Abstract
Colorectal cancer is the second most leading cause of cancer death among adult Americans. Two autosomal dominant hereditary forms of the disease, familial adenomatous polyposis and hereditary nonpolyposis colorectal cancer, together account for perhaps 5% of all cases. However, in ≈20% of additional colon cancer cases, the affected individuals report a family history of colon cancer in a first-degree relative. Similar familial clusters of colon cancer and early-onset colon adenomas have also been reported. To determine whether such familial aggregations arise by chance or reflect a hereditary colon cancer susceptibility, we conducted a whole genome scan to test for genetic linkage in 53 kindreds in which two or more siblings were affected by age 65 or younger with colon cancer or with advanced colon adenomas that were >1 cm in size or that showed high-grade dysplasia. In this cohort we found genetic linkage of disease (P = 0.00045) to chromosomal region 9q22.2-31.2 in a pattern consistent with autosomal dominant disease alleles. These data suggest that a single locus can contribute to disease susceptibility in a subset of patients with nonsyndromic forms of familial colorectal neoplasia.
Keywords: colorectal cancer, colon adenomas, genetic linkage, familial cancers
Colorectal cancer is the second most leading cause of cancer death among adult Americans (1). Strongly penetrant autosomal dominant hereditary forms of the disease, familial adenomatous polyposis (FAP) and hereditary nonpolyposis colorectal cancer (HNPCC), together account for perhaps 5% of all cases (2–4). However, in ≈20% of additional colon cancer cases the affected individuals report a family history of colon cancer in a first-degree relative, and a genetic basis for these familial disease clusters has also been hypothesized (5).
Colorectal cancers themselves develop from precursor colon adenomas (3). Thus, the inherited FAP syndrome, which is caused by inactivating mutations in the adenomatous polyposis coli (APC) gene, is marked by development of a profusion of hundreds of colon adenomas that then confer a near 100% risk of colon cancer development by an average age of 40 (2, 3). A subtler adenoma and cancer phenotype is associated with an APC I1307K polymorphism present in the Ashkenazi Jewish population (2). The APC I1307K variant DNA sequence demonstrates enhanced vulnerability to development of inactivating somatic APC mutations, resulting in a slightly less than 2-fold increased risk of colon adenoma and cancer development (2). The existence of additional disease genes associated with increased risk of developing colon adenomas and cancers is suggested by observations of an ≈3-fold increased risk of colon cancer among first-degree relatives of individuals who develop colon adenomas before age 60 (6), and by one study that suggested that 19% of persons in the general population might carry autosomal dominant disease alleles conferring susceptibility for colon adenoma or cancer development (7). However, more recent endoscopic-based studies have suggested that small colon adenomas are common in the human population (8). Rather, it appears that it is individuals harboring adenomas that are “advanced” in size or histology who are particularly at risk of developing future colon cancers (9, 10) and future advanced colon adenomas (11), and are most strongly associated with the occurrence of frank colon cancer in a first-degree relative (12).
Accordingly, to attempt to identify loci harboring susceptibility alleles that could give rise to familial colorectal neoplasia, we accrued a population of kindreds demonstrating familial clustering of colon cancers and/or advanced adenomas. We analyzed this population by employing the sibling pair method of linkage analysis (13, 14). In this cohort, we found genetic linkage of disease (P = 0.00045) to chromosomal region 9q22.2-31.2 in a pattern consistent with autosomal dominant disease alleles. These data suggest that a single locus can contribute to disease susceptibility in a subset of patients with nonsyndromic forms of familial colorectal neoplasia.
Materials and Methods
Ascertainment and Collection of Familial Colorectal Neoplasia Kindreds. Kindreds were enrolled in the Colon Neoplasia Sibling Study after review and approval by the Institutional Review Board of University Hospitals of Cleveland of the study design and of all informed consent documents. Initial ascertainment was as described (15), wherein we enrolled families if at least one sibling was affected with colorectal cancer or adenomatous polyps at or before the age of 65, and there was a second living sibling who was willing to participate. From that sample we selected for this study kindreds meeting the following criteria: (i) the presence of an index case and a full sibling who were both diagnosed with colorectal cancer, or a colon adenomatous polyp ≥1 cm, or a colon adenomatous polyp displaying high-grade dysplasia, by age 65 years; (ii) histologic verification of colonic adenomatous polyps or colorectal cancer; (iii) no histologic evidence for inflammatory bowel disease; (iv) no evidence for FAP or HNPCC as described below; and (v) donation of a blood sample for genetic analysis from two or more affected siblings. In addition, we requested blood samples from all available living parents and additional siblings.
Kindreds with known hereditary forms of colorectal cancer, such as FAP and HNPCC, were excluded from this analysis by a combination of pedigree review and molecular testing. Thus, complete kindreds were excluded if a family member displayed clinical evidence of FAP as documented upon review of clinical records by finding a single individual being affected by >100 adenomatous polyps, or by a physician letter documenting FAP in other family members (16). To exclude HNPCC, we tested colon cancer cases for microsatellite instability (MSI) and excluded all kindreds in whom an MSI-high colon cancer was found (17). APC I1307K testing was performed on DNA samples from all kindreds of Ashkenazi Jewish heritage, with exclusion of all kindreds testing positive for the I1307K variant (18). In total, these criteria excluded 21 kindreds because of likely HNPCC, three kindreds because of FAP, and two kindreds because of I1307K.
Sample Extraction and Tumor Analysis. Between 15 and 20 ml of whole blood was obtained from each consenting participant, and DNA was extracted by using standard procedures (Puregene, Gentra Systems). Extraction of DNA from paraffin-embedded colon tissues and typing for microsatellite instability was performed as described (19).
Genome Scan. Genotyping was performed at the Center for Inherited Disease Research (CIDR) by using a modification of the version 9 marker set. A total of 389 markers were used with an average spacing of 9 centimorgans (cM); zero gaps of >20 cM; and an average heterozygosity of 0.76. Allele calls were reviewed by two technicians, blind to family structure and to phenotype. Binning of all allele sizes for each marker was performed by using the FASTCLUS procedure in SAS (SAS Institute, Cary, NC). Calls falling outside of bins and Mendelian inconsistencies, identified in GAS, were reviewed for obvious laboratory errors. Four blind duplicates, four positive, and two negative control samples were run with every 86 study samples. The error rate for the genome scan, based on paired genotypes from these blind duplicates, was 0.012%. The overall missing data rate was 3.5%. The rate of remaining Mendelian inconsistencies was 0.22%. Further details on the marker set and genotyping methods are available from CIDR (www.cidr.jhmi.edu).
Multipoint Linkage Analysis. Genotypes from all siblings and their available parents were used to estimate the proportion of alleles shared identical by descent (IBD) at 2-cM intervals in a multipoint analysis using GENIBD in the program package STATISTICAL ANALYSIS FOR GENETIC EPIDEMIOLOGY (S.A.G.E.) (20). Marker allele frequencies used in this analysis were obtained, using the program FREQ in S.A.G.E., as the maximum likelihood estimates of the founder allele frequencies. Strength of linkage was determined by analysis of the proportion of alleles IBD by the mean tests and by the Haseman–Elston regression approach as implemented in the SIBPAL program in S.A.G.E. (20), which is based on the methods of Idury, Elston, and Haseman (20–22). Under the null hypothesis of no linkage, the allele-sharing from different pairs in a sibship are pairwise independent and so can be treated as independent events for the mean tests (13, 23). Decomposition of the total genetic variance for linkage at the D9S1786 region into additive and nonadditive components was done as described (24, 25). In this analysis, significance for the additive genetic component of variance, and nonsignificance for the nonadditive genetic component of variance, is suggestive of linkage reflecting one or more infrequent dominant alleles. In contrast, significance of the nonadditive component of variance would be indicative of linkage reflecting either recessive alleles or dominant alleles that are so common in the population as to be frequently carried by both parents of affected individuals. Although extensions (26) of the Haseman–Elston test have recently been proposed that are asymptotically more powerful than the original test, employing the original version in this analysis resulted in pooling the 19 concordantly unaffected sibling pairs with the 74 concordantly affected sibling pairs, so that approximately equal numbers of concordant (93) and discordant (94) pairs are contrasted by using a robust t statistic (27) in a sample of sufficient size for asymptotic calculations of P values to apply. These P values for linkage were additionally confirmed by comparison to a Monte Carlo sample of the permutation distribution created by permuting the allele sharing values relative to the pair labels (concordant or discordant). Permutations were done both across sibships of the same size and within sibships, to create 216,583 permutations, sufficient to assure with 95% confidence that the estimated P value was within 5% of the true P value. We note that such a permutation analysis does not require independence among sibling pairs. Additional details are provided in the supporting information, which is published on the PNAS web site, www.pnas.org.
Results and Discussion
Kindreds were enrolled in this analysis if they contained an affected sibling pair in which two full siblings were at age 65 or younger diagnosed with either a colorectal cancer or an advanced colorectal adenoma as characterized by high-grade dysplasia or a size of 1 cm or greater. Affection status of all individuals was confirmed by medical record review and by central pathology review of pathology reports and, where indicated, of diagnostic tissue sections. Kindreds with family members displaying clinical evidence of FAP, HNPCC, or carriage of the I1307K APC variant (2–4, 18) were excluded. In total, 53 kindreds with 116 affected individuals were enrolled in the study (see supporting information). These individuals had a mean age at disease diagnosis of 51 years (range 32–65). We additionally enrolled from these kindreds “unaffected” individuals of any age who had undergone an endoscopic colon examination with no finding of either colon cancer or adenomas. This yielded 43 unaffected individuals, with a mean age of 53 (range 35–81) at the time of their negative colon screening. We note that these unaffected siblings constitute a population somewhat older than their affected siblings, and, moreover, that having a negative colon endoscopy places an individual at low risk of developing colon cancer or advanced colon adenomas for from 5 to 10 years subsequent to the negative examination (28–30) (supporting information). In total, the enrolled individuals from these 53 kindreds defined 74 concordantly affected sibling pairs, 94 discordantly affected pairs, and 19 concordantly unaffected sibling pairs (see supporting information).
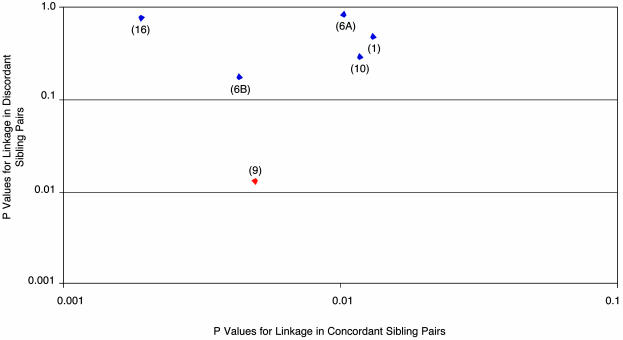
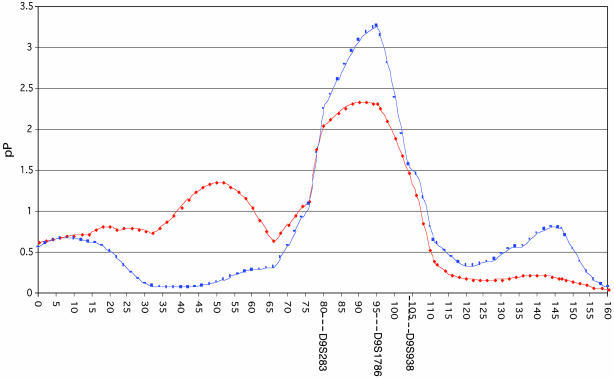
DNA samples obtained from all affected and unaffected individuals were used for a whole genome scan employing 389 microsatellite markers with average spacing of 9.0 cM. To help improve the estimate of allele sharing between siblings, better estimates of those parental genotypes not available were obtained by using the genotypic information from 67 other first-degree relatives of unknown affection status. As shown in Table 1, multipoint analysis using the programs FREQ, GENIBD, and SIBPAL in S.A.G.E. (20) identified six genomic regions showing excess allele sharing among concordantly affected sibling pairs significant at a P ≤ 0.016 level (asymptotically equivalent to a logarithm of odds score of 1 or higher). This cutoff was selected because modeling a two-stage genome scan using the DESPAIR program within S.A.G.E. (see supporting information) suggested that this level of statistical significance at the first stage would provide 80% power to detect a true linkage, although it could in addition yield up to six or seven false positive linkages. Accordingly, to winnow further these initial six candidate regions, we next examined these six loci, which had shown the most extreme allele sharing between pairs of affected siblings, for evidence of deficient allele sharing among the 94 discordant sibling pairs. A region of true linkage would be predicted to show not only significant excess allele sharing >0.5 among concordantly affected sibling pairs, but also to show deficient allele sharing <0.5 among pairs containing one affected and one unaffected sibling. Moreover, under the null hypothesis of no linkage, these discordant sibling pairs are equivalent to a population that is uncorrelated with the initially tested pairs of concordantly affected siblings. As shown in Table 1 and Fig. 1, among our six candidate loci, this analysis highlighted as particularly significant a peak on chromosome 9 at 95 cM. Among concordantly affected sibling pairs, this peak showed allele sharing of 0.59, with a P = 0.0049. Among discordant sibling pairs, the chromosome 9 peak showed deficient allele sharing of 0.42, with a P = 0.0135. In contrast, none of the other six candidate regions showed a P value of <0.18 for linkage among the discordant sibling pairs. The original Haseman–Elston regression method (22), which contrasts the allele sharing between the concordant and discordant sibling pairs (27), was used to calculate an overall combined P value for linkage at each of the six candidate regions, and yielded P = 0.00055 for linkage of disease to chromosome 9 at 95 cM. As shown in Fig. 2, the linkage on chromosome 9 peaks at 95 cM (marker D9S1786) and is broad, being flanked by markers D9S283 (80 cM) and D9S938 (104 cM). To show that the estimates of marker allele frequencies (used in the S.A.G.E. program GENIBD) were not critical to the analysis, we demonstrated that slightly stronger P values for linkage would be obtained under alternate scenarios in which allele frequencies are all set to be equal (P = 0.0003), or in which allele frequencies F are all replaced by values proportional to their complements (1 - F) (P = 0.0003) (see supporting information). Moreover, the significance of the observed linkage at D9S1786 was additionally confirmed by comparison of the observed values of excess allele sharing in concordant sibling pairs versus deficient sharing in discordant sibling pairs in a Monte Carlo sample of the permutation distribution created by permuting the allele sharing values relative to the pair labels (concordant or discordant). This analysis, which is the most valid method for calculating a P value, yielded a confirmatory, and slightly stronger, P value of 0.00045 for linkage of disease to the D9S1786 locus. The addition of discordant sibling pairs to this affected sibling pair linkage analysis both increases the statistical significance of the linkage result obtained, and is also of value in identifying as false positives any regions that show increased allele sharing among siblings independent of disease status. Moreover, it also enables a permutations test to be performed as a robust method for calculating and confirming the P values of linkages that are obtained.
Table 1. Chromosomal regions with a logarithm of odds score ≥1 in 53 kindreds with multiple colon cancer and/or advanced colon adenoma cases.
| Concordantly affected sibling pairs (n = 74)
|
Discordantly affected sibling pairs (n = 94)
|
Concordantly unaffected sibling pairs (n = 19)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome location (cM)* | Allele sharing | SE | P value | Allele sharing | SE | P value | Allele sharing | SE | P value | H-E P value |
| 1 (164) | 0.5827 | 0.0364 | 0.013 | 0.5000 | 0.0335 | 0.49 | 0.5479 | 0.0816 | 0.28 | 0.052 |
| 6 (119) | 0.5803 | 0.0339 | 0.010 | 0.5357 | 0.0331 | 0.86 | 0.5076 | 0.0798 | 0.46 | 0.31 |
| 6 (146) | 0.5913 | 0.0339 | 0.0043 | 0.4687 | 0.0342 | 0.18 | 0.5404 | 0.0980 | 0.34 | 0.0059 |
| 9 (95) | 0.5903 | 0.0341 | 0.0049 | 0.4246 | 0.0336 | 0.014 | 0.5085 | 0.0868 | 0.46 | 0.0006 |
| 10 (0) | 0.5831 | 0.0359 | 0.012 | 0.4862 | 0.0259 | 0.30 | 0.4895 | 0.0874 | 0.55 | 0.025 |
| 16 (60) | 0.6021 | 0.0341 | 0.0019 | 0.5279 | 0.0332 | 0.80 | 0.3795 | 0.0562 | 0.98 | 0.37 |
The six chromosomal regions listed were selected because the P value associated with the allele sharing for the concordantly affected sibling pairs was ≤0.016, which is asymptotically equivalent to a logarithm of odds score ≥1. For these regions, the table lists the P values for linkage as calculated by mean tests for each of the three classes of sibling pairs. Also listed are P values for linkage as calculated by the Haseman-Elston method (H-E), which contrasts allele sharing between concordant and discordant sibling pairs. Boldface indicates values associated with the most significant linkage region.
Distances in cM were calculated relative to the first typed marker on each chromosome: D1S2845, F13A1 (chromosome 6), D9S2169, D10S1435, D16S2616. Closest markers defining each region are: chromosome 1, D1S1679 to D1S1677; chromosome 6 (119 cM), D6S1040 to D6S1009; chromosome 6 (146 cM), D6S1009 to D6S2436; chromosome 9, D9S283 to D9S938; chromosome 10, D10S1435; chromosome 16, D16S403 to D16S3091
Fig. 1.
Comparisons of P values for linkage among concordantly and discordantly affected sibling pairs. Shown on the x axis are the P values for linkage among 74 concordantly affected sibling pairs for the six chromosomal regions listed in Table 1 that show the most extreme allele sharing among concordant sibling pairs. Shown on the y axis are the P values for linkage at these same six chromosomal regions as determined by analysis of the degree of deficient allele sharing among 94 discordant sibling pairs. Red diamond designates the chromosome 9 linkage region marked by D9S1786. Other chromosomal locations are indicated in parentheses, with two loci noted that map to chromosome 6.
Fig. 2.
Linkage of the D9S1786 region of chromosome 9 to development of familial colorectal neoplasia. Chromosome 9, in cM, is depicted along the x axis. Distances of the markers were calculated relative to the first typed marker on the chromosome, D9S2169. The y axis plots pP = (-log10[P value for linkage]). Red symbols depict pP values for linkage as determined by the mean test among concordantly affected sibling pairs. Blue symbols depict pP values for linkage as determined by the Haseman–Elston method employing all affected and unaffected siblings. pP values have not been corrected for multiple hypothesis testing; a pP value of 4 corresponds to a corrected logarithm of odds score of 3.
Further analysis of the pattern of linkage in the D9S1786 region, by using a model that allowed for both additive and nonadditive components of variance, showed significance of only the additive genetic component of variance. This pattern is most consistent with this linkage reflecting a dominant mode of inheritance (24, 25) (supporting information). To estimate the fraction of our cohort that could potentially be linked to a dominant disease allele at D9S1786, we modeled our cohort as composed of two groups of sibling pairs: one group in which the affected pairs arose by inheritance of a rare autosomal dominant disease allele at D9S1786 and a second group that was not linked to this locus. Affected sibling pairs obligately sharing a putative dominant disease allele on one of their two copies of chromosome 9 would, on average, demonstrate allele sharing of 0.75 across all D9S1786 marker alleles (supporting information); whereas sibling pairs not linked to this locus would on average show only the randomly expected allele sharing value of 0.50. In this model, the value of 0.59 for sharing of D9S1786 marker alleles observed among our cohort of affected sibling pairs is equivalent to that expected from a mixed population consisting of 36% percent of sibling pairs linked to a dominant disease allele admixed with 64% of pairs that are not linked [i.e., 0.59 = (0.36)(0.75) + (0.64)(0.50)]. However, the 36% value for the linked subset suggested by this model should be regarded as only an estimate, as the model further provides a 95% confidence interval for this value ranging from 9% to 63% (supporting information).
Review of the 38 affected individuals in the sibships demonstrating the strongest linkage to D9S1786 showed that 19 of these individuals were affected by colorectal cancer, with an average age of cancer diagnosis of 52, and that 19 of these individuals were affected with advanced adenomas, with an average age of diagnosis of 53. Thus, these individuals demonstrated onset by the early 6th decade of life of severe colon neoplasia as defined by frank colorectal cancer or by advanced colorectal adenomas. A secondary analysis of our 53 kindreds performed subsequent to this initial study shows that small adenomas that are also present in these kindreds are unlikely to have arisen from the same disease allele(s) that are accounting for the colon cancers and advanced adenomas. In particular, including as affected in the linkage analysis individuals demonstrating the less severe phenotype of developing small colorectal adenomas of <1 cm reduces by 10-fold the P value for linkage to the putative disease locus on 9q22.2-31.2 (supporting information).
The chromosomal interval between D9S283 and D9S938, corresponding to 9q22.2-31.2, has not previously been associated with risk for developing human colorectal cancer. However, this interval contains numerous candidate genes that might mediate such an association. Among these we note the tumor suppressor gene Patched (PTCH) (31, 32), the DNA repair gene XPA (33), and the tyrosine kinase SYK (34).
In summary, in a whole genome scan we have observed linkage of chromosome 9q22.2-31.2 (P = 0.00045) to risk of developing colorectal cancer or advanced colon adenomas by age 65 or younger. This linkage is supported both by excess allele sharing among concordantly affected sibling pairs and by deficient allele sharing among discordant sibling pairs. These data suggest that a single locus can contribute to disease susceptibility in a subset of patients with nonsyndromic forms of familial colorectal neoplasia. In addition to the well characterized inherited colon cancer syndromes of FAP and HNPCC, a further 20% of individuals with colorectal cancer report a family history of having a first-degree relative also affected with this disease. Of these nonsyndromic familial clusters, only a small fraction are accounted for by other recently described colon cancer susceptibility genes. For example, the I1307K APC variant associated with a 2-fold increased risk of colon cancer appears to be exclusive to a 6% subpopulation within the Ashkenazi Jewish community (2). Recessive null alleles of the MYH gene on chromosome 1 have recently been shown to give rise to an FAP-like phenotype, characterized by on average developing 55 adenomas per affected individual and by an increased risk of colorectal cancer, but such MYH linked disease appears at present to be restricted to ≈5% of cases within the group of individuals having the uncommon multiple polyposis phenotype (35). Lastly, linkage of colon neoplasia to chromosome 15q13-14 has been described, but thus far has been restricted to several Ashkenazi Jewish kindreds with a distinctive phenotype of “serrated” adenomas and increased colon cancer risk (36). Among the cohort reported in this current study, the linkage to D9S1786 is seemingly a relatively frequent cause of colorectal neoplasia development. At present, this linkage must be regarded as specific to the population sample in which it was observed. However, the criteria defining the cohort selected for this study would apply to in excess of 20% of all individuals with colon cancer (5). Moreover, the potential for this locus to play a major role in determining colon cancer risk is consistent with twin studies that have estimated that 35% of colon cancer risk may be heritable (37, 38). Accordingly, we look forward to future studies in additional populations that will help determine the generality of the linkage we have observed, as well as to studies that will ultimately provide identification of the putative disease gene that underlies this linkage.
Supplementary Material
Acknowledgments
We gratefully acknowledge the individuals and families who generously participated in this study, and the substantial assistance in reaching these individuals provided by the National Colon Cancer Research Alliance (NCCRA), the Entertainment Industry Foundation, the NBC “Today Show,” and Ms. Katie Couric. We additionally acknowledge the contribution of individual kindreds to this study by Dr. Henry Lynch (Creighton University, Omaha, NE) and Dr. Robert Bresalier (University of Texas M. D. Anderson Cancer Center, Houston). This work was supported by U.S. Public Health Service (PHS) Grants U01 CA82901 (to S.D.M.), P30CA43703 (to the Case Western Reserve University/Ireland Comprehensive Cancer Center), K23 1K23CA81308 (to G.L.W.), P41 RR03655 (to R.C.E.), GM28356 (to R.C.E.), and M01 RR00080 (to the General Clinical Research Center), by Department of Defense Grant DAMD17-03-1-0289 (to D.D.), and by a grant from the NCCRA (to S.D.M.). S.D.M. is an investigator of the Howard Hughes Medical Institute. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through federal Contract NO1-HG-65403 from the National Institutes of Health to Johns Hopkins University.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FAP, familial adenomatous polyposis; HNPCC, hereditary nonpolyposis colorectal cancer; APC, adenomatous polyposis coli.
References
- 1.Greenlee, R. T., Hill-Harmon, M. B., Murray, T. & Thun, M. (2001) CA Cancer J. Clin. 51, 15-36. [DOI] [PubMed] [Google Scholar]
- 2.Kinzler, K. & Vogelstein, B. (2002) in The Genetic Basis of Human Cancer, eds. Vogelstein, B. & Kinzler, K. (McGraw–Hill, New York), pp. 583-612.
- 3.Markowitz, S., Dawson, D. M., Willis, J. & Willson, J. K. (2002) Cancer Cell 1, 233-236. [DOI] [PubMed] [Google Scholar]
- 4.Kolodner, R. (1996) Genes Dev. 10, 1433-1442. [DOI] [PubMed] [Google Scholar]
- 5.Burt, R. & Peterson, G. M. (1996) in Prevention and Early Detection of Colorectal Cancer, eds. Young, G., Rozen, P. & Levin, B. (Saunders, Philadelphia), pp. 171-194.
- 6.Winawer, S. J., Zauber, A. G., Gerdes, H., O'Brien, M. J., Gottlieb, L. S., Sternberg, S. S., Bond, J. H., Waye, J. D., Schapiro, M., Panish, J. F., et al. (1996) N. Engl. J. Med. 334, 82-87. [DOI] [PubMed] [Google Scholar]
- 7.Cannon-Albright, L., Skolnick, M., Bishop, T., Lee, R. & Burt, R. (1988) N. Engl. J. Med. 319, 533-537. [DOI] [PubMed] [Google Scholar]
- 8.Imperiale, T. F., Wagner, D. R., Lin, C. Y., Larkin, G. N., Rogge, J. D. & Ransohoff, D. F. (2000) N. Engl. J. Med. 343, 169-174. [DOI] [PubMed] [Google Scholar]
- 9.Atkin, W. S., Morson, B. C. & Cuzick, J. (1992) N. Engl. J. Med. 326, 658-662. [DOI] [PubMed] [Google Scholar]
- 10.Otchy, D. P., Ransohoff, D. F., Wolff, B. G., Weaver, A., Ilstrup, D., Carlson, H. & Rademacher, D. (1996) Am. J. Gastroenterol. 91, 448-454. [PubMed] [Google Scholar]
- 11.Martinez, M. E., Sampliner, R., Marshall, J. R., Bhattacharyya, A. K., Reid, M. E. & Alberts, D. S. (2001) Gastroenterology 120, 1077-1083. [DOI] [PubMed] [Google Scholar]
- 12.Pariente, A., Milan, C., Lafon, J. & Faivre, J. (1998) Gastroenterology 115, 7-12. [DOI] [PubMed] [Google Scholar]
- 13.Blackwelder, W. & Elston, R. C. (1985) Genet. Epidemiol. 2, 85-98. [DOI] [PubMed] [Google Scholar]
- 14.Lander, E. S. & Schork, N. J. (1994) Science 265, 2037-2048. [DOI] [PubMed] [Google Scholar]
- 15.Wiesner, G., Platzer, P., Buxbaum, S., Lewis, S., MacMillen, M., Olechnowicz, J., Willis, J., Chakravarti, A., Elston, R. & Markowitz, S. (2001) J. Natl. Cancer Inst. 93, 635-639. [DOI] [PubMed] [Google Scholar]
- 16.Sieber, O. M., Tomlinson, I. P. & Lamlum, H. (2000) Mol. Med. Today 6, 462-469. [DOI] [PubMed] [Google Scholar]
- 17.Boland, C. R. (2002) in The Genetic Basis of Human Cancer, eds. Vogelstein, B. & Kinzler, K. (McGraw–Hill, New York), pp. 307-322.
- 18.Laken, S., Petersen, G., Gruber, S., Oddoux, C., Ostrer, H., Giardiello, F., Hamilton, S., Hampel, H., Markowitz, A., Klimstra, D., et al. (1997) Nat. Genet. 17, 79-83. [DOI] [PubMed] [Google Scholar]
- 19.Grady, W., Rajput, A., Myeroff, L., Liu, D., Willis, J., Kwon, K. & Markowitz, S. (1998) Cancer Res. 58, 3101-3104. [PubMed] [Google Scholar]
- 20.Statistical Solutions (2003) S.A.G.E.: Statistical Analysis for Genetic Epidemiology, Computer Program Package (Statistical Solutions, Cork, Ireland), Release 4.3.
- 21.Idury, R. M. & Elston, R. C. (1997) Hum. Hered. 47, 197-202. [DOI] [PubMed] [Google Scholar]
- 22.Haseman, J. K. & Elston, R. C. (1972) Behav. Genet. 2, 3-19. [DOI] [PubMed] [Google Scholar]
- 23.Hodge, S. (1984) Genet. Epidemiol. 1, 109-122. [DOI] [PubMed] [Google Scholar]
- 24.Elston, R. C., Buxbaum, S., Jacobs, K. B. & Olson, J. M. (2000) Genet. Epidemiol. 19, 1-17. [DOI] [PubMed] [Google Scholar]
- 25.Tiwari, H. K. & Elston, R. C. (1997) Annu. Hum. Genet. 61, 253-261. [DOI] [PubMed] [Google Scholar]
- 26.Shete, S., Jacobs, K. B. & Elston, R. C. (2003) Hum. Hered. 55, 79-85. [DOI] [PubMed] [Google Scholar]
- 27.Elston, R. C., Kringlen, E. & Namboodiri, K. K. (1973) Behav. Genet. 3, 101-106. [DOI] [PubMed] [Google Scholar]
- 28.Selby, J. V., Friedman, G. D., Quesenberry, C. P., Jr., & Weiss, N. S. (1992) N. Engl. J. Med. 326, 653-657. [DOI] [PubMed] [Google Scholar]
- 29.Rex, D. K., Cummings, O. W., Helper, D. J., Nowak, T. V., McGill, J. M., Chiao, G. Z., Kwo, P. Y., Gottlieb, K. T., Ikenberry, S. O., Gress, F. G., et al. (1996) Gastroenterology 111, 1178-1181. [DOI] [PubMed] [Google Scholar]
- 30.Rex, D. K., Johnson, D. A., Lieberman, D. A., Burt, R. W. & Sonnenberg, A. (2000) Am. J. Gastroenterol. 95, 868-877. [DOI] [PubMed] [Google Scholar]
- 31.Johnson, R. L., Rothman, A. L., Xie, J., Goodrich, L. V., Bare, J. W., Bonifas, J. M., Quinn, A. G., Myers, R. M., Cox, D. R., Epstein, E. H., Jr., & Scott, M. P. (1996) Science 272, 1668-1671. [DOI] [PubMed] [Google Scholar]
- 32.Hahn, H., Wicking, C., Zaphiropoulous, P. G., Gailani, M. R., Shanley, S., Chidambaram, A., Vorechovsky, I., Holmberg, E., Unden, A. B., Gillies, S., et al. (1996) Cell 85, 841-851. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka, K., Miura, N., Satokata, I., Miyamoto, I., Yoshida, M. C., Satoh, Y., Kondo, S., Yasui, A., Okayama, H. & Okada, Y. (1990) Nature 348, 73-76. [DOI] [PubMed] [Google Scholar]
- 34.Muller, B., Cooper, L. & Terhorst, C. (1994) Immunogenetics 39, 359-362. [DOI] [PubMed] [Google Scholar]
- 35.Sieber, O. M., Lipton, L., Crabtree, M., Heinimann, K., Fidalgo, P., Phillips, R. K., Bisgaard, M. L., Orntoft, T. F., Aaltonen, L. A., Hodgson, S. V., et al. (2003) N. Engl. J. Med. 348, 791-799. [DOI] [PubMed] [Google Scholar]
- 36.Jaeger, E. E., Woodford-Richens, K. L., Lockett, M., Rowan, A. J., Sawyer, E. J., Heinimann, K., Rozen, P., Murday, V. A., Whitelaw, S. C., Ginsberg, A., et al. (2003) Am. J. Hum. Genet. 72, 1261-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lichtenstein, P., Holm, N. V., Verkasalo, P. K., Iliadou, A., Kaprio, J., Koskenvuo, M., Pukkala, E., Skytthe, A. & Hemminki, K. (2000) N. Engl. J. Med. 343, 78-85. [DOI] [PubMed] [Google Scholar]
- 38.Hemminki, K. & Mutanen, P. (2001) Mutat. Res. 473, 11-21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.