Abstract
The hyperthermophile Nanoarchaeum equitans is an obligate symbiont growing in coculture with the crenarchaeon Ignicoccus. Ribosomal protein and rRNA-based phylogenies place its branching point early in the archaeal lineage, representing the new archaeal kingdom Nanoarchaeota. The N. equitans genome (490,885 base pairs) encodes the machinery for information processing and repair, but lacks genes for lipid, cofactor, amino acid, or nucleotide biosyntheses. It is the smallest microbial genome sequenced to date, and also one of the most compact, with 95% of the DNA predicted to encode proteins or stable RNAs. Its limited biosynthetic and catabolic capacity indicates that N. equitans' symbiotic relationship to Ignicoccus is parasitic, making it the only known archaeal parasite. Unlike the small genomes of bacterial parasites that are undergoing reductive evolution, N. equitans has few pseudogenes or extensive regions of noncoding DNA. This organism represents a basal archaeal lineage and has a highly reduced genome.
The discovery and cultivation of Nanoarchaeum equitans (1), probably representing a novel archaeal kingdom, raised new questions about the evolution of the Archaea. These hyperthermophiles grow only in coculture with another archaeon, Ignicoccus sp., and phylogenetic analysis of their 16S rRNA sequences suggests that they diverged early in the archaeal lineage, before the emergence of the Euryarchaeota and Crenarchaeota. The unknown nature of this symbiosis and the remarkable evolutionary divergence of N. equitans raised the question of whether this archaeon is “primitive” or has been extensively modified by reductive evolution (2, 3).
Organisms undergoing reductive evolution usually have a surplus of pseudogenes and noncoding DNA in their genomes (4). Although these vestiges of a more complex ancestral genome implicate organisms currently undergoing reductive evolution, the process must eventually stop in a free-living organism and the evidence of gene loss may erode. The assertion that an organism branches deeply from the tree of life is even more controversial, and the potential for artifacts of phylogenetic inference and genetic exchange is well known (2). Even organisms believed to be deeply branching have acquired some genetic material by horizontal transfer (5). To determine whether the small genome of N. equitans is the product of reductive evolution driven by its symbiosis or whether the organism represents the “primitive” archaeal ancestor, we have sequenced its genome.
Materials and Methods
Library Construction and DNA Sequencing. N. equitans was grown in a 300-liter fermenter in a coculture with Ignicoccus sp. and the N. equitans cells were purified from Ignicoccus as described (1). The cell pellet was lysed by enzymatic and chemical digestion, followed by the isolation and purification of genomic DNA (6–8). Genomic DNA was either digested with restriction enzymes or sheared to provide clonable fragments. Two plasmid libraries were made by subcloning randomly sheared fragments of this DNA into a high-copy number vector (≈2.8 kbp library) or low-copy number vector (≈6.3 kbp library). DNA sequence was obtained from both ends of plasmid inserts to create “mate-pairs,” pairs of reads from single clones that should be adjacent to one another in the genome. Library construction, DNA sequencing, and assembly methods were essentially as described (9–11). The assembly procedures resulted in a single scaffold of four contigs comprising 489,082 base pairs. The gaps between the four contigs were then sequenced, resulting in a single circular sequence.
Annotation. A set of computational methods was applied to the N. equitans genome. Two gene prediction programs, GLIMMER (12) and CRITICA (13), were run on the assembled sequences. The results of the two programs were merged to generate a unique set of genes. When the two programs selected different start codons for genes with the same stop codon, the longer gene was included in the set for further analysis. Additional genes were identified in the intergenic regions by using TBLASTN to compare DNA sequences with protein sequences from other archaeal genomes. The unique set of genes was then translated into amino acid sequences and subjected to BLASTP searches (with an E value cutoff of 10-10) against the nonredundant amino acid protein database (http://ncbi.nlm.nih.gov) (14). The predicted protein set was searched against the InterPro database release 3.1 (15) by using software modified from the original iprscan programs provided by InterPro. The predicted protein set was also searched against the NCBI Clusters of Orthologous Groups database mid-2001 update (16). Finally, gene family analysis was performed by using the NCBI BLASTCLUST program. Protein sets from the main scaffold and small scaffolds were compared with the protein sequences from all finished genomes deposited in the GenBank by using the BLASTP program. Inteinlike regions of split genes were identified by using the BLASTP program to search the InBase database (17).
tRNA genes were identified by the TRNASCAN-SE program (18) or the RNAMOTIF program by using customized motifs (19). Ribosomal RNAs were identified by searching the genomic sequences against a set of known rRNAs with BLASTN and verified by profile alignment to multiple alignments from known rRNA sequences. Small nucleolar-like RNAs (snoRNAs) were identified by using a profile-hidden Markov model constructed from an alignment of Pyrococcus furiosus snoRNAs by the HMMER program (20, 21). A new model was constructed from an alignment of N. equitans snoRNAs and used to iteratively search for additional snoRNAs.
Preparation of Alanyl–tRNA Synthetases and Aminoacylation Assay. The methods were adapted from Ahel et al. (22). N. equitans alaS1 (NEQ547), N. equitans alaS2 (NEQ211) and Methanococcus jannaschii alaS genes were amplified by PCR from the respective genomic DNA and cloned into the pCR2.1 TOPO vector (Invitrogen). Correct sequences were subcloned into pET11a (Invitrogen) for expression of the proteins in the E. coli BL21-Codon Plus (DE3)-RIL strain (Stratagene). Cultures were grown at 37°C in Luria–Bertani medium supplemented with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol. Expression of the recombinant proteins was induced for 3 h at 30°C by addition of 1 mM isopropyl α-D-thiogalactopyrano-side before cell harvesting. Cells were resuspended in buffer containing 50 mM Tris·HCl, pH 7.5, and 300 mM NaCl, and broken by sonication. S-100 fractions were extensively flocculated at 70°C for 45 min, and then centrifuged for 30 min at 20,000 × g. Supernatants were collected and stored at 4°C before use in aminoacylation assays.
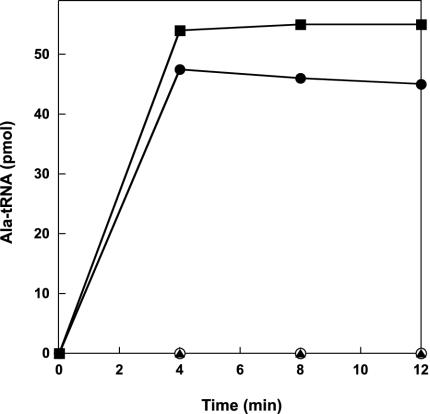
Aminoacylation was performed in a 0.1 ml reaction at 70°C in 50 mM Hepes (pH 7.2), 50 mM KCl, 10 mM ATP, 50 μM [3H]alanine (52 Ci/ml; 1 Ci = 37 GBq), 15 mM MgCl2, 5 mM 2-mercaptoethanol, 3 mg/ml unfractionated M. jannaschii tRNA, and the different alanyl–tRNA synthetases (100 nM). Aliquots of 20 μl were removed at the time intervals indicated in Fig. 2, and radioactivity was measured as described (22).
Fig. 2.
Alanylation of unfractionated M. jannaschii tRNA by alanyl–tRNA synthetases. The purification and aminoacylation procedures were adapted from Ahel et al. (22) and are detailed in Materials and Methods. The enzymes used are M. jannaschii AlaRS (filled squares), N. equitans AlaRS1 N-terminal part (open circles), N. equitans AlaRS2 C-terminal part (filled triangles), and N. equitans AlaRS1 + AlaRS2 (filled circles).
Phylogenetic Analysis. A concatenated alignment of 35 ribosomal proteins was obtained from Matte-Tailliez (23). To this alignment we added the N. equitans, Methanopyrus kandleri, and eukaryotic outgroup sequences (Arabidopsis thaliana and Saccharomyces cerevisiae). The alignment was then recalculated with CLUSTALW (24) and optimized by hand with BIOEDIT (25). The program RASA was used to evaluate the alignment for the presence of long branches (26). Alignments can be obtained by request from the authors. Maximum likelihood analysis was performed with PROML from the PHYLIP package (version 3.6a2.3) (27) by using the Jones–Taylor–Thornton model, the default program parameters, and a randomized input order of sequences with three jumbles. One hundred bootstrap resamplings were performed to assess the support for individual branches. Bayesian analysis of the data set was done with the MRBAYES software (28). Four simultaneous chains were run, using the Markov chain Monte Carlo method, for 200,000 generations after the convergence of the likelihood values, using the default settings of the program. A 50% majority-rule consensus tree was generated based on the resulting 2,000 trees and the bipartition values (percentage representation of a particular clade) were recorded at the nodes. The program PAUP* (29) was used for parsimony analysis. The alignment was sampled for 500 bootstrap replicates. Each bootstrap replicate was analyzed with 10 random addition sequence replicates with tree bisectionreconnection branch swapping and equal weighting for all sites.
Results and Discussion
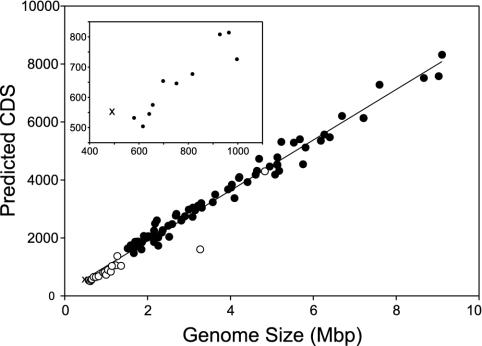
The genome of N. equitans (GenBank accession no. AACL01000000) consists of a single, circular chromosome of 490,885 bp and has an average G+C content of 31.6%. All 61 sense codons are used, but in line with the low G+C content the third codon position is mainly A or T. We identified 552 coding DNA sequences (CDS) with an average length of 827 bp. No extrachromosomal elements could be detected (either by biochemical methods or during sequencing). Despite having the smallest genome of a cellular organism sequenced to date, this archaeon has an unusually high gene density, with CDS and stable RNA sequences covering ≈95% of the genome. There is a remarkably good correlation between microbial genome size and the predicted number of CDS in a genome, an average of one gene per 1,090 bp (Fig. 1). However, the N. equitans genome contains even more predicted CDS than the genome of Buchnera aphidicola str. Sg, which is 23% larger (30). In contrast to the other small microbial genomes that are undergoing reductive evolution, N. equitans has little noncoding DNA and few recognized pseudogenes.
Fig. 1.
Correlation between microbial genome size and the number of predicted coding DNA sequences CDS. Bacterial genomes predicted to be undergoing reductive evolution are indicated by open circles, whereas other genomes are indicated by filled circles. The N. equitans genome is marked by “x”.(Inset) An expansion of the data from small microbial genomes with the abscissa shown in genome size units of kbp.
Functional roles could be assigned to two-thirds of the annotated genes. Among the CDS of unknown function, only 18.3% have homologs in other organisms, whereas the remaining ones are unique to N. equitans (see Supporting Data Set 1, which is published as supporting information on the PNAS web site, www.pnas.org). Predicted protein sequences from 39 genes grouped into 37 clusters of archaeal genome signature proteins, proteins that are believed to function uniquely in the Archaea (31). Three-quarters of these signature clusters include both euryarchaeal and crenarchaeal homologs and may be fundamental to the archaeal cell type.
Genes encoding single copies of 5S, 16S, and 23S rRNA and 38 tRNAs were identified along with at least 14 sno-like RNAs. These noncoding RNAs exhibit much higher G+C content (65–80%) than the rest of the genome and are readily identified by their base composition, as observed for other AT-rich hyperthermophiles (32). The rRNA genes were not found in an operon. Gene clusters (putative operons), although less common in archaea than in bacteria, are rarely conserved between N. equitans and other archaeal genomes. Even ribosomal proteins that are clustered together in bacterial, euryarchaeal, and crenarchaeal genomes are dispersed in this genome (33).
Unlike its Ignicoccus host, which gains energy by using hydrogen to reduce elemental sulfur, N. equitans has no genes to support a chemolithoautotrophic physiology. However the genome does encode two enzymes for amino acid oxidative deamination: a branched-chain amino acid aminotransferase (NEQ190) and glutamate dehydrogenase (NEQ077). There are also a limited number of enzymes that could catalyze electron transfer reactions. The genome encodes five subunits of an archaeal A1A0-type ATP synthase, including the major A and B subunits, subunit D, subunit I (required for ion translocation), and proteolipid subunit K. This minimal ATPase is much simpler than the prototypical nine-subunit ATPase purified from the euryarchaeon M. jannaschii (34). Because knowing the bioenergetics of N. equitans metabolism will be essential to understanding its symbiotic relationship with Ignicoccus, future studies must test whether N. equitans can produce ATP by electron transport phosphorylation or whether it derives energy from its host.
Given its small genome size, it is not surprising that N. equitans lacks the metabolic capacity to synthesize many cell components. This organism lacks almost all known genes that are required for the de novo biosyntheses of amino acids, nucleotides, cofactors, and lipids. Exceptions include a multifunctional prephenate dehydrogenase/chorismate mutase/prephenate dehydratase (NEQ192), similar to the Archaeoglobus fulgidus enzyme used in aromatic amino acid biosynthesis, the archaeal Glu-tRNAGln amidotransferase (GatDE; NEQ126 and NEQ245+NEQ396) and the Asp-tRNAAsn/Glu-tRNAGln amidotransferase (GatABC; NEQ360, NEQ185, and NEQ513) (35). Also missing are genes for glycolysis/gluconeogenesis, the pentose phosphate pathway, the tricarboxylic acid cycle, and other known pathways for carbon assimilation. This absence of metabolic capacity necessitates the transport of most cellular metabolites from the host Ignicoccus.
Although a number of putative transporters have been identified in this genome, this set of membrane proteins appears insufficient to import all of the metabolites required by N. equitans. Transporters include a mechanosensitive ion channel, three ATP-binding cassette-type transporter systems, a CaCAtype Na+/Ca2+ antiporter, a metal ion transporter and a member of the tellurite/dicarboxylate transporter family. In addition, this genome encodes the SecYE, SecDF and signal peptidase components of a protein translocase, although it appears to lack the protein and RNA components of the signal recognition particle found in other archaea. This system could be used to export the surface-layer protein that forms the N. equitans cell wall. One gene (NEQ300) encodes a potential S-layer protein containing a canonical N-terminal secretory signal peptide. Although there is no evidence that N. equitans cells are motile or encode flagellar proteins, this organism has two members (NEQ169 and NEQ425) of the type II/type IV protein export system used for pili or archaeal flagella biosynthesis. One such appendage has been observed in electron micrographs of N. equitans and could be used to attach to its Ignicoccus host (3).
N. equitans may acquire its lipids directly from its host Ignicoccus: a striking feature of this organism is the vast formation of vesicles at its cytoplasmic membrane (36), which may be part of a mechanism to supply cell components to N. equitans. Because similar features are found in parasites, the genome of N. equitans points to a parasitic lifestyle. In addition to housekeeping chaperones and proteases, the genome encodes 11 proteases and peptidases that could be used to hydrolyze proteins derived from either the environment or the host cell. Although low densities of N. equitans may not significantly affect the growth of Ignicoccus (1), the presence of several N. equitans cells per host cell prevented multiplication of the Ignicoccus (U. Jahn, personal communication).
In contrast to many obligate bacterial parasites, N. equitans has an extensive repertoire of DNA repair enzymes. Base and nucleotide excision repair enzymes include endonucleases III, I V, and V (NEQ126a, NEQ398, NEQ077a, NEQ368, NEQ346a), flap endonuclease-1 (NEQ088), and Rad25 DNA helicase (NEQ369). A dUTP diphosphatase (NEQ329) reduces uridine misincorporation into DNA, which uracil DNA glycosylase (NEQ372) would otherwise remove. Genes encoding the recombination protein RadA (NEQ426), DNA double-strand break repair protein Rad50 (NEQ256), single-strand DNAbinding protein (NEQ199), and a Holliday-junction resolvase (NEQ424) suggest that N. equitans can undergo homologous recombination. The presence of a full set of archaeal DNA repair and recombination enzymes differentiates N. equitans from other organisms with small genomes and may be required to repair DNA damage that is likely to occur in its hightemperature habitat. It is noteworthy that many organisms with small genomes have lost recombination/repair enzymes even though these losses have significant negative consequences. Organisms that lack these enzymes are at best evolutionarily stable (30) and at worst subject to the negative effects of increased occurrence and fixation of deleterious mutations (37).
In contrast to its paucity of metabolic genes, N. equitans possesses a large and reasonably complete set of components for information processing (replication, transcription, and translation) and completion of the cell cycle. For transcription, a DNA-dependent RNA polymerase consisting of 14 subunits and the archaeal genre proteins involved in transcription initiation, elongation, and termination could be identified. The gene sets for DNA replication and cell cycle are similar to those found in the Euryarchaeota and contain several components usually absent from the Crenarchaeota (e.g., DNA polymerase II, two copies of FtsZ and histones). Thymidine biosynthesis is catalyzed by archaeal-type bifunctional dCTP deaminase/dUTP diphosphatase and flavin-dependent thymidylate synthase enzymes. Deoxyribonucleotides may be produced by an anaerobic ribonucleotide reductase enzyme using electrons from thioredoxin and thioredoxin reductase.
The translational machinery of N. equitans is similar to other archaea. However, three tRNA genes (for glutamate, histidine, and tryptophan) were not identified in the genome. These tRNA species may have an unusual sequence or structure causing them to be missed by standard search algorithms (18). Some tRNA fragments, widely separated on the chromosome, are observed; they could be joined to form active tRNAs. Alternatively, tRNA import from the host, the possibility of a tRNA species with dual function (based on different nucleotide modification), or of an anticodon change by RNA editing are also plausible. Four tRNA species (for serine, tyrosine, isoleucine, and methionine) contain single introns in the expected position. These introns could be excised by two copies of the archaeal-type splicing endoribonuclease (EndA: NEQ261, NEQ205) (38, 39).
Surprising for an organism with a compact minimal genome and no homolog of S-adenosylmethionine synthetase, N. equitans encodes an extensive set of RNA-modifying enzymes (40). tRNA methyltransferases (NEQ108, NEQ228, NEQ337, NEQ440, and NEQ522), rRNA methyltransferases (NEQ053, NEQ238, and NEQ384), pseudouridine synthases (NEQ293, NEQ333, and NEQ454) and the archaeosine/queosine insertion enzyme (NEQ124+NEQ305) were all predicted from the genome. In addition, homologs of the guide RNA-directed modification complex subunits were detected: fibrillarin (NEQ125) and NOP56 (NEQ342). N. equitans has at least 14 sno-like RNAs that direct site-specific 2′ O-methylation, primarily in rRNAs as observed in other archaea (21). Thus, guide RNA-directed modification may be an ancient characteristic of archaea and eukaryotes that was present in a predecessor of all known archaeal phyla. Overall, most of the information processing systems in N. equitans resemble their euryarchaeal counterparts, although they share some features specific to the crenarchaeal systems.
An unusual characteristic of the N. equitans genome is the high number of split genes, whose gene product is encoded by two unlinked CDS (Table 1). A majority of available archaeal genome sequences encode fused versions of these genes. In N. equitans, the split sites for most of these genes lie between functional domains of the encoded proteins; thus, it seems likely that the two separated genes are expressed to form subunits of a functional enzyme. The genes for two subunits of alanyl–tRNA synthetase are separated by half of the chromosome. These genes provided the opportunity to test the idea that the individual protein parts are catalytically inactive, but that they reconstitute activity when combined (41). Only a combination of both parts of the split protein yielded a fully active enzyme as checked by the standard aminoacylation assay (Fig. 2); thus, in this case, covalent linkage is not a prerequisite for enzyme activity.
Table 1. Split noncontiguous genes in N. equitans.
| Gene | CDS encoding N-terminal part | CDS encoding C-terminal part |
|---|---|---|
| Large helicase-related protein | NEQ003 | NEQ409 |
| Topoisomerase I | NEQ045 | NEQ324 |
| DNA polymerase I* | NEQ068 | NEQ528 |
| Archaeosine tRNA—guanine transglycosylase† | NEQ124 | NEQ305 |
| RNA polymerase subunit B‡ | NEQ173 | NEQ156 |
| Glu—tRNAGln amidotransferase (gatE) | NEQ245 | NEQ396 |
| Reverse gyrase§ | NEQ434 | NEQ318 |
| Hypothetical RNA-binding protein | NEQ438 | NEQ506 |
| Hypothetical protein | NEQ495 | NEQ096 |
| Alanyl—tRNA synthetase | NEQ547 | NEQ211 |
Also split in Methanothermobacter thermautotrophicus
Also split in Methanopyrus kandleri, the Methanosarcinales, A. fulgidus, the extreme halophiles and crenarchaea
Also split in methanogens, A. fulgidus, and extreme halophiles
Also split in Methanopyrus kandleri (different site)
Many archaeal DNA processing and replication genes contain inteins, intervening protein sequences that self-splice from nascent polypeptides. A split gene with remnants of an intein encodes the N. equitans DNA-directed polymerase I (Table 1). The C-terminal part of NEQ068 contains the A and B motifs for protein cleavage, whereas the N-terminal region of NEQ528 encodes the F and G motifs. Together they form a mini-intein, although the two genes are separated by 83 kbp on the chromosome. We predict that the two parts of the DNA polymerase are expressed separately and then covalently linked after the reassembled intein has been excised by a transsplicing mechanism, as observed in the DnaE protein from Synechocystis sp (42). Topoisomerase I and reverse gyrase are also known to contain inteins in some archaea; however, no intein sequences were detected in these split genes from N. equitans.
The N. equitans reverse gyrase is split into two distinct genes encoding a helicase (NEQ434) and a topoisomerase (NEQ318) domain. Reverse gyrase appears to be the fusion product of a helicase and a topoisomerase domain, and catalyzes positive supercoil formation in DNA (43, 44). Because this complex enzyme is present only in hyperthermophiles, it was concluded that hyperthermophily appeared secondarily in the evolution of life (45). In light of the presence of independent helicase and topoisomerase domains in the deep-rooted N. equitans, the evolution of hyperthermophily may have been a very early event in agreement with the view of a hot primeval earth (46).
Assuming that multidomain proteins evolved from the fusion of simple domains, then split genes could reflect the multisubunit ancestral state of the proteins (47, 48). Alternatively, genes may be split by DNA mutation, insertion, integration, or chromosomal rearrangement events (49). In the parasitic Rickettsia spp., gene degradation has produced a large number of split, colinear ORFs (50). Whereas most of these split genes are contiguous on the genome and are in the process of becoming pseudogenes (51), the split genes found in N. equitans are widely scattered about the chromosome and have few deletions or other evidence of deterioration. Two CDS (NEQ023 and NEQ455) are preceded by short sequences that may encode N-terminal regions of the proteins in alternative reading frames, deduced by their similarity to homologs in other archaea. Because the structures and functions are unknown for both genes, it is unclear whether these ORFs are in fact pseudogenes. The genetic conservation of split genes, along with the paucity of pseudogenes and a minimum of noncoding DNA (<5% of the chromosome) suggests that the N. equitans genome is evolutionarily stable compared with many bacterial parasites.
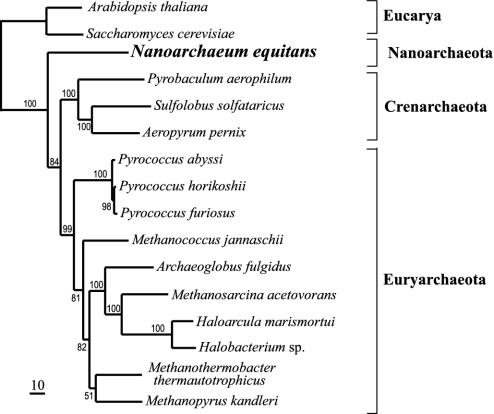
To shed light on the phylogenetic relationship of N. equitans among the Archaea, we concatenated and aligned the amino acid sequences of 35 ribosomal proteins. N. equitans was placed with high support at the most deeply branching position within the Archaea in the maximum likelihood, maximum parsimony, and Bayesian trees (Fig. 3), suggesting that the Nanoarchaeota diverged early within the Archaea. This result is consistent with the small subunit rRNA phylogeny reported previously (1).
Fig. 3.
Phylogenetic position of N. equitans within the Archaea. The tree was determined by the maximum likelihood method, based on 35 concatenated ribosomal protein sequences. Numbers indicate percentage of bootstrap resamplings. The scale bar corresponds to 10 estimated substitutions per 100 amino acid positions.
N. equitans is the first obligate archaeal symbiont, thus far cultured only in association with Ignicococcus sp. Two 16S rRNA sequences from Uzon Caldera (Kamchatka, Russia) and Yellowstone National Park (U.S.) exhibited 83% sequence similarity to N. equitans, and therefore represent a distinct group within the Nanoarchaeota (52). Light microscopy and fluorescence in situ hybridization reveal that these novel Nanoarchaeota are tiny cocci-like N. equitans attached to other archaeal species (M.J.H., unpublished data). No free-living Nanoarchaeota have been detected. Therefore, symbiosis may be widespread or even ubiquitous within the Nanoarchaeota. The minimal gene complement of N. equitans implies that this organism behaves parasitically: it must derive lipids, nucleotides, amino acids, cofactors, and possibly energy from its host. Although N. equitans has considerable proteolytic capacity for peptide degradation and may release ammonia from the oxidative deamination of amino acids, it is unclear whether these cells ever benefit their chemolithoautotroph hosts under environmental conditions. In contrast, Ignicoccus cells grow at least as well in pure culture as in symbiosis with N. equitans. Too high a burden of N. equitans cells inhibits Ignicoccus growth. This evidence suggests that N. equitans has a parasitic behavior.
Although N. equitans diverged relatively early in the archaeal lineage, it is not primitive (53). It contains complete versions of the modern archaeal-genre replication, transcription and translation systems, including a number of archaeal-specific innovations. Its genome encodes a significant number of archaeal signature proteins. Its few biosynthetic, DNA repair and RNA modification genes are characteristically archaeal. But this genome lacks the genes for central metabolism, primary biosynthesis and bioenergetic apparatus that are expected to have been present in an archaeal ancestor. As a highly modified, derived organism, N. equitans does not fit the stereotype of a microbial parasite undergoing genomic degradation. It has a highly compact genome with few pseudogenes or long regions of noncoding DNA. Consequently, we suggest that this microbe is a derived, but genomically stable parasite that diverged anciently from the archaeal lineage. The complexity of its information processing systems and the simplicity of its metabolic apparatus suggests an unanticipated world of organisms to be discovered.
Supplementary Material
Acknowledgments
We thank Ulrike Jahn for providing unpublished data. This work was partly supported by the Diversa Corporation and the Fonds der Chemischen Industrie (to K.O.S.), and by grants from the Department of Energy (to D.S.), the National Institute of General Medical Sciences (to D.S.), and the National Science Foundation (to D.E.G.).
Abbreviations: CDS, coding DNA sequences; snoRNA, small nucleolar-like RNA.
Data deposition: This Whole Genome Shotgun project has been deposited in the DDBJ/EMBL/GenBank databases under the project accession no. AACL00000000. The version described in this paper is the first version (accession no. AACL01000000).
References
- 1.Huber, H., Hohn, M. J., Rachel, R., Fuchs, T., Wimmer, V. C. & Stetter, K. O. (2002) Nature 417, 63-67. [DOI] [PubMed] [Google Scholar]
- 2.Boucher, Y. & Doolittle, W. F. (2002) Nature 417, 27-28. [DOI] [PubMed] [Google Scholar]
- 3.Huber, H., Hohn, M. J., Stetter, K. O. & Rachel, R. (2003) Res. Microbiol. 154, 165-171. [DOI] [PubMed] [Google Scholar]
- 4.Silva, F. J., Latorre, A. & Moya, A. (2001) Trends Genet. 17, 615-618. [DOI] [PubMed] [Google Scholar]
- 5.Ochman, H., Lawrence, J. G. & Groisman, E. A. (2000) Nature 405, 299-304. [DOI] [PubMed] [Google Scholar]
- 6.Zhou, J., Bruns, M. A. & Tiedje, J. M. (1996) Appl. Environ. Microbiol. 62, 316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robertson, D. E., Mathur, E. M., Swanson, R. V., Marrs, B. L. & Short, J. M. (1996) Soc. Indust. Microbiol. News 46, 3-8. [Google Scholar]
- 8.Short, J. M. (1997) Nat. Biotechnol. 15, 1322-1323. [DOI] [PubMed] [Google Scholar]
- 9.Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., Scherer, S. E., Li, P. W., Hoskins, R. A., Galle, R. F., et al. (2000) Science 287, 2185-2195. [DOI] [PubMed] [Google Scholar]
- 10.Myers, E. W., Sutton, G. G., Delcher, A. L., Dew, I. M., Fasulo, D. P., Flanigan, M. J., Kravitz, S. A., Mobarry, C. M., Reinert, K. H., Remington, K. A., et al. (2000) Science 287, 2196-2204. [DOI] [PubMed] [Google Scholar]
- 11.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304-1351. [DOI] [PubMed] [Google Scholar]
- 12.Delcher, A. L., Harmon, D., Kasif, S., White, O. & Salzberg, S. L. (1999) Nucleic Acids Res. 27, 4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Badger, J. H. & Olsen, G. J. (1998) Mol. Biol. Evol. 16, 512-524. [DOI] [PubMed] [Google Scholar]
- 14.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Apweiler, R., Attwood, T. K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., Bucher, P., Cerutti, L., Corpet, F., Croning, M. D., et al. (2001) Nucleic Acids Res. 29, 37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tatusov, R. L., Natale, D. A., Garkavtsev, I. V., Tatusova, T. A., Shankavaram, U. T., Rao, B. S., Kiryutin, B., Galperin, M. Y., Fedorova, N. D. & Koonin, E. V. (2001) Nucleic Acids Res. 29, 22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perler, F. B. (2002) Nucleic Acids Res. 30, 383-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowe, T. M. & Eddy, S. R. (1997) Nucleic Acids Res. 25, 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsui, V., Macke, T. & Case, D. A. (2003) RNA 9, 507-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eddy, S. R. (2003) hmmer (Washington University, St. Louis), Version 2.3.1.
- 21.Omer, A. D., Lowe, T. M., Russell, A. G., Ebhardt, H., Eddy, S. R. & Dennis, P. P. (2000) Science 288, 517-522. [DOI] [PubMed] [Google Scholar]
- 22.Ahel, I., Stathopoulos, C., Ambrogelly, A., Sauerwald, A., Toogood, H., Hartsch, T. & Söll, D. (2002) J. Biol. Chem. 277, 34743-34748. [DOI] [PubMed] [Google Scholar]
- 23.Matte-Tailliez, O., Brochier, C., Forterre, P. & Philippe, H. (2002) Mol. Biol. Evol. 19, 631-639. [DOI] [PubMed] [Google Scholar]
- 24.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall, T. (1999) Nucleic Acids Symp. Ser. 41, 95-98. [Google Scholar]
- 26.Lyons-Weiler, J., Hoelzer, G. A. & Tausch, R. J. (1996) Mol. Biol. Evol. 13, 749-757. [DOI] [PubMed] [Google Scholar]
- 27.Felsenstein, J. (2001) phylip: Phylogeny Inference Package (Department of Genetics, University of Washington, Seattle), Version 3.6a2.1.
- 28.Huelsenbeck, J. P. & Ronquist, F. (2001) Bioinformatics 17, 754-755. [DOI] [PubMed] [Google Scholar]
- 29.Swofford, D. (1996) paup*: Phylogenic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Version 4.0b.
- 30.Tamas, I., Klasson, L., Canback, B., Näslund, A. K., Eriksson, A.-S., Wernegreen, J. J., Sandström, J. P., Moran, N. A. & Andersson, S. G. E. (2002) Science 296, 2376-2379. [DOI] [PubMed] [Google Scholar]
- 31.Graham, D. E., Overbeek, R., Olsen, G. J. & Woese, C. R. (2000) Proc. Natl. Acad. Sci. USA 97, 3304-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein, R. J., Misulovin, Z. & Eddy, S. R. (2002) Proc. Natl. Acad. Sci. USA 99, 7542-7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wächtershäuser, G. (1998) Syst. Appl. Microbiol. 187, 483-494. [Google Scholar]
- 34.Lingl, A., Huber, H., Stetter, K. O., Mayer, F., Kellermann, J. & Müller, V. (2003) Extremophiles 7, 249-257. [DOI] [PubMed] [Google Scholar]
- 35.Tumbula, D. L., Becker, H. D., Chang, W.-Z. & Söll, D. (2000) Nature 407, 106-110. [DOI] [PubMed] [Google Scholar]
- 36.Huber, H., Burggraf, S., Mayer, T., Wyschkony, I., Rachel, R. & Stetter, K. O. (2000) Int. J. Syst. Evol. Microbiol. 50, 2093-2100. [DOI] [PubMed] [Google Scholar]
- 37.Moran, N. A. (1996) Proc. Natl. Acad. Sci. USA 93, 2873-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lykke-Andersen, J. & Garrett, R. A. (1997) EMBO J. 16, 6290-6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleman-Leyer, K., Armbruster, D. W. & Daniels, C. J. (1997) Cell 89, 839-847. [DOI] [PubMed] [Google Scholar]
- 40.Andachi, Y., Yamao, F., Muto, A. & Osawa, S. (1989) J. Mol. Biol. 209, 37-54. [DOI] [PubMed] [Google Scholar]
- 41.Burbaum, J. J. & Schimmel, P. (1991) Biochemistry 30, 319-324. [DOI] [PubMed] [Google Scholar]
- 42.Wu, H., Hu, Z. & Liu, X.-Q. (1998) Proc. Natl. Acad. Sci. USA 95, 9226-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krah, R., Kozyavkin, S., Slesarev, A. & Gellert, M. (1996) Proc. Natl. Acad. Sci. USA 93, 106-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forterre, P. (2002) Trends Genet. 18, 236-237. [DOI] [PubMed] [Google Scholar]
- 45.Forterre, P., de la Tour, C. B., Philippe, H. & Duguet, M. (2000) Trends Genet. 16, 152-154. [DOI] [PubMed] [Google Scholar]
- 46.Stetter, K. O. (1997) in Commentarii Pontificia Academica Scientiarium (Vatican City), Vol. IV, pp. 261-283. [Google Scholar]
- 47.Doolittle, W. F. (1978) Nature 272, 581-582. [Google Scholar]
- 48.Gilbert, W. (1987) Cold Spring Harbor Symp. Quant. Biol. 52, 901-905. [DOI] [PubMed] [Google Scholar]
- 49.Stoltzfus, A., Logsdon, J. M., Jr., Palmer, J. D. & Doolittle, W. F. (1997) Proc. Natl. Acad. Sci. USA 94, 10739-10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogata, H., Audic, S., Renesto-Audiffren, P., Fournier, P.-E., Barbe, V., Samson, D., Roux, V., Cossart, P., Weissenbach, J., Claverie, J.-M., et al. (2001) Science 293, 2093-2098. [DOI] [PubMed] [Google Scholar]
- 51.Andersson, J. O. & Andersson, S. G. E. (2001) Mol. Biol. Evol. 18, 829-839. [DOI] [PubMed] [Google Scholar]
- 52.Hohn, M. J., Hedlund, B. P. & Huber, H. (2002) Syst. Appl. Microbiol. 25, 551-554. [DOI] [PubMed] [Google Scholar]
- 53.Woese, C. (1998) Proc. Natl. Acad. Sci. USA 95, 6854-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.