Abstract
By using a reverse genetics system that is based on the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV), we have identified the arenavirus small RING finger Z protein as the main driving force of virus budding. Both LCMV and Lassa fever virus (LFV) Z proteins exhibited self-budding activity, and both substituted efficiently for the late domain that is present in the Gag protein of Rous sarcoma virus. LCMV and LFV Z proteins contain proline-rich motifs that are characteristic of late domains. Mutations in the PPPY motif of LCMV Z severely impaired the formation of virus-like particles. LFV Z contains two different proline-rich motifs, PPPY and PTAP, which are separated by eight amino acids. Mutational analysis revealed that both motifs are required for efficient LFV Z-mediated budding. Both LCMV and LFV Z proteins recruited to the plasma membrane Tsg101, which is a component of the class E vacuolar protein sorting machinery that has been implicated in budding of HIV and Ebola virus. Targeting of Tsg101 by RNA interference caused a strong reduction in Z-mediated budding. These results indicate that Z is the arenavirus functional counterpart of the matrix proteins found in other negative strand enveloped RNA viruses. Moreover, members of the vacuolar protein sorting pathway appear to play an important role in arena-virus budding. These findings open possibilities for antiviral strategies to combat LFV and other hemorrhagic fever arenaviruses.
Arenaviruses include Lassa fever virus (LFV) and the South American hemorrhagic fever (HF) viruses. These viruses cause severe human disease, and they pose a threat as agents of bioterrorism (1). The prototypic arenavirus, lymphocytic choriomeningitis virus (LCMV), is an important model with which to study both acute and persistent viral infection (2). In addition, LCMV provides an excellent system with which to study basic aspects of the molecular and cell biology of HF arenaviruses.
LCMV is an enveloped virus, whose genome consists of two negative-sense, single-stranded RNA segments, called L (7.2 kb) and S (3.4 kb). Each segment uses an ambisense coding organization to direct the synthesis of two gene products in opposite orientation, and each is separated by an intergenic region (3) The S RNA encodes the nucleoprotein (NP), and the two surface virion glycoproteins (GPs), GP-1 and GP-2, which are derived by proteolytic cleavage of a precursor polypeptide, GP-C (4). GP-1 and GP-2 form the spikes on the virion envelope and mediate cell entry by interaction with the host cell surface receptor (5). The L RNA directs the synthesis of the virus RNA-dependent RNA polymerase (L protein), and a small RING finger protein called Z (11 kDa) (6). The NP associates with the viral genomic RNA species and L to form the viral ribonucleoprotein (RNP) core that is competent in transcription and RNA replication, and constitutes the minimal infectious unit (7). The role of Z in the virus life cycle is poorly understood, and homologues of Z are not found in other negative-strand (NS) RNA viruses. Z is a structural component of the virus (8), and in infected cells, Z has been reported to interact with several cellular factors, including promyelocytic leukemia protein (9), and the eukaryotic translation initiation factor 4E, the latter of which has been proposed to repress CAP-dependent translation (10, 11). In addition, early studies suggested a role of Z in viral transcriptional regulation (12).
We have developed a reverse genetic system for LCMV that allows for intracellular reconstitution of viral transcription and RNA replication, as well as for assembly and budding of infectious particles. By using this system, we showed that Z is not required for RNA synthesis mediated by the LCMV polymerase (13), but, rather, Z exhibits a dose-dependent inhibitory effect on both LCMV transcription and RNA replication (7). Similar findings have been subsequently reported for the Z protein of Tacaribe virus (TV), a New World arenavirus (14).
We have also shown that production of infectious virus-like particles (VLPs) requires both GP and Z (15). Arenaviruses bud from the plasma membrane, which involves the association of the virus RNP core with host-derived membrane that is highly enriched with the viral GP (16). For many NS RNA viruses, this process is mediated mainly by the virus matrix (M) protein (17). The Gag and M proteins of retroviruses and NS RNA viruses, respectively, contain late (L) domains that play an essential role in the last steps of virus budding (18, 19). Three types of motifs have been so far defined within viral L domains: P(T/S)AP, PPxY, and YxxL (18, 19), where “x” is any amino acid. L domains are highly conserved and have been shown to mediate interaction with host cell proteins required for virus budding. Thus, Tsg101, a component of the class E vacuolar protein sorting (VPS) cellular machinery, has been reported to interact with the PTAP motif of Ebola virus VP40 and HIV-Gag proteins. This interaction recruits Tsg101 to sites of particle assembly, which is required for efficient formation of HIV-1 virions and Ebola VLPs (20, 21). On the other hand, the WW domains of the cellular ubiquitin ligase, Nedd4, interact with the PPxY motifs that are present on Rous sarcoma virus (RSV) Gag, Ebola VP40, and VSV M proteins (22). This interaction results in covalent addition of ubiquitin to the viral Gag and M proteins and subsequent particle formation (18, 19).
Arenaviruses do not have a counterpart of the M protein that is found in many other NS RNA viruses. The requirement of Z for VLP production suggested its possible role in arenavirus budding. Here, we present evidence that Z is the main driving force of arenavirus budding. We also show that both LCMV and LFV Z proteins have characteristic features of bona fide budding viral proteins, including self-budding activity and the ability to substitute for an unrelated L domain (18, 19). Arenavirus Z proteins contain conserved proline motifs at their C terminus, which are similar to those that are present within the L domains of several Gag and M proteins. The PPPY motif present at the C terminus of LCMV Z was strictly required for efficient assembly and budding of VLP, whereas both nonoverlapping L domains, PTAPP and PPPY, were required for LFV Z-mediated budding. Both LCMV and LFV Z proteins colocalized with Tsg101 at the plasma membrane, and targeting of Tsg101 by RNA interference (RNAi) caused a strong inhibition of VLP production. These results indicate that Z is a functional counterpart of the M protein present in enveloped NS RNA viruses that bud from the plasma membrane, and that Tsg101 plays an important role in arenavirus budding. Our findings raise the possibility of targeting arenavirus budding as an antiviral strategy to combat HF arenaviruses.
Materials and Methods
Viruses, Cells, Transfection Procedures, and Western Blots. LCMV strain Armstrong (ARM) 53b was grown in baby hamster kidney (BHK)-21 cells as described (23). vTF7–3 was provided by B. Moss (National Institutes of Health, Bethesda) (24). Human embryonic kidney (HEK)-293T and African green monkey kidney COS-1 cells were maintained in high-glucose DMEM supplemented with 2 mM glutamine and 10% heat-inactivated FCS. BHK-21 cells were maintained in DMEM supplemented with 10% FCS, 2 mM glutamine, 1× tryptose phosphate broth, 1 mM sodium pyruvate, and 0.5% glucose. HEK-293T and COS-1 cells were transfected with Lipofectamine (Invitrogen) as described (15).
For Western blot analysis, cells were harvested in sample buffer (50 mM Tris·HCl, pH 8.0/62.5 mM EDTA/1% Nonidet P-40/0.4% deoxycholate). Cell lysates (CE) were analyzed by Western blotting as described (25). Immunodetection of the proteins was done by using the Boehringer Mannheim chemiluminescence kit (Roche Molecular Biochemicals). We used rabbit polyclonal antibodies against hemagglutinin (HA) (Santa Cruz Biotechnology) and GFP (Molecular Probes).
Plasmids. Plasmids pC-L, pC-NP, pC-Z, and pC-GP express the polymerase, NP, Z, and GP of LCMV, respectively, and have been described (15). Plasmids pC-VSV G (26) and pC-T7 (15) express the VSV G and the bacteriophage T7 RNA polymerase (T7RP), respectively. The backbone and DNA polymerase II promoter in plasmid pC are the same promoters present in plasmid pCAGGS (27). LCMV and LFV Z cDNAs were tagged at their C termini with an influenza HA epitope by using a patch PCR strategy (28), and were then cloned between the EcoRI and BglII sites of pCAGGS, or between the NcoI and BamHI sites of pUCIRES. Mutations in the proline-rich motifs were introduced by using the Stratagene site-directed mutagenesis kit.
Plasmid pMG#7Δ2G was generated by deleting the two 3′ termini G residues of the T7RP promoter in plasmid pMG#7 (29). In cells expressing T7RP, pMG#7Δ2G allows for synthesis of a minigenome (MG) RNA consisting of the 5′ and 3′ UTR and intergenic region of the LCMV ARM S segment, and a chloramphenicol acetyltransferase (CAT) reporter gene substituting for the NP ORF. The 5′ end of the MG#7Δ2G RNA starts with the nontemplate G residue reported to be present at the 5′ termini of arenavirus genomic and antigenomic RNA species (3).
Plasmids pC-Tsg101myc (30) and pSUPER (31) were obtained from S. N. Cohen (Stanford University School of Medicine, Stanford, CA) and R. Agami (The Netherlands Cancer Institute, Amsterdam), respectively. Plasmids pS-EGFP, a gift from C. H. McGowan (The Scripps Research Institute) and pS-Tsg101 allow for intracellular expression mediated by an H1 RNA promoter of small interfering (si)RNA that specifically targets enhanced GFP (EGFP) and Tsg101, respectively.
To generate pS-Tsg101, two annealed 64-bp oligonucleotides, each encoding two 19-nt reverse complements homologous to nucleotides 415–433 of Tsg101 coding sequence separated by a short spacer region, were cloned between the BglII and HindIII cloning sites of pSUPER. The 19-nt sequence used had been reported to efficiently promote degradation of Tsg101by RNAi (20). To generate plasmid pS.EGFP, the selection of the target region (nucleotides 11–30) was completed by using the program OLIGOENGINE WORKSTATION.
Plasmids pSV.Myr, pSV.RSV Gag, and pSV.VSV M-GagN have been described (32). To generate the LCMV or LFV Z RSV Gag chimeras, the corresponding ORF was amplified by PCR to introduce an NdeI site at the AUG, and a SpeI site after the last codon of each Z ORF. This PCR product was ligated to a 130-nt SstI–NdeI fragment bearing the 5′ UTR from pSV.Myr. The ligated fragments were reamplified by using a 5′ primer, which spans the upstream SstI site and the 3′ SpeI-containing primer. This cDNA was cut with SstI and SpeI and cloned into pSV.Myr, and was digested with the same enzymes.
Plasmid constructs and mutations were verified by DNA sequencing. Sequences of the primers used for cloning and mutagenesis, and detailed experimental conditions are available on request.
Generation and Infectivity of VLPs. HEK-293T cells (1.5 × 106) growing in 35-mm-diameter wells were transfected with plasmids encoding NP (0.8 μg), L (1 μg), GP (0.4 μg), Z (0.1 μg), T7RP (1 μg), and MG#7Δ2G (0.5 μg). Control transfections lacking any of the support plasmids received empty pCAGGS plasmid to keep the total amount of DNA transfected in each well constant. All plasmid DNA was prepared by using Qiagen (Valencia, CA) technology. At 72 h posttransfection, supernatants (SP) (1.5 ml) were saved, and CE were prepared for CAT assay as described (29). Aliquots of the clarified SP (600 μl) were used to infect fresh monolayers of BHK-21 cells in 12-well plates. After 4 h of adsorption, the inoculum was removed, and LCMV ARM helper virus was added at a multiplicity of infection (moi) of 3. After adsorption for 2 h, the inoculum was removed and fresh medium was added. Seventy-two hours later, CE were prepared and were analyzed in a CAT assay.
Isolation and Analysis of RNA Present in VLP. At 72 h posttransfection, VLP-containing SPs were collected and clarified at low speed. An aliquot (0.9 ml) was layered on top of a 20% sucrose cushion, and was centrifuged at 50,000 rpm for 90 min at 4°C on a TLS55 rotor. RNA was extracted from the VLP-containing pellet by using Tri reagent (Molecular Research Center, Cincinnati). Before the reverse transcription reaction with SuperScript II and BsmICAT primer, possible contaminant DNA was removed by using the DNA-free kit (Ambion, Austin, TX). PCR was performed by using Taq polymerase (Roche Diagnostics, Indianapolis) and specific primers BsmICAT (5′-GATGAATGCTCATCCGGAATTCCG-3′) and PfmlICAT (5′-CCCAGGGATTGGCTGAGACAAAAAACATATTC-3′) to amplify a segment of 280 base pairs within the CAT ORF.
Budding Assays. To analyze self-budding properties of Z, COS-1 cells were infected with vTF7–3 at an moi of 10, and were then transfected with the indicated T7 promoter-expression plasmids. Cells were metabolically labeled with [35S]Met/[35S]Cys, starting at 2 h posttransfection, and, 10 h later, cells and culture SPs were harvested as described (33), and were analyzed by immunoprecipitation as described (34), by using a guinea pig serum against LCMV, an α-LCMV Z rabbit polyclonal serum, or the α-HA rabbit polyclonal serum.
A budding assay of chimera Gag proteins was performed in COS-1 cells as described (32). At 48 h posttransfection, cells were labeled for 2.5 h with [35S]Met/[35S]Cys and Gag-related proteins in the medium, and CEs were analyzed by immunoprecipitation by using an anti-RSV serum that recognizes the Gag protein and its cleavage products.
Immunofluorescence. BHK-21 cells growing on coverslips were transfected with the indicated plasmids. Forty-eight hours later, cells were fixed with methanol:acetone and incubated with α-HA, α-myc (9E10), or both antibodies. As secondary antibodies, we used fluorescein-conjugated antibodies against mouse IgG or rhodamine X-conjugated antibodies against rabbit IgG (The Jackson Laboratory). Stains were visualized by using a Bio-Rad MRC1024 confocal microscope and oil-immersion ×63 objective. All 2D confocal images illustrate a single Z section captured at a position approximating the midline of the cell.
Results
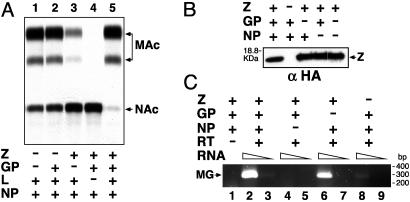
The Small RING Finger Z Protein Is the Main Driving Force of Arenavirus Budding. LCMV GP and Z proteins are required for production of infectious LCMV-like particles (VLPs) (15). The requirement of GP was expected, due to its role in receptor recognition and virus entry (35), whereas the need for Z suggested a possible role of this protein in virus assembly and budding. We hypothesized that Z could be the arenavirus-functional counterpart of the M proteins that mediate budding in other NS RNA viruses. To test this hypothesis, we examined the requirement of LCMV proteins for efficient production of VLPs. For this examination, we transfected HEK-293T cells with different combinations of plasmids (Fig. 1A). LCMV MG expression in transfected cells was determined, based on levels of CAT activity in CE (Fig. 1A). Consistent with our previous findings (7), L and NP were sufficient for efficient MG expression, whereas Z exhibited an inhibitory effect on LCMV MG expression (Fig. 1A, compare lanes 1 and 3). Coexpression of GP diminished the inhibitory effect of Z (Fig. 1A, compare lanes 3 and 5). Moreover, in several independent experiments we consistently observed that lysates from transfected cells that contained Z and GP, in addition to the minimal transacting viral factors L and NP, had higher levels of CAT activity (Fig. 1, compare lanes 1 and 5). GP and NP were not required for the release of Z-containing vesicles into the SP of transfected cells (Fig. 1B). We next examined whether Z was able to mediate the generation of VLPs containing bona fide viral nucleocapsides (NCs). For this purpose, VLPs present in culture SP of transfected cells were pelleted, and their associated RNA was isolated and subjected to RT-PCR by using specific primers to amplify a region within the CAT ORF of the LCMV MG RNA (Fig. 1C). As predicted, the lack of NP prevented the formation of MG-containing NCs that could be incorporated into VLPs (Fig. 1C, lanes 4 and 5). The absence of GP resulted in only a slight decrease in VLP production (Fig. 1C, compare lanes 2 and 6). In contrast, absence of Z caused a dramatic reduction on VLP production (Fig. 1C, lanes 8 and 9). Together, these results indicated that Z is the main driving force of LCMV budding.
Fig. 1.
LCMV Z is strictly required for production of VLP. (A) CAT expression by an LCMV MG in the presence of different combinations of LCMV proteins. Nonacetylated (NAc) and monoacetylated (MAc) chloramphenicol. (B) Release of vesicles containing Z protein. VLPs present in SP of transfected cells were harvested by ultracentrifugation through a 20% sucrose cushion and were analyzed by Western blotting. (C) Detection by RT-PCR of MG RNA associated with VLPs, which are present in the SP of transfected cells.
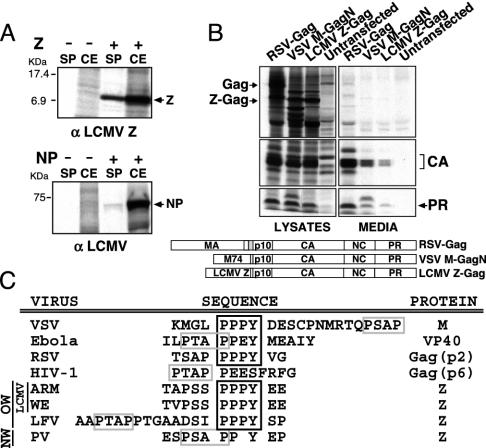
LCMV Z Has Features of a Bona Fide Budding Protein and Contains a Proline-Rich Motif (PPPY) That Is Necessary for Z-Mediated Budding. We first examined whether LCMV Z has the self-budding activity characteristic of bona fide viral budding proteins. Cells infected with vTF7–3 and transfected with Z or, as controls, with NP or empty plasmid, were radiolabeled with [35S]Met and equal amounts of CE or culture SP analyzed by immunoprecipitation (IPP) (Fig. 2A). Approximately 25% of the total Z protein synthesized in transfected cells was released into the SP (Fig. 2A Upper), whereas <2% of NP was detected in the SP (Fig. 2A Lower), which likely reflects minor cell damage due to the experimental conditions associated with cell transfection. Another feature that is characteristic of viral budding proteins is the flexibility of their L domains. One L domain substitutes for another in promoting virion release (18). To investigate this issue, we examined the budding properties of Z-Gag chimeric proteins, where Z was fused to an RSV Gag protein that lacked both its membrane targeting and binding signal (M domain) and L domain (32). Budding of Gag particles into the SP was monitored by the appearance of the cleavage products of correctly processed Gag protein: the capsid protein and the protease. Z efficiently substituted for the M and L domains present in the Gag protein of RSV (Fig. 2B, lane 7). As an additional control, we used a Gag protein in which its M and L domains had been replaced by the first 74 amino acids of VSV M, resulting in a chimeric VSV M-GagN protein. As reported (32), this chimera was also competent in promoting budding (Fig. 2B, lane 6).
Fig. 2.
Z has features of a bona fide budding protein. (A) Z has self-budding activity. Z was immunoprecipitated from the SP of transfected cells, and was metabolically labeled with [35S]Met/[35S]Cys. As a control, cells were transfected with a plasmid encoding for LCMV NPs. (B) Z can substitute for the RSV L domain. Proteins present in lysates and SP of cells transfected with the indicated construct and metabolically labeled with [35S]Met/[35S]Cys were immunoprecipitated to detect Gag cleavage products. (C) Comparison of the proline-rich motifs present in arenavirus Z and in different viral Gag and M proteins.
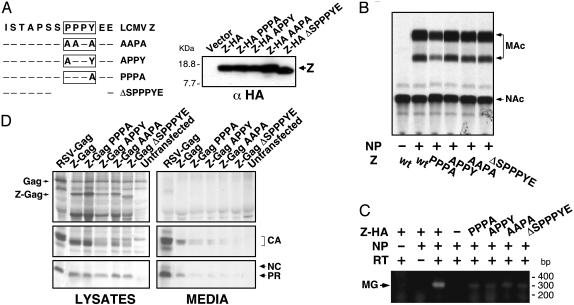
Arenavirus Z contains proline-rich motifs similar to those present in the L domains of Gag and M proteins of several viruses (Fig. 2C). The Z protein of LCMV contains a single PPPY motif, whereas the Z protein of the highly pathogenic arenavirus, LFV, possesses both PTAP and PPPY motifs separated by eight amino acids. To determine whether the PPPY motif was essential for LCMV Z-mediated budding, we introduced alanine substitutions, either singly or in various combinations within the PPPY motif (Fig. 3A). In the case of the ΔSPPPYE mutant, the entire motif was removed. To facilitate the detection of these proteins, they were tagged at their C termini with an HA epitope. All mutants were expressed to similar levels (Fig. 3A). Mutations in the PPPY motif did not affect transcription and replication levels of the MG RNA (Fig. 3B), but all of them resulted in lower levels of VLPs containing MG NCs (Fig. 3C). Moreover, mutant Z proteins were impaired in their ability to substitute for the L domain of RSV Gag protein (Fig. 3D). These results indicated that the PPPY motif present on the LCMV Z protein is a functional L domain.
Fig. 3.
The PPPY motif is responsible for the budding activity of LCMV Z. (A) Sequence and expression of Z proteins with mutated PPPY motifs is shown. Identity is represented by a dashed line. Proteins were detected by Western blot by using an α-HA antibody. (B) Mutant proteins had no effect on MG-derived CAT activity. (C) Mutations in the PPPY motif caused a dramatic reduction in VLP production. MG RNA associated with VLP was amplified by RT-PCR. (D) Z-containing mutations in the PPPY failed to restore the budding properties of RSV Gag protein lacking its L domain. Proteins present in the lysates and SP of transfected cells were analyzed by IPP. CA, capsid protein; PR, protease.
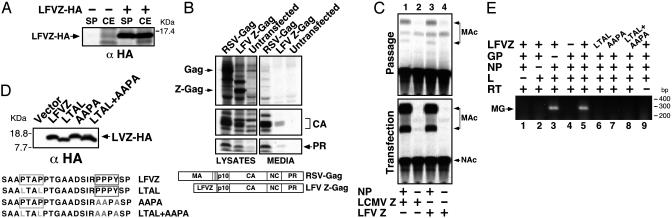
LFV Z PTAP and PPPY Motifs Are Both Required for Efficient Budding of VLP. We next investigated whether the Z protein of the highly pathogenic arenavirus, LFV, had also properties of a bona fide budding protein. We first examined whether LFV Z had self-budding activity and could also substitute for the L domain of the RSV Gag protein. COS-1 cells infected with vTF7–3 and transfected with pUC-LFVZHA were radiolabeled with [35S]Met/ [35S]Cys, and both the SP and CE were analyzed by IPP using an anti-HA antibody. Approximately 40% of the total LFV Z protein synthesized in transfected cells was found in the SP (Fig. 4A). In addition, LFV Z was able to restore the budding properties of an RSV Gag mutant protein that lacked its M and L domains (Fig. 4B). Furthermore, LFV Z substituted very efficiently for its LCMV counterpart in the VLP formation assay (Fig. 4C).
Fig. 4.
Both PTAP and PPPY motifs are required for efficient budding mediated by LFV Z. (A) LFV Z has self-budding activity. IPP of LFV Z from the SP of transfected cells labeled with [35S]Met/[35S]Cys. (B) LFV Z can functionally substitute for the L domain of RSV Gag. IPP of proteins present in the lysates and SP of transfected cells revealed the presence of Gag cleavage products. (C) LFV Z can efficiently substitute for LCMV Z in formation of infectious VLPs. (D) The sequence and expression of LFV Z with mutated proline-rich motifs. Mutated residues are gray. Intact motifs are represented by a box. Proteins were detected by Western blot by using an α-HA antibody. (E) Mutations in both the PTAP and the PPPY motif impaired production of VLP. MG RNA associated with VLPs was amplified by RT-PCR.
LFV Z contains two L domain motifs: PTAP and PPPY, which are separated by eight amino acids. To assess the contribution of each individual motif to budding, we mutated each motif individually and simultaneously (Fig. 4D) in an LFV Z protein tagged at its C terminus with an HA epitope. Mutant LFV Z proteins were efficiently expressed in transfected cells (Fig. 4D), and all of them were impaired in their budding activity, based on undetectable levels of VLP-associated MG RNA in the SPs of transfected cells (Fig. 4E). These results indicated that both proline-rich domains are required for efficient budding mediated by LFV Z.
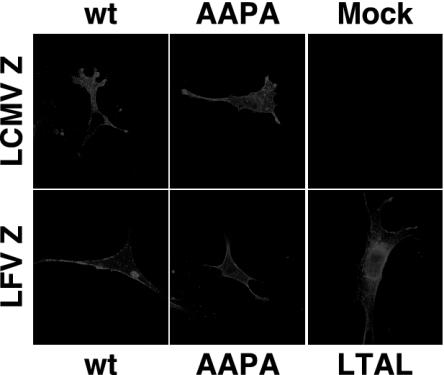
Intracellular Localization of Wild-Type and Mutant Z Proteins of LCMV and LFV. Impaired budding activity of mutant Z proteins could be explained by their mislocalization within the cell and their inability to localize to the cell membrane. To address this question, we used indirect immunofluorescence and confocal microscopy to analyze cells expressing either wild-type or mutant Z proteins (Fig. 5). Both wild-type and mutant Z proteins exhibited the same pattern of staining that included a clear association with the plasma membrane. Cells transfected with empty vector alone served as a negative control. Therefore, the inability of mutant Z proteins to promote viral assembly and budding was not due to their mislocalization, but, rather, was related to an impaired L domain function.
Fig. 5.
Wild-type and mutant LCMV and LFV Z proteins have the same pattern of intracellular distribution when expressed in BHK-21 cells. AAPA, Z proteins whose PPPY domain has been mutated to AAPA; LTAL, Z proteins whose PTAP motif has been mutated to LTAL.
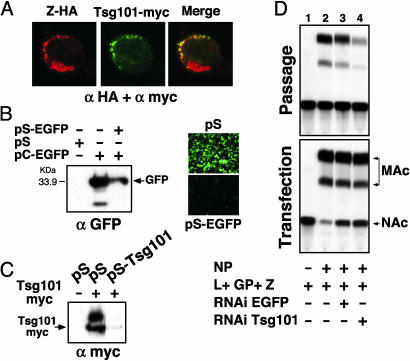
The Role of Tsg101 in Arenavirus Budding. Tsg101 has been implicated in the budding of HIV-1 and Ebola virus through its interaction with the PTAP motif present in the L domains of Gag and VP40, respectively (36, 37). The PTAP motif present in LFV Z suggested its possible interaction with Tsg101. Confocal microscopy studies of cells cotransfected with LFV Z-HA, or LCMV Z-HA, and Tsg101-myc, showed that both LFV and LCMV Z proteins colocalized with Tsg101 (Fig. 6A). To assess a possible functional role of Tsg101 in arenavirus budding, we examined the effect of reducing the cellular levels of Tsg101 on the budding of VLPs. The lack of cell lines deficient for Tsg101 led us to consider the use of RNAi. For this purpose, we generated plasmid pSTsg101. This plasmid contains, between the H1 promoter and terminator sequences, a 19-nt sequence derived from the Tsg101 gene, which is separated by a short spacer sequence from the reverse complement of the same 19 nucleotides. As a control, we used plasmid pS-EGFP that contains sequences that allow for intracellular production of siRNA that specifically targets the EGFP mRNA. Cells cotransfected with pS-EGFP and pC-EGFP exhibited a dramatic reduction in the expression levels of EGFP (Fig. 6B). Cotransfection of pS-Tsg101, but not pS-EGFP control, caused a severe reduction in the levels of Tsg101-myc expression (Fig. 6C). We then cotransfected HEK-293T cells with all of the plasmid required for VLP formation in the presence of pS-Tsg101 or control plasmid pS-EGFP. The inclusion of pS-EGFP in the transfection mix did not affect VLP production. In contrast, cotransfection of pS-Tsg101 resulted in a significant reduction in VLP production, which was measured as a decrease in the CAT activity transmitted to fresh BHK-21 monolayers (Fig. 6D Upper, compare lanes 3 and 4). The same effect was observed when LCMV Z was replaced by LFV Z (data not shown).
Fig. 6.
(A) Z coexpression of LCMV Z-HA (red) and Tsg101-myc (green) in BHK-21 cells results in colocalization of both proteins. (B) Transfection of a plasmid that directs the synthesis of siRNA specific for EGFP (pS-EGFP) resulted in decreased levels of EGFP protein, as detected by Western blot or direct fluorescence. (C) Cotransfection of cells with plasmids Tsg101-myc and pS-Tsg101 causes a severe reduction in the expression levels of Tsg101-myc protein. (D) Intracellular expression of siRNA specific for Tsg101 results in impaired VLP formation.
Discussion
The mechanisms responsible for arenavirus budding have not been previously addressed. The M protein found in most enveloped NS RNA viruses plays a key role in viral assembly and budding (16). However, arenaviruses do not appear to have an M protein, and crosslinking studies have suggested a possible direct interaction between GP2 and the virus RNP core without a need for an M-like protein.
Arenaviruses encode for a small RING finger protein Z without an apparent counterpart in other NS RNA viruses. The role of Z in the virus life cycle is poorly understood. In this study, we have shown that Z is the main driving force of arenavirus budding (Fig. 1). Hence, Z can be considered as the arenavirus-functional counterpart of the M protein found in many other enveloped NS RNA viruses. Z displayed properties characteristic of bona fide budding proteins, including self-budding (Fig. 2A) and the capacity to substitute for the unrelated L domain of RSV (Fig. 2B). The Z-Gag chimeric constructs used in our studies lacked both Gag M and L domains (32). Hence, budding of Z-Gag particles required that Z was also capable of providing membrane targeting and binding activities. A conserved myristoylation sequence motif present at their N termini may provide Z proteins with membrane-targeting properties. Consistent with this idea, mutation of G2 to A resulted in a Z protein defective in budding activity (M.P. and J.C.d.l.T., unpublished results). Consistent with their budding properties, the C termini of Z proteins contain proline-rich motifs (PPxY and PT/SAP) found in L domains (ref. 18 and Fig. 2C). The integrity of these motifs was strictly required for efficient Z-mediated budding of VLP (Figs. 3 and 4 C–F). Wild-type Z and Z proteins with mutated L domain motifs were expressed to similar levels and exhibited similar intracellular expression patterns (Figs. 3A,4D, and 5), suggesting that the lack of budding activity of the mutant Z proteins was related to a defect in the last steps of virus egress.
Comparison of known arenavirus Z proteins revealed that TV is the only member of the family whose Z protein does not contain either PPxY or PT/SAP tetrapeptide motif. Instead, TV Z contains the YxxL motif found in the L domain of the retrovirus equine immune anemia virus (38). In addition, TV Z contains an ASAP sequence that partially mimics the L domain, PSAP. Budding mediated by PSAP-containing L domains appears to require interaction with the cellular protein Tsg101 (18). The change, P to A, in the first position of this L domain, reduced its binding to Tsg101 only 3-fold (20). Therefore, it is possible that the ASAP motif of TV Z might still support interaction with Tsg101. Likewise, the Z of the WE strain of LCMV was reported to have the sequence LPPY, instead of PPPY found in other LCMV strains. However, after revisiting the sequence of the WE Z gene, we determined that amino acid position 85 was P, instead of L, and, hence, WE Z also has the PPPY motif.
LFV, an HF arenavirus, contains both PTAP and PPPY motifs, which are separated by eight amino acids. Interestingly, both PTAP and PPPY motifs are also found overlapping within amino acids 7–13 (7PTAPPEY13) in the budding protein VP40 of Ebola virus (18, 19, 39), another causative agent of severe HF. In the case of Ebola VP40, each motif alone was active as an L domain (39). In contrast, LFV Z-containing mutations that disrupted either PTAP or PPPY motifs were severely impaired in their ability to promote VLP production (Fig. 4 C–E). This finding suggests, that, in the case of LFV Z, each motif may engage different host factors, with all of them being required for efficient LFV Z-mediated budding.
LCMV and LFV Z proteins are colocalized with Tsg101 in the proximity of the plasma membrane (Fig. 6A). Compelling evidence indicates that budding of viruses with a PTAP-containing L domain requires interaction between Tsg101 and the tetrapeptide PTAP (18), which is present in LFV Z. However, the L domain found in LCMV Z is of the class PPxY, suggesting that recruitment of Tsg101 by LCMV Z may not be due to a direct interaction, but, rather, is mediated by a third protein capable of binding to both of them. In this regard, the ubiquitin ligase, Nedd4 protein, has been involved in the budding of several viruses through its interaction with PPxY motifs, but Nedd4 also interacts with Tsg101 (40).
The arenavirus Z is, to our knowledge, the first example of a RING finger protein as the key regulator of budding of an NS RNA virus. RING finger domains mediate protein–protein interactions, which could contribute to Z recruitment of cellular proteins required for arenavirus budding. Consistent with such a view, a mutant Z protein, in which the first loop of the RING finger was disrupted by mutations to A of the conserved C in positions 32 and 35, failed to promote budding of VLP (not shown).
Increasing evidence indicates that virus budding from the plasma membrane involves a complex interplay between viral proteins and members of the VPS cellular machinery, including Tsg101, VPS4, and Nedd 4 (18). Reduced intracellular expression of Tsg101 by the use of RNAi caused a dramatic reduction in budding mediated by either LCMV or LFV Z proteins, suggesting that Tsg101 plays a fundamental role in arenavirus budding. Additional cellular proteins are likely to contribute to arenavirus budding. The identification these cellular proteins, together with the characterization of their interactions with Z, will open the possibility of targeting arenavirus budding as an antiviral strategy to combat highly pathogenic HF arenaviruses, including LFV.
Acknowledgments
We thank S. N. Cohen for the c-Myc-tagged Tsg101 construct, and C. Wilson and P. Lam for excellent technical assistance. This work was supported by National Institutes of Health Grants AI47140 (to J.C.d.l.T.) and R37 CA47482 (to R.C.C.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LCMV, lymphocytic choriomeningitis virus; LFV, Lassa fever virus; TV, Tacaribe virus; L, late; VLP, virus-like particle; HF, hemorrhagic fever; NP, nucleoprotein; GP, glycoprotein; NS, negative strand; M, virus matrix; RSV, Rous sarcoma virus; RNAi, RNA interference; BHK, baby hamster kidney; HEK, human embryonic kidney; HA, hemagglutinin; MG, minigenome; EGFP, enhanced GFP; CAT, chloramphenicol acetyltransferase; SP, supernatant; CE, cell lysate; NC, nucleocapside; IPP, immunoprecipitation.
References
- 1.Borio, L., Inglesby, T., Peters, C. J., Schmaljohn, A. L., Hughes, J. M., Jahrling, P. B., Ksiazek, T., Johnson, K. M., Meyerhoff, A., O'Toole, T., et al. (2002) J. Am. Med. Assoc. 287 2391-2405. [DOI] [PubMed] [Google Scholar]
- 2.Oldstone, M. B. (2002) in Arenaviruses, ed. Oldstone, M. B. (Springer, Berlin), Vol. 263, pp. 83-118. [Google Scholar]
- 3.Buchmaier, M. J., Bowen, M. D. & Peters, C. J. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Williams & Wilkins, Philadelphia), Vol. 2, pp. 1635-1668. [Google Scholar]
- 4.Beyer, W. R., Popplau, D., Garten, W., von Laer, D. & Lenz, O. (2003) J. Virol. 77 2866-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, W., Henry, M. D., Borrow, P., Yamada, H., Elder, J. H., Ravkov, E. V., Nichol, S. T., Compans, R. W., Campbell, K. P. & Oldstone, M. B. (1998) Science 282 2079-2081. [DOI] [PubMed] [Google Scholar]
- 6.Salvato, M. S. & Shimomaye, E. M. (1989) Virology 173 1-10. [DOI] [PubMed] [Google Scholar]
- 7.Cornu, T. I. & de la Torre, J. C. (2001) J. Virol. 75 9415-9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salvato, M. S., Schweighofer, K. J., Burns, J. & Shimomaye, E. M. (1992) Virus Res. 22 185-198. [DOI] [PubMed] [Google Scholar]
- 9.Borden, K. L., Campbell Dwyer, E. S. & Salvato, M. S. (1998) J. Virol. 72 758-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell Dwyer, E. J., Lai, H., MacDonald, R. C., Salvato, M. S. & Borden, K. L. (2000) J. Virol. 74 3293-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kentsis, A., Dwyer, E. C., Perez, J. M., Sharma, M., Chen, A., Pan, Z. Q. & Borden, K. L. (2001) J. Mol. Biol. 312 609-623. [DOI] [PubMed] [Google Scholar]
- 12.Garcin, D., Rochat, S. & Kolakofsky, D. (1993) J. Virol. 67 807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, K. J., Novella, I. S., Teng, M. N., Oldstone, M. B. & de la Torre, J. C. (2000) J. Virol. 74 3470-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez, N., Jacamo, R. & Franze-Fernandez, M. T. (2001) J. Virol. 75 12241-12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, K. J., Perez, M., Pinschewer, D. D. & de la Torre, J. C. (2002) J. Virol. 76 6393-6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garoff, H., Hewson, R. & Opstelten, D. J. E. (1998) Microbiol. Mol. Biol. Rev. 62 1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mebatsion, T., Konig, M. & Conzelmann, K. K. (1996) Cell 84 941-951. [DOI] [PubMed] [Google Scholar]
- 18.Freed, E. O. (2002) J. Virol. 76 4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pornillos, O., Garrus, J. E. & Sundquist, W. I. (2002) Trends Cell Biol. 12 569-579. [DOI] [PubMed] [Google Scholar]
- 20.Garrus, J. E., von Schwedler, U. K., Pornillos, O. W., Morham, S. G., Zavitz, K. H., Wang, H. E., Wettstein, D. A., Stray, K. M., Cote, M., Rich, R. L., et al. (2001) Cell 107 55-65. [DOI] [PubMed] [Google Scholar]
- 21.Martin-Serrano, J., Zang, T. & Bieniasz, P. D. (2001) Nat. Med. 7 1313-1319. [DOI] [PubMed] [Google Scholar]
- 22.Harty, R. N., Brown, M. E., McGettigan, J. P., Wang, G., Jayakar, H. R., Huibregtse, J. M., Whitt, M. A. & Schnell, M. J. (2001) J. Virol. 75 10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salvato, M., Shimomaye, E. M., Southern, P. & Oldstone, M. B. (1988) Virology 164 517-522. [DOI] [PubMed] [Google Scholar]
- 24.Fuerst, T. R., Niles, E. G., Studier, F. W. & Moss, B. (1986) Proc. Natl. Acad. Sci. USA 83 8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 26.Takada, A., Robison, C., Goto, H., Sanchez, A., Murti, K. G., Whitt, M. A. & Kawaoka, Y. (1997) Proc. Natl. Acad. Sci. USA 94 14764-14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niwa, H., Yamamura, K. & Miyazaki, J. (1991) Gene 108 193-199. [DOI] [PubMed] [Google Scholar]
- 28.Cornu, T. I. & de la Torre, J. C. (2002) J. Virol. 76 6678-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez, M. & de la Torre, J. C. (2003) J. Virol. 77 1184-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, L., Liao, J., Ruland, J., Mak, T. W. & Cohen, S. N. (2001) Proc. Natl. Acad. Sci. USA 98 1619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Science 296 550-553. [DOI] [PubMed] [Google Scholar]
- 32.Craven, R. C., Harty, R. N., Paragas, J., Palese, P. & Wills, J. W. (1999) J. Virol. 73 3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Justice, P. A., Sun, W., Li, Y., Ye, Z., Grigera, P. R. & Wagner, R. R. (1995) J. Virol. 69 3156-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Barreno, B., Delgado, T. & Melero, J. A. (1996) J. Virol. 70 801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunz, S., Borrow, P. & Oldstone, M. B. (2002) Curr. Top. Microbiol. Immunol. 262 111-137. [DOI] [PubMed] [Google Scholar]
- 36.Freed, E. O. (2003) Trends Microbiol. 11 56-59. [DOI] [PubMed] [Google Scholar]
- 37.Timmins, J., Schoehn, G., Ricard-Blum, S., Scianimanico, S., Vernet, T., Ruigrok, R. W. & Weissenhorn, W. (2003) J. Mol. Biol. 326 493-502. [DOI] [PubMed] [Google Scholar]
- 38.Puffer, B. A., Parent, L. J., Wills, J. W. & Montelaro, R. C. (1997) J. Virol. 71 6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Licata, J. M., Simpson-Holley, M., Wright, N. T., Han, Z., Paragas, J. & Harty, R. N. (2003) J. Virol. 77 1812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carter, C. A. (2002) Trends Microbiol. 10 203-205. [DOI] [PubMed] [Google Scholar]