels of CTL are present in CD28-negative cells.
Optimally, both the identity of immunodominant HSV-2 epitopes and their relative numerical prevalence in the circulation would be confirmed with more than one assay. The immunodominant epitopes detected among CLA-expressing cells by using clonal analysis and CTL readout were generally the same as those eliciting the largest responses in IFN-γ ELISPOT. The CLA sorting/CTL method is not as sensitive as IFN-γ ELISPOT in detecting minor specificities (Table 5). Although all CTL clones tested also secrete IFN-γ, we do not know whether the cells detected by IFN-γ ELISPOT also have CTL activity.
Our data indicate that HLA genotype is not the most important factor in determining immunodominance. Within our group of HLA A*02-bearing persons, A*0201-restricted clones were commonly detected in some subjects, such as 1 and 4, but were not detected in subject 3 or 5. Similarly, B*0702 was internally dominant in subject 5, but no B*07-restricted clones were detected in the panel of CTL from subject 7. In animal models, several factors other than multihistocompatibility complex haplotype can influence immunodominance (39–43). HLA haplotypes at the nonrestricting allele can influence the CD8 response (44). Crossreactivity occurs between self HLA/HSV-2 peptide and allogeneic HLA molecules (9, 20); self antigens may therefore modify the CD8 response to HSV-2.
The identification of immunodominant HSV-2 CD8 CTL antigens may have practical applications. HSV-2 causes serious infections (45) and may double the risk of HIV-1 acquisition (46). A vaccine eliciting antibody and CD4 responses had only partial efficacy, limited to HSV-1 and -2 dually seronegative women (47). Overall, our data indicate that in the context of chronic viral infection, CD8 responses specific for a limited number of epitopes and antigens become immunodominant among cells expressing a pathogen-appropriate homing-associated receptor, and that the specificity of the numerically immunodominant clones cannot be predicted from HLA typing. Further studies of possible correlations among homing receptor expression, abundance, and specificity of circulating HSV-2-specific CD8 cells exhibiting CTL or other functional activities and HSV-2 disease severity are required to fully justify vaccine strategies targeting CD8 responses. The methods and specificities in this report may contribute to these efforts.
Supplementary Material
Acknowledgments
B. Rouse (University of Tennessee, Knoxville) provided vaccinia-ICP27; Dr. S. Riddell (Fred Hutchinson Cancer Research Center) supplied purified mAb W6/32; A. Sutherland, L. Misher, and G. Spies assisted with vaccinia-UL47; and S. Selke provided database support. This work was supported by National Institutes of Health Grants AI50132, AI30731, and AI48135.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HSV-2, herpes simplex virus type 2; CTL, cytotoxic T lymphocyte; CLA, cutaneous lymphocyte-associated antigen; PBMC, peripheral blood mononuclear cells; ICP, infected cell protein; UL, unique long part (HSV genome with ORF number); US, unique short part (HSV genome with ORF number); VP, virion protein; ELISPOT, enzyme-linked immunospot.
Footnotes
Posavad, C. M., Wald, A., Kuntz, S., Huang, M.-L., Zeh, J., Selke, S. & Corey, L. (2001) 26th International Herpesvirus Workshop, July 2001, Regensberg, Germany (abstr.).
References
- 1.Fleming, D. T., McQuillan, G. M., Johnson, R. E., Nahmias, A. J., Aral, S. O., Lee, F. K. & St Louis, M. E. (1997) N. Engl. J. Med. 337 1105–1111. [DOI] [PubMed] [Google Scholar]
- 2.Koelle, D. M., Posavad, C. M., Barnum, G. R., Johnson, M. L., Frank, J. M. & Corey, L. (1998) J. Clin. Invest. 101 1500–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Posavad, C. M., Koelle, D. M., Shaughnessy, M. F. & Corey, L. (1997) Proc. Natl. Acad. Sci. USA 94 10289–10294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sercarz, E. E., Lehmann, P. V., Ametani, A., Benichou, G., Miller, A. & Moudgil, K. (1993) Annu. Rev. Immunol. 11 729–766. [DOI] [PubMed] [Google Scholar]
- 5.Picker, L. J., Michie, S. A., Rott, L. S. & Butcher, E. C. (1990) Am. J. Pathol. 136 1053–1068. [PMC free article] [PubMed] [Google Scholar]
- 6.Koelle, D. M., Liu, Z., McClurkan, C. M., Topp, M., Riddell, S. R., Pamer, E. G., Johnson, A. S., Wald, A. & Corey, L. (2002) J. Clin. Invest. 110 537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashley, R. A., Militoni, J., Lee, F., Nahmias, A. & Corey, L. (1988) J. Clin. Microbiol. 26 662–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wald, A., Corey, L., Cone, R., Hobson, A., Davis, G. & Zeh, J. (1997) J. Clin. Invest. 99 1092–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koelle, D. M., Chen, H. B., McClurkan, C. M. & Petersdorf, E. W. (2002) Blood 99 3844–3847. [DOI] [PubMed] [Google Scholar]
- 10.Koelle, D. M., Tigges, M. A., Burke, R. L., Symington, F. W., Riddell, S. R., Abbo, H. & Corey, L. (1993) J. Clin. Invest. 91 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koelle, D. M., Corey, L., Burke, R. L., Eisenberg, R. J., Cohen, G. H., Pichyangkura, R. & Triezenberg, S. J. (1994) J. Virol. 68 2803–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koelle, D. M., Chen, H., Gavin, M. A., Wald, A., Kwok, W. W. & Corey, L. (2001) J. Immunol. 166 4049–4058. [DOI] [PubMed] [Google Scholar]
- 13.Manickan, E., Francotte, M., Kuklin, N., Dewerchin, M., Molitor, C., Gheysen, D., Slaoui, M. & Rouse, B. T. (1995) J. Virol. 69 4711–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kit, S., Kit, M., Qavi, H., Trkula, D. & Otsuka, H. (1983) Biochim. Biophys. Acta 741 158–170. [DOI] [PubMed] [Google Scholar]
- 15.Earl, P. L. & Moss, B. (1993) in Current Protocols in Molecular Biology, eds. Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. M., Seidman, J. G., Smith, J. A. & Struhl, K. (Wiley, New York), Vol. 2, pp. 16.17.1–16.17.6. [Google Scholar]
- 16.Chakrabarti, S., Brechling, K. & Moss, B. (1985) Mol. Cell. Biol. 5 3403–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spruance, S. L. & Chow, F. S. (1980) J. Infect. Dis. 142 671–675. [DOI] [PubMed] [Google Scholar]
- 18.Koelle, D. M. (2003) Methods 29 213–226. [DOI] [PubMed] [Google Scholar]
- 19.Brodie, S. J., Lewinsohn, D. A., Patterson, B. K., Jiyamapa, D., Krieger, J., Corey, L., Greenberg, P. D. & Riddell, S. R. (1999) Nat. Med. 5 34–41. [DOI] [PubMed] [Google Scholar]
- 20.Tigges, M. A., Koelle, D. M., Hartog, K., Sekulovich, R. E., Corey, L. & Burke, R. L. (1992) J. Virol. 66 1622–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tigges, M. A., Leng, S., Johnson, D. C. & Burke, R. L. (1996) J. Immunol. 156 3901–3910. [PubMed] [Google Scholar]
- 22.Dolan, A., Jamieson, F. E., Cunningham, C., Barnett, B. C. & McGeoch, D. J. (1998) J. Virol. 72 2010–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willard, M. (2002) J. Virol. 76 5220–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nozawa, N., Diakoku, T., Yamauchi, Y., Takakuwa, H., Goshima, F., Yoshikawa, T. & Nishiyama, Y. (2002) Virus Genes 24 257–266. [DOI] [PubMed] [Google Scholar]
- 25.Ogasawara, M., Suzutani, T., Yoshida, I. & Azuma, M. (2001) J. Virol. 75 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. (2001) J. Mol. Biol. 305 567–580. [DOI] [PubMed] [Google Scholar]
- 27.Marsh, S. G. E., Parham, P. & Barber, L. D. (2000) The HLA FactsBook (Academic, San Diego).
- 28.MacLean, A. R. (1998) in Methods in Molecular Medicine: Herpes Simplex Virus Protocols, eds. Brown, S. M. & MacLean, A. R. (Humana, Totowa, NJ), Vol. 10, pp. 19–25. [Google Scholar]
- 29.Mikloska, Z. & Cunningham, A. L. (1998) J. Gen. Virol. 79 353–361. [DOI] [PubMed] [Google Scholar]
- 30.Mikloska, Z., Ruckholdt, M., Ghadiminejad, I., Dunckley, H., Denis, M. & Cunningham, A. L. (2000) J. Immunol. 164 5167–5176. [DOI] [PubMed] [Google Scholar]
- 31.Posavad, C. M., Koelle, D. M. & Corey, L. C. (1996) J. Virol. 70 8165–8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGeoch, D. J., Dalrymple, M. A., Davison, A. J., Dolan, A., Frame, M. C., McNab, D., Perry, L. J., Scott, J. E. & Taylor, P. (1988) J. Gen. Virol. 69 1531–1574. [DOI] [PubMed] [Google Scholar]
- 33.McGeoch, D. J., Dolan, A., Donald, S. & Brauer, D. H. (1986) Nucleic Acids Res. 14 1727–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roizman, B. & Knipe, D. M. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), Vol. 2, pp. 2399–2459. [Google Scholar]
- 35.Wagner, R., Leschonsky, B., Harrer, E., Paulus, C., Weber, C., Walker, B. D., Buchbinder, S., Wolf, H., Kalden, J. R. & Harrer, T. (1999) J. Immunol. 162 3727–3734. [PubMed] [Google Scholar]
- 36.Appay, V., Dunbar, P. R., Callan, M., Klenerman, P., Gillespie, G. M., Papagno, L., Ogg, G. S., King, A., Lechner, F., Spina, C. A., et al. (2002) Nat. Med. 8 379–385. [DOI] [PubMed] [Google Scholar]
- 37.Catalina, M. D., Sullivan, J. L., Brody, R. M. & Luzuriaga, K. (2002) J. Immunol. 168 4184–4191. [DOI] [PubMed] [Google Scholar]
- 38.Hislop, A. D., Gudgeon, N. H., Callan, M. F., Fazou, C., Hasegawa, H., Salmon, M. & Rickinson, A. B. (2001) J. Immunol. 167 2019–2029. [DOI] [PubMed] [Google Scholar]
- 39.Tourdot, S. & Gould, K. G. (2002) J. Immunol. 169 5615–5621. [DOI] [PubMed] [Google Scholar]
- 40.Kedl, R. M., Rees, W. A., Hildeman, D. A., Schaefer, B., Mitchell, T., Kappler, J. & Marrack, P. (2000) J. Exp. Med. 192 1105–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neisig, A., Roelse, J., Sijts, A. J., Ossendorp, F., Feltkamp, M. C., Kast, W. M., Melief, C. J. & Neefjes, J. J. (1995) J. Immunol. 154 1273–1279. [PubMed] [Google Scholar]
- 42.Yewdell, J. W. & Bennink, J. R. (1999) Annu. Rev. Immunol. 17 51–88. [DOI] [PubMed] [Google Scholar]
- 43.Brehm, M. A., Pinto, A. K., Daniels, K. A., Schneck, J. P., Welsh, R. M. & Selin, L. K. (2002) Nat. Immunol. 3 627–634. [DOI] [PubMed] [Google Scholar]
- 44.Burrows, S. R., Silins, S. L., Moss, D. J., Khanna, R., Misko, I. S. & Argaet, V. P. (1995) J. Exp. Med. 182 1703–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corey, L. & Wald, A. (1999) in Sexually Transmitted Diseases, eds. Holmes, K. K., Sparling, P. F., Mardh, P. A., Lemon, S. M., Stamm, W. E., Piot, P. & Wasserheit, J. M. (McGraw–Hill, New York), pp. 285–312.
- 46.Wald, A. & Link, K. (2002) J. Infect. Dis. 185 45–52. [DOI] [PubMed] [Google Scholar]
- 47.Stanberry, L. R., Spruance, S., Cunningham, A. L., Bernstein, D. I., Mindel, A., Sacks, S., Tyring, S., Aoki, F. Y., Slaoui, M., Denes, M., et al. (2002) N. Engl. J. Med. 347 1652–1661. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.