Abstract
Worldwide, 90% of HIV-1 infections are transmitted heterosexually. Because the genital mucosa are the sites of initial contact with HIV-1 for most exposed individuals, study of the virus from the genital tract is critical for the development of vaccines and therapeutics. Previous analyses of HIV-1 in various tissues have documented compartmentalization of viral genomes. Whether compartmentalization was associated with viral phenotypic differences or immune status, however, was not well understood. We compared HIV-1 gp120 env sequences from the genital tract and plasma of 12 women. Eight women displayed compartmentalized HIV-1 RNA genomes, with viral sequences from each site that were clearly discrete, yet phylogenetically related. The remaining four exhibited env sequences that were intermingled between the two sites. Women with compartmentalized HIV-1 genomes had higher CD4+ cell counts than those displaying intermingled strains (P = 0.02). Intrapatient HIV-1 recombinants comprising sequences that were characteristic of both sites were identified. We next compared viral phenotypes in each compartment. HIV-1 coreceptor usage was often compartmentalized (P ≤ 0.01). The number of N-linked glycosylation sites, associated with neutralization resistance, also differed between compartments (P < 0.01). Furthermore, disparities between the density of gp120 glycosylations in each compartment correlated with higher CD4+ counts (P = 0.03). These data demonstrate that the genital tract and plasma can harbor populations of replicating HIV-1 with different phenotypes. The association of higher CD4+ cell counts with compartmentalization of viral genomes and density of gp120 glycosylations suggests that the immune response influences the development of viral genotypes in each compartment. These findings are relevant to the prevention and control of HIV-1 infection.
Women constitute half of the world's HIV-1-infected adults, and 90% of infection is spread by heterosexual transmission (1). For the vast majority of individuals exposed to HIV-1, the genital mucosa are the sites of initial contact. Investigation of HIV-1 in the female genital tract, therefore, is critical to the development of vaccines, therapies, and strategies to block heterosexual and mother-to-child transmission. HIV-1 infection is characterized by tremendous genetic variation. Even within a single individual, the virus exists as a population of highly related but genetically distinct variants called quasispecies (2). Although much variation stems from mutations introduced by the errorprone reverse transcriptase, viral recombination produces leaps in genetic evolution as well (2). Recent work (3) has suggested that intrapatient HIV-1 recombination may occur frequently, but recombination between quasispecies from different body sites within the same individual has not been described in detail.
Substantial research has focused on HIV-1 variation in separate compartments within the same person. Much of the work has examined viral reservoirs that persist after antiviral therapy (4). Compartmentalization, the occurrence of distinct yet phylogenetically related HIV-1 genotypes within different anatomic sites, has been well documented in treated and untreated individuals alike. Compartmentalization occurs in diverse tissues, including plasma, brain, and lung (5–7). Pioneering studies of both women and men have also revealed viral variants in the genital tract that differed from those in blood (8–12).
The forces leading to compartmentalization are not well delineated. Compartmentalization may stem from a founder effect, with seeding of tissues, followed by localized viral evolution. Selective pressures, however, may also shape the distinct viral populations (7), and immune pressures are likely to play a role. Local conditions such as sexually transmitted infections (STIs) or coreceptors on host cells may also exert selective pressures (13, 14). To address the formation of viral compartments, we compared HIV-1 in the genital tract and plasma of women with a broad range of HIV-1 risks and disease.
Apart from considerations of drug treatment and resistance, the biologic consequences of compartmentalized viral genomes are not well understood. We therefore compared phenotypic traits of viruses from the two sites, focusing on traits encoded by the env gene. HIV-1 coreceptor usage is a determinant of viral tropism and pathogenesis. HIV-1 transmitted in vivo generally uses the coreceptor CCR5 (R5) (15), but later CXCR4 (X4) quasispecies may emerge, usually in a mixture with R5 strains. Emergence of X4 strains heralds CD4+ cell depletion and clinical deterioration (16). Previous analyses (6, 10–12) of coreceptor usage in various anatomic sites showed that it can differ between compartments. This article extends these findings by quantifying and comparing HIV-1 coreceptor usage in the genital tract and plasma.
The second phenotypic trait we studied, N-linked glycosylation of gp120, has been connected by multiple studies to the creation of a “glycan shield” enabling HIV-1 to escape neutralization (17–19). Despite marked variation in HIV-1 generally, the number of glycosylation sites found in gp120 envelopes globally is conserved at ≈25, suggesting strong selective pressures to maintain that number (17). Density of gp120 N-linked glycosylation is a phenotypic trait that may be relevant to humoral immune response and vaccine design, and the distribution of such sites on viruses in the female genital tract vs. blood may differ (8). To examine HIV-1 genotypes and phenotypes in different compartments, we focused on gp120 sequences in the plasma and female genital tract.
Methods
Study Population. We studied 12 participants in the Bronx-Manhattan and Brooklyn, New York sites of the Women's Interagency HIV Study (WIHS), a multicenter, prospective study of HIV-1 infection of women (15, 20). Participants were interviewed and examined semiannually; blood and gynecologic specimens including cervicovaginal lavage (CVL) were collected. The institutional review boards at each site approved the investigation and each woman provided informed consent.
To study HIV-1 sequences in CVL and plasma, we identified 12 women with viral loads of ≥1,000 copies per ml in samples obtained from both compartments simultaneously. Seeking women with a spectrum of HIV-1 risks and disease, we selected them from a WIHS substudy of illicit drug use and HIV-1 infection (20). We classified participants according to the following three patterns of drug use reported by the women at WIHS visits one and two: (i) Group I. No history of either injection drug use (IDU) or noninjecting use of crack, cocaine, or heroin at any time; (ii) Group II. Noninjecting drug use of crack, cocaine, or heroin in the 6 months preceding visits one or two, but no history of IDU; and (iii) Group III. IDU in the 6 months preceding one or both visits. HIV-1 risks were categorized as IDU, heterosexual, or no known risk. Women reporting IDU may also have had heterosexual risks, but WIHS classified their exposure as IDU (15, 21).
Sample Collection and Analysis. CVL was collected as described (14) and was cryopreserved at -80°C. CVL specimens were tested for blood by using a DiaScreen dipstick (Chronimed, Minneapolis), and for semen by using the SEMA test (Humagen Fertility Diagnostics, Charlottesville, VA) or the OneStep ABAcard p30 test (Abacus Diagnostics, West Hills, CA). Specimens contaminated by blood or semen were not studied. We quantitated HIV-1 RNA in plasma and CVL by using NucliSens (BioMérieux, Durham, NC), with a lower limit of quantification of ≈80 copies per ml. CCR5 genotypes were determined as described (15).
RT-PCR, Cloning, and Sequencing. RNA was extracted from plasma and CVL as described (22, 23). The V1–V5 region of the HIV-1 gp120 env gene was amplified by using PCR and nested primers, seen in Table 3, which is published as supporting information on the PNAS web site, www.pnas.org. The PCR product was then directly cloned into a TA vector (pCR2.1-TOPO; Invitrogen). Both strands of the DNA templates were sequenced by using an automated DNA sequencer (Applied Biosystems). Problems associated with long RT-PCR include recombination between molecules and underestimates of sequence diversity due to PCR amplification. To minimize the potential for PCR-mediated recombination, we controlled the reaction conditions by using methods we described (22, 23). Although we generally employ limiting dilution before long PCR amplification (22, 23), V1–V5 sequences obtained from CVL could be amplified from undiluted cDNA only. To analyze CVL and plasma specimens in parallel, we cloned and sequenced env genes from undiluted cDNA samples, then performed full analyses of the 160 unique clones exhibiting ≥0.3% intrapatient diversity. To specifically investigate putative intrapatient recombinants, we did, however, perform limiting dilutions of selected cDNA samples as described (23).
Computational Analyses. Sequences were aligned by using the program BIOEDIT, VERSION 5.0.6, obtained from Tom Hall (North Carolina State University, Raleigh, which can be accessed at www.mbio.ncsu.edu/BioEdit/bioedit.html), then were manually edited and checked for unusual mutations, insertions, or deletions. Phylogenetic tree analyses included sequences of representative subtypes in the Los Alamos HIV sequence database (http://hivweb.lanl.gov), as well as the 160 sequences described here. After stripping gaps, phylogenetic trees were constructed by using the neighbor-joining method and Kimura's two-parametric distance estimates. Clones suspected to be recombinants based on phylogenetic analysis were investigated further by using SIMPLOT (24). Each putative recombinant clone was chosen as a query sequence, and representative sequences from the plasma and CVL of the same patient were used as references. A graph depicted the degree of similarity of each reference strain to the query sequence. The recombination break point was identified by comparing alignments of CVL and plasma sequences. Potential N-linked glycosylation sites were identified by using the N-Glycosite tool from the Los Alamos HIV sequence database.
Prediction of HIV-1 Coreceptor Usage. Envelope sequences were used to predict coreceptor usage as described (16). Evaluation of the predictive value of the V3 sequence, in comparison to phenotypic analyses of 199 env genes in our laboratory, showed that the sensitivity of the predictive method was 97.8% and the specificity 98.4%. To quantify the proportion of virus in a specimen using each coreceptor, we predicted the phenotype of each clone derived from that specimen, and used a mathematical model derived in our previous study (16).
Statistical Analyses. The Wilcoxon rank sum test was used to compare the characteristics of women in the three drug use groups and to analyze the association of CD4+ cell count and different numbers of glycosylation sites in CVL vs. plasma. Numbers of glycosylations in each compartment were compared by using the t test and the proportion of virus that used R5 by using the binomial proportions test. We analyzed the relationship of patient characteristics in Table 1 and the number of clones per patient to the presence of intermingled HIV-1 sequences by stratifying the population into intermingled and compartmentalized groups, and then comparing the patient variables by using the median test, Wilcoxon rank sum test, Fisher's exact test, or logistic regression as appropriate.
Table 1. Virologic, immunologic, and clinical characteristics of women.
| HIV-1 RNA, log copies per ml
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Age/ethnicity | Plasma | CVL | CD4+ count, cells per mm3 | Pattern of HIV-1 sequence variation: CVL vs. plasma | ART | Reported drug use, last 6 mo* | Lifetime male partners, N | HIV risk | Lower genital infections |
| Group I: No history of snorting or smoking crack, cocaine, or heroin or IDU at entry to WIHS | ||||||||||
| 15 | 38/AA | 2.99 | 4.30 | 263 | Compartmentalized | AZT, d4T, 3TC | Crack | 4 | NR | Vag trich |
| 16 | 43/AA | 4.67 | 5.48 | 188 | Compartmentalized | AZT, ddl | — | 4 | Het | — |
| 17 | 61/Lat | 5.99 | 5.66 | 11 | Intermingled | AZT, 3TC, ABC | — | 2 | Het | — |
| 18 | 36/AA | 5.15 | 4.08 | 110 | Compartmentalized | — | — | 15 | Het | BV, vag candida, warts |
| 19 | 33/Lat | 4.82 | 4.32 | 304 | Compartmentalized | AZT | — | 1 | Het | — |
| Group II: Recent history of snorting or smoking crack, cocaine, or heroin, but no history of IDU at entry to WIHS | ||||||||||
| 21 | 39/AA | 4.78 | 3.65 | 20 | Intermingled | — | Crack | 21 | Het | BV, vag candida |
| 22 | 43/AA | 4.69 | 3.40 | 82 | Compartmentalized | — | Crack, cocaine | 5 | Het | — |
| 23 | 30/AA | 5.56 | 4.15 | 234 | Compartmentalized | — | Crack | 522 | Het | Vag candida, warts |
| 25 | 41/AA | 5.46 | 3.61 | 6 | Intermingled | — | — | 121 | Het | — |
| Group III: History of IDU at entry to WIHS | ||||||||||
| 26 | 57/Lat | 4.78 | 4.57 | 142 | Intermingled | — | — | 40 | IDU | — |
| 27 | 45/AA | 4.86 | 5.64 | 217 | Compartmentalized | — | — | 30 | IDU | — |
| 28 | 41/AA | 3.97 | 4.44 | 519 | Compartmentalized | — | — | 10 | IDU | — |
ART, antiretroviral therapy (d4T, stavudine; AZT, zidovudine; 3TC, lamivudine; ddl, didanosine; ABC, abacavir); AA, African American; NR, no known risk; vag trich, vaginal trichomoniasis; Het, known heterosexual partner with HIV-1 infection or high HIV-1 risk; Lat, Latina; BV, bacterial vaginosis; vag candida, vaginal candidiasis.
Participants' report of recreational drug use including smoking, snorting, or injecting of crack, cocaine, or heroin
Results
Patient Population. We studied 12 women whose virologic, immunologic, and clinical characteristics are presented in Table 1. The women displayed a broad range of HIV-1 RNA loads in both plasma and CVL and a spectrum of CD4+ cell counts. Although four women in Group I received antiretroviral therapy, viral loads in both compartments did not differ significantly among women from the three drug use groups. All 12 women carried the homozygous wild-type genotype for the coreceptor CCR5. Heterosexual exposure was the HIV-1 risk for eight of the nine women who lacked a history of IDU; one woman had no known risk. Although the women in Group III were at risk for acquiring infection through IDU, their report of multiple sexual partners (Table 1) suggests that heterosexual spread was also possible (21). Illicit drug use of any type was strongly associated with a history of multiple sexual partners; women in Groups II and III had significantly larger numbers of lifetime male partners than those in Group I, who had no history of drug use (P = 0.01). Four women had infections of the lower genital tract, but there were no genital ulcers.
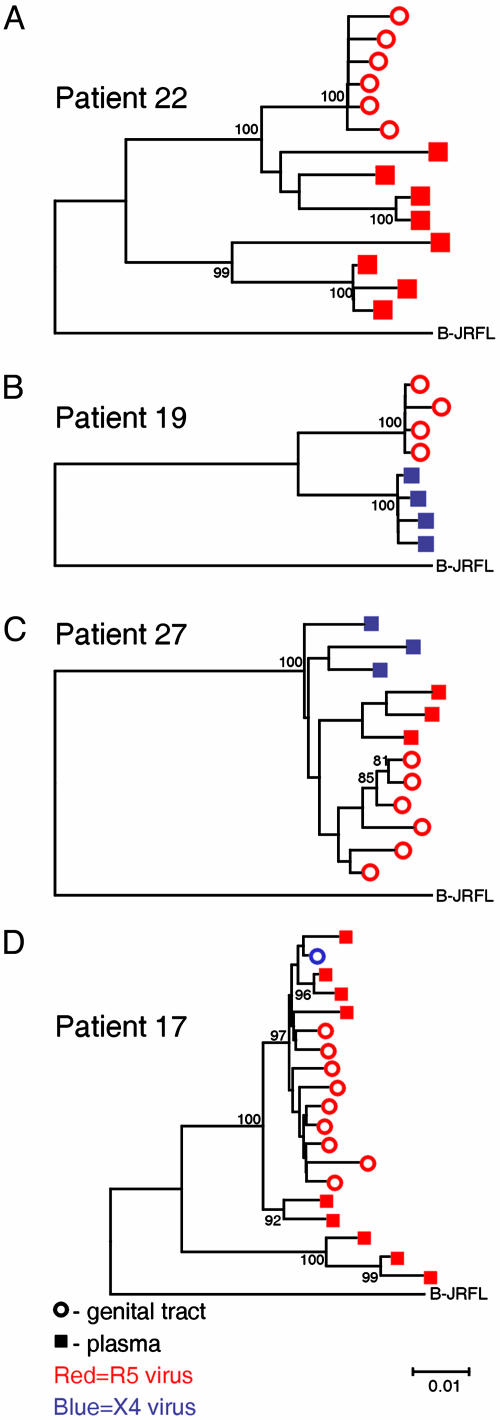
Sequence Analyses. We compared HIV-1 RNA sequences of gp120 env genes obtained from the female genital tract, specifically the CVL and plasma. One-hundred sixty clones of V1–V5 (≈1200 base pairs) were sequenced (GenBank accession nos. AY316363–AY316522). Computational analyses, including a BLAST search, showed no evidence of contamination, and ORFs were intact, with no significant deletions, alterations, or nonsense mutations. All sequences were identified as subtype B. The phylogenetic tree constructed from these genomes demonstrated that sequences from CVL and plasma of each patient clustered in a single branch. Analyses also revealed compartmentalization of HIV-1 sequences in 8 of 12 women. In these eight women, viral genomes from the genital tract and plasma were clearly distinct, yet sufficiently related phylogenetically to suggest evolution from a single ancestor (Fig. 1 A–C and Table 1). The remaining four women exhibited quasispecies that were intermingled between the two sites. Intermingled strains were phylogenetically related and distributed in both sites (Fig. 1D and Table 1). A tree depicting sequences from all 12 women is available (see Fig. 3, which is published as supporting information on the PNAS web site). We then asked whether patient characteristics were associated with compartmentalization. All four women with intermingled sequences showed CD4+ cell depletion, with three exhibiting CD4+ cell counts ≤20 cells per mm3 (Table 1). Women displaying compartmentalized HIV-1 genomes, by contrast, had significantly higher CD4+ counts than those with intermingled strains (P = 0.02). The pattern of sequence variation between the genital tract and plasma observed in these 12 women did not correlate with the number of unique clones analyzed from each patient, nor with risk of HIV-1 acquisition, drug use, or STIs.
Fig. 1.
Phylogenetic trees of V1–V5 env sequences from the plasma and genital tract of representative patients. Each tree depicts the plasma and genital tract (CVL) sequences derived from a single patient, and the subtype B reference strain, JRFL. Numbers at branch points represent bootstrap values. ○, genital tract clones; ▪, plasma clones. The R5 virus is red, and the X4 virus is blue. (A–C) Compartmentalized sequences. (D) Intermingling of sequences.
HIV-1 Coreceptor Usage. We next asked whether phenotypic traits encoded by the env gene were compartmentalized. After predicting HIV-1 coreceptor usage for each clone derived from a specimen, we calculated λ, the proportion of virus in the specimen using R5. If λ = 1, all of the virus in a population uses R5; if λ = 0, all of the viruses use X4 (16). In 5 of 12 women, HIV-1 coreceptor usage in the two sites differed significantly (P < 0.01, Table 2 and Fig. 1). Sequences from Patient 22 were strongly compartmentalized between CVL and plasma, yet all of the strains used R5 (Fig. 1 A). Patient 19, by contrast, not only demonstrated compartmentalized sequences, but also showed complete compartmentalization of HIV-1 coreceptor usage (Fig. 1B); all of the plasma strains from this patient used X4 and all of the CVL strains used R5. Sequences from patient 27 were highly compartmentalized, with plasma genomes clustering on two large branches of a phylogenetic tree; the plasma-derived virus on each of the two large branches used a different coreceptor (Fig. 1C). For four of the five patients in whom HIV-1 coreceptor usage differed significantly, the proportion of R5 strains in CVL exceeded that in plasma (Table 2). Strains that used R5, however, were not significantly more prevalent in the CVL than in the plasma among the group of 12 women overall. To examine whether the sequences encoding HIV-1 coreceptor usage, the V3 loop, determined the compartmentalized patterns seen in the evolutionary trees, we stripped the V3 sequences from the data set. Construction of additional phylogenetic trees from sequences lacking the V3 loop resulted in the same patterns of evolution and compartmentalization observed in the first analyses (data not shown). We next asked whether λ values were associated with drug use or HIV-1 risk (Table 2). Comparison of HIV-1 coreceptor usage between the three groups revealed that women in Group III had significantly higher proportions of X4 strains (expressed as lower λ values) in both plasma and CVL than the women who had not injected drugs (P = 0.03).
Table 2. Characteristics of HIV-1 gp120 in plasma vs. CVL.
| HIV-1 coreceptor Usage, λ*
|
N-linked glycosylation sites V1—V5, mean (SD)
|
||||
|---|---|---|---|---|---|
| Patient | Plasma | CVL | CD4+ count, cells per mm3 | Plasma | CVL |
| Group I: No history of illicit drug use | |||||
| 15 | 1 | 1 | 263 | 27.0 (0.0)† | 23.8 (0.8) |
| 16 | 1 | 1 | 188 | 26.4 (1.2) | 27.0 (0.0) |
| 17 | 1 | 0.92 | 11 | 24.3 (1.0) | 24.5 (0.9) |
| 18 | 1 | 1 | 110 | 25.1 (1.7) | 25.0 (0.0) |
| 19 | 0‡ | 1 | 304 | 24.0 (0.0)† | 26.8 (0.5) |
| Group II: Snorting or smoking of crack, cocaine, or heroin, no IDU | |||||
| 21 | 0.44‡ | 1 | 20 | 22.7 (0.7)† | 24.0 (0.0) |
| 22 | 1 | 1 | 82 | 22.4 (1.1) | 21.8 (0.4) |
| 23 | 1 | 1 | 234 | 20.2 (1.7)† | 25.7 (0.8) |
| 25 | 1 | 1 | 6 | 25.1 (0.6) | 24.7 (0.5) |
| Group III: History of IDU | |||||
| 26 | 0.88‡ | 0.22 | 142 | 23.3 (1.3) | 22.0 (2.3) |
| 27 | 0.57‡ | 1 | 217 | 22.0 (0.0)† | 25.2 (0.8) |
| 28 | 0‡ | 0.36 | 519 | 22.0 (0.0)† | 19.9 (1.1) |
λ, Proportion of virus that uses CCR5
Differences in mean number of glycosylation sites between CVL and plasma were statistically significant (P < 0.01); correlation of significant differences in glycosylation densities in the two compartments and CD4+ cell count >200 cells per mm3 was also statistically significant (P = 0.03)
Differences in λ between CVL and plasma were statistically significant (P ≤ 0.01)
N-linked Glycosylation Sites. We also deduced the density of gp120 N-linked glycosylations from the sequences. After identifying the glycosylation sites in each clone, we determined the mean number of glycosylations of all of the clones derived from each specimen. Although the number of such sites is highly conserved among all HIV-1 subtypes, the mean number of glycosylations differed significantly between the genital tract and plasma in 6 of 12 women (P < 0.01, Table 2). Furthermore, significant disparities between the density of gp120 glycosylations in the two compartments correlated with higher CD4+ counts (P = 0.03), suggesting that the immune response may influence the evolution of viral genotypes within separate compartments.
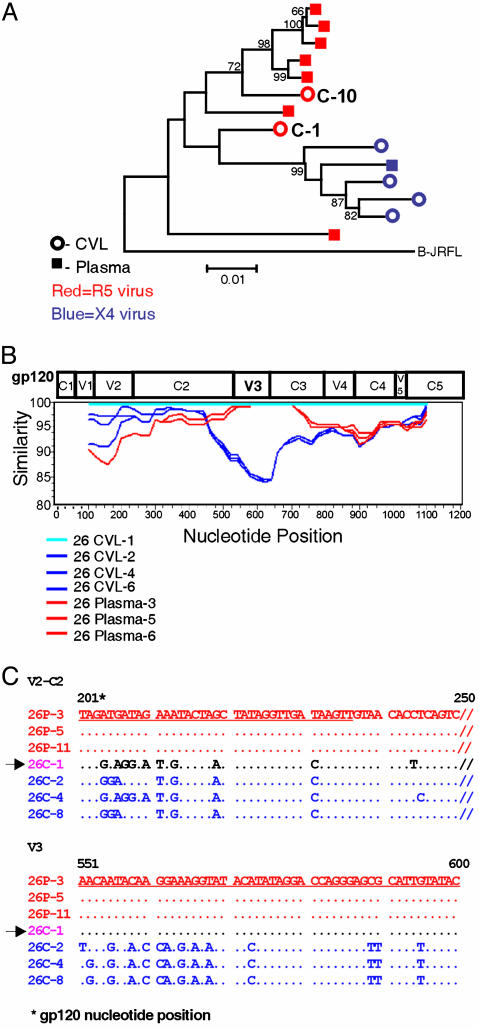
Recombinant HIV-1 Genomes. Compartmentalization of HIV-1 quasispecies between plasma and CVL made it possible to distinguish sequences derived from each site. We thereby identified intrapatient recombination between HIV-1 genomes from CVL and plasma. Recombinants were first identified in two CVL clones from patient 26, and two plasma clones from patient 23. The phylogenetic tree depicting patient 26's sequences (Fig. 2A) displayed intermingling, and also illustrated that two of the six CVL clones, C-1 and C-10, were distinguishable from the others; they were the only CVL clones from patient 26 bearing the R5 phenotype, and they clustered phylogenetically with the majority of plasma clones. A SIM-PLOT comparing C-1 to six other clones from patient 26 (Fig. 2B) revealed that the 5′ portion of clone C-1 encoding V1–C2 most closely resembled the CVL-derived clones. The V3–C3 portion of clone C-1, however, was almost identical to the plasma-derived clones. Sequence alignments depicted in Fig. 2C confirmed that the V2-C2 fragment of clone C-1 closely resembled the other CVL-derived sequences, whereas the V3 region of the C-1 clone resembled the plasma-derived clones. These analyses of HIV-1 sequences derived from patient 26's genital tract strongly suggested that the V3 sequences, which confer the R5 phenotype, were acquired through recombination with a viral genome resembling those from the patient's plasma.
Fig. 2.
Recombinant genomes derived from patient 26. (A) A phylogenetic tree of V1–V5 env sequences from plasma and CVL of patient 26. Clones C-1 and C-10 were identified as recombinant genomes obtained from the CVL. (B) A simplot comparing clone C-1 (26 CVL-1) to clones derived contemporaneously from CVL and plasma of the same patient. (C) The alignment of CVL and plasma sequences from patient 26. Sequences depict a portion of V2/C2 and V3 regions from clones compared in B. Recombinant clone 26C-1, noted by an arrow, is purple. C denotes CVL, with other CVL clones in blue; P denotes plasma clones, which are red. Nucleotides differing from 26P-3 are shown; if identical to 26P-3, they are depicted as dots. These sequences demonstrate that 26C-1 resembles CVL-derived clones in the V2/C2 region and plasma sequences in V3.
To control for possible PCR-mediated recombination as a source of the intrapatient recombinants, we performed end-point dilutions of the cDNA's yielding the recombinant clones. By diluting cDNA from patient 23's plasma, we found results very similar to those obtained by using undiluted PCR products, and identified a third recombinant. The recovery of a recombinant genome after endpoint dilution strongly supports the idea that the recombination event occurred in the patient under study, rather than during PCR. Dilution of cDNA from patient 26's CVL precluded successful PCR amplification. To review the handling of specimens from patients 23 and 26 for possible opportunities for crosscontamination, we checked laboratory records. RNA extraction and RT-PCR of plasma from patient 26 took place a full year after investigation of the CVL sample from the same patient. For patient 23, the CVL and plasma specimens were extracted separately, and were handled by two different investigators. These control measures make it very unlikely that the five intrapatient recombinants we observed were the result of PCR-mediated recombination.
Discussion
This analysis of viral sequence variation in the female genital tract and plasma led to three findings that are directly relevant to control of HIV-1 infection. Previous reports rigorously documented HIV-1 compartmentalization (4–12). This study went on to examine whether a variety of characteristics of the infected individual contribute to selecting and maintaining distinct compartments. By doing so, it demonstrated a significant association between higher CD4+ cell counts and compartmentalization of both viral genomes and density of gp120 glycosylation sites, suggesting that the immune response influences the development of viral genotypes in each compartment. By documenting that HIV-1 coreceptor usage and number of glycosylations may differ significantly in genital tract vs. plasma, this report indicated that each site may harbor viral populations with different characteristics. Finally, it identified recombinant HIV-1 genomes composed of sequences from both the genital tract and blood of the same person.
The CD4+ lymphocyte plays a central role in coordinating immune responses to HIV-1, providing help to the cellular and humoral arms of the immune system (25, 26). Complex cellular and humoral responses to HIV-1 occur in both the blood and genital tract (25–31). The data presented here suggest that during much of the course of chronic infection, distinct HIV-1 variants are compartmentalized within different anatomic sites. Progression to immune deficiency, with a decline in both the number and function of CD4+ cells, usually ensues over time. The immune system loses control of HIV-1 replication and, as suggested here, may also lose the ability to influence the evolution of viral strains in different compartments.
Half of the women whom we studied displayed significantly different numbers of gp120 N-linked glycosylations in each compartment. Because the number of glycosylations is highly conserved in HIV-1 strains worldwide, and sequence changes modulating potential N-linked glycosylation play an important role in conferring escape from immune recognition (17–19), differences in glycosylation density imply the evolution of two distinct sets of sequences under distinct immune pressures. What bolsters this conclusion is the correlation of differences in glycosylation densities and higher CD4+ cell counts. Together, these findings suggest that the compartmentalization of HIV-1 quasispecies in the genital tract reflects localized disparities in immune pressure. The next step in this line of inquiry is to determine experimentally the cellular and humoral responses in the genital tract and blood of patients with various patterns of compartmentalization. These studies are likely to provide insights into the role of the host immune response in controlling HIV-1 infection.
The findings discussed here build on previous observations. One study (7) of sequence diversity in different tissues during acute vs. chronic infection showed that compartmentalization appears during chronic infection, suggesting that a common viral variant disseminates during primary infection and then diversifies under selective pressures. A study of HIV-1 in the blood and genital tract of African women (9) showed that compartmentalization and sequence evolution were already present soon after seroconversion. Significant differences have been seen in the frequency of CTL epitope variants among HIV-1 quasispecies in different compartments, suggesting different immune pressures in these sites (5, 32). Finally, studies have shown that specific immune responses to HIV-1 wane as disease progresses (26, 33, 34). CD4+ cell help is crucial, and although memory CD8+ T cells are produced in the absence of CD4+ cells, their activity is diminished when compared with the CD8+ cells produced in the presence of CD4+ T cells (34).
This study found that compartmentalization correlated with the stage of HIV-1 disease progression, rather than with transmission risk or local characteristics. This report complements a larger WIHS study (20), which found that immune activation was driven by HIV-1 infection, and not by illicit drug use. Conditions affecting HIV-1 replication in local compartments such as cell type, cytokine profile, or inflammation, may indeed exert selective pressures leading to compartmentalization, and additional in vivo analyses are needed (13, 14). Similarly, antiretroviral therapy may present a powerful selective pressure for compartmentalization (4). It is unlikely, however, to have influenced the findings on sequence variation described here because few patients with compartmentalized genomes were treated, viremia was not suppressed, and the gene we analyzed coded for the gp120 envelope, which was not the target of any of the therapies used.
This study documented significant differences in the mean number of glycosylations on viruses derived from the genital tract and plasma, underscoring the importance of considering HIV-1 and immune response in the genital tract when designing vaccines. In addition, by quantifying the proportion of R5 and X4 viruses in each site, we found that coreceptor usage often varied significantly between them. HIV-1 coreceptor usage not only plays a pivotal role in disease progression but may also be relevant to therapeutic response. Our previous work (16) demonstrated preferential suppression of X4 strains by antiviral therapy, perhaps contributing to the clinical efficacy of treatment. In addition, agents aimed at blocking HIV-1 coreceptors are likely to join the therapeutic armamentarium. This report shows that the proportion of X4 strains in one compartment does not necessarily reflect coreceptor usage in another (Table 2), suggesting that measuring coreceptor usage in the genital tract and blood may aid in monitoring disease progression and response to therapy.
In the women studied here, a history of IDU correlated with larger proportions of X4 strains in plasma and CVL than those observed without an IDU history (Table 2). An explanation for this finding, however, is not clear. Introduction of HIV-1 into the bloodstream may select for X4 strains, based on greater availability of X4 receptors in blood as compared with the genital tract (13). Although R5 strains are thought to initiate infection by all routes, data suggest that X4 strains may also be transmitted but suppressed, reemerging later in the course of infection (15, 25). Two caveats are relevant to the patients studied here. First, IDU's may have acquired HIV-1 heterosexually (21). Second, genital inflammation can stimulate the expression of R5 receptors dramatically, conferring a selective advantage on R5 virus in the genital tract of women with STIs (13). The relationship of coreceptor usage to route of transmission is relevant to the initiation of infection, and needs to be pursued further.
This study documented intrapatient recombination between quasispecies from different compartments in the same individual. Predicted by recognition of recombinants between HIV-1 subtypes, this finding is relevant to the formation and dissemination of viral variants within infected individuals. Because recombination results in evolutionary leaps, it can lead to the rapid accumulation of multiple genetic traits such as drug resistance that confer an advantage on the new viral strain. A recombinant strain described here was obtained from the CVL, but bears a V3 loop that confers the R5 phenotype and closely resembles those of plasma viruses from the same patient. It is likely that this recombinant had a selective advantage in the genital tract, where R5 coreceptors outnumber X4. The data presented here underscore the necessity of investigating the interaction of HIV-1 and the immune system in the female genital tract and blood.
Supplementary Material
Acknowledgments
We thank the Wadsworth Center Molecular Genetics Core for oligonucleotide synthesis and DNA sequence analysis and the study patients for their participation. This work was supported by National Institutes of Allergy and Infectious Diseases Drug Abuse and Child Health and Development Grants UO1-AI-35004, UO1-HD-032632, UO1-AI-31834, UO1-AI-34993, and RO1-AI-52015.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: WIHS, Women's Interagency HIV Study; CVL, cervicovaginal lavage; IDU, injection drug use.
Data Deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY316363–AY316522).
References
- 1.Joint United Nations Programme on HIV/AIDS (UNAIDS), World Health Organization (WHO)/AIDS Epidemic Update: December 2002 (UNAIDS, Geneva).
- 2.Malim, M. & Emerman, M. (2001) Cell 104, 469-472. [DOI] [PubMed] [Google Scholar]
- 3.Jung, A., Maier, R., Vartanian, J., Bocharov, G., Jung, V., Fischer, U., Meese, E., Wain-Hobson, S. & Meyerhans, A. (2002) Nature 418, 144. [DOI] [PubMed] [Google Scholar]
- 4.Cavert, W., Notermans, D., Staskus, K., Wietgrefe, S., Zupancic, M., Gebhard, K., Henry, K., Zhang, Z., Mills, R., McDade, H., et al. (1997) Science 276, 960-964. [DOI] [PubMed] [Google Scholar]
- 5.Wong, J., Ignacio, C., Torriani, F., Havlir, D., Fitch, N. & Richman, D. (1997) J. Virol. 71, 2059-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh, A., Besson, G., Mobasher, A. & Collman, R. (1999) J. Virol. 73, 6680-6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang, L., Rowe, L., He, T., Chung, C., Yu, J., Yu, W., Talal, A., Markowitz, M. & Ho, D. (2002) J. Virol. 76, 9465-9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Overbaugh, J., Anderson, R., Ndinya-Achola, J. & Kreiss, J. (1996) AIDS Res. Hum. Retroviruses 12, 107-115. [DOI] [PubMed] [Google Scholar]
- 9.Poss, M., Rodrigo, A., Gosink, J., Learn, G., de Vange Panteleeff, D., Martin, H., Jr., Bwayo, J., Kreiss, J. & Overbaugh, J. (1998) J. Virol. 72, 8240-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellerbrock, T., Lennox, J., Clancy, K., Schinazi, R., Wright, T., Pratt-Palmore, M., Evans-Strickfaden, T., Schnell, C., Pai, R., Conley, L., et al. (2001) J. Infect. Dis. 184, 28-36. [DOI] [PubMed] [Google Scholar]
- 11.Delwart, E., Mullins, J., Gupta, P., Learn, G., Jr., Holodniy, M., Katzenstein, D., Walker, B. & Singh, M. (1998) J. Virol. 72, 617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ping, L., Cohen, M., Hoffman, I., Vernazza, P., Moiseiwitsch, F., Chakraborty, H., Kazembe, P., Zimba, D., Maida, M., Fiscus, S., et al. (2000) J. Virol. 74, 8946-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patterson, B., Landay, A., Andersson, J., Brown, C., Behbahani, H., Jiyamapa, D., Burki, Z., Stanislawski, D., Czerniewski, M. & Garcia, P. (1998) Am. J. Pathol. 153, 481-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovacs, A., Wasserman, S., Burns, D., Wright, D., Cohn, J., Landay, A., Weber, K., Cohen, M., Levine, A., Minkoff, H., et al. (2001) Lancet 358, 1593-1601. [DOI] [PubMed] [Google Scholar]
- 15.Philpott, S., Weiser, B., Tarwater, P., Vermund, S., Kleeberger, C., Gange, S., Anastos, K., Cohen, M., Greenblatt, R., Kovacs, A., et al. (2003) J. Infect. Dis. 187, 569-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Philpott, S., Weiser, B., Anastos, K., Kitchen, C., Robison, E., Meyer, W., III, Sacks, H., Mathur-Wagh, U., Brunner, C. & Burger, H. (2001) J. Clin. Invest. 107, 431-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei, X., Decker, J., Wang, S., Hui, H., Kappes, J., Wu, X., Salazar-Gonzalez, J., Salazar, M., Kilby, J., Saag, M., et al. (2003) Nature 422, 307-312. [DOI] [PubMed] [Google Scholar]
- 18.Back, N., Smit, L., De Jong, J., Keulen, W., Schutten, M., Goudsmit, J. & Tersmette, M. (1994) Virology 199, 431-438. [DOI] [PubMed] [Google Scholar]
- 19.Cheng-Mayer, C., Brown, A., Harouse, J., Luciw, P. & Mayer, A. (1999) J. Virol. 73, 5294-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landay, A., Benning, L., Bremer, J., Weiser, B., Burger, H., Nowicki, M. & Kovacs, A. (2003) J. Infect. Dis. 188, 209-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strathdee, S., Galai, N., Safaiean, M., Celentano, D., Vlahov, D., Johnson, L. & Nelson, K. (2001) Arch. Intern. Med. 161, 1281-1288. [DOI] [PubMed] [Google Scholar]
- 22.Fang, G., Weiser, B., Visosky, A., Townsend, L. & Burger, H. (1996) J. Acquired Immune Defic. Syndr. Hum. Retrovirol. 12, 352-357. [DOI] [PubMed] [Google Scholar]
- 23.Fang, G., Zhu, G., Burger, H., Keithly, J. & Weiser, B. (1998) J. Virol. Methods 76, 139-148. [DOI] [PubMed] [Google Scholar]
- 24.Lole, K., Bollinger, R., Paranjape, R., Gadkari, D., Kulkarni, S., Novak, N., Ingersoll, R., Sheppard, H. & Ray, S. (1999) J. Virol. 73, 152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rowland-Jones, S., Pinheiro, S. & Kaul, R. (2001) Cell 104, 473-476. [DOI] [PubMed] [Google Scholar]
- 26.Moir, S., Ogwaro, K., Malaspina, A., Vasquez, J., Donoghue, E., Hallahan, C., Liu, S., Ehler, L., Planta, M., Kottilil, S., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 6057-6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaul, R., Plummer, F., Kimani, J., Dong, T., Kiama, P., Rostron, T., Njagi, E., MacDonald, K., Bwayo, J., McMichael, A., et al. (2000) J. Immunol. 164, 1602-1611. [DOI] [PubMed] [Google Scholar]
- 28.Kaul, R., Trabattoni, D., Bwayo, J., Arienti, D., Zagliani, A., Mwangi, F., Kariuki, C., Ngugi, E., MacDonald, K., Ball, T., et al. (1999) AIDS 13, 23-29. [DOI] [PubMed] [Google Scholar]
- 29.Haigwood, N. & Zolla-Pazner, S. (1998) AIDS 12, Suppl. A, S121-S132. [PubMed] [Google Scholar]
- 30.Steinman, R., Granelli-Piperno, A., Pope, M., Trumpfheller, C., Ignatius, R., Arrode, G., Racz, P. & Tenner-Racz, K. (2003) Curr. Top. Microbiol. Immunol. 276, 1-30. [DOI] [PubMed] [Google Scholar]
- 31.Shibata, R., Igarashi, T., Haigwood, N., Buckler-White, A., Ogert, R., Ross, W., Willey, R., Cho, M. & Martin, M. (1999) Nat. Med. 5, 204-210. [DOI] [PubMed] [Google Scholar]
- 32.Shacklett, B., Cu-Uvin, S., Beadle, T., Pace, C., Fast, N., Donahue, S., Caliendo, A., Flanigan, T., Carpenter, C. & Nixon, D. (2000) AIDS 14, 1911-1915. [DOI] [PubMed] [Google Scholar]
- 33.Lee, S., Xu, Z., Lieberman, J. & Shankar, P. (2002) J. Clin. Invest. 110, 1339-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaech, S. & Ahmed, R. (2003) Science 300, 263-265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.