Abstract
Despite the importance of tuberculosis as a public health problem, we know relatively little about the molecular mechanisms used by the causative organism, Mycobacterium tuberculosis, to persist in the host. To define these mechanisms, we have mutated virtually every nonessential gene of M. tuberculosis and determined the effect disrupting each gene on the growth rate of this pathogen during infection. A total of 194 genes that are specifically required for mycobacterial growth in vivo were identified. The behavior of these mutants provides a detailed view of the changing environment that the bacterium encounters as infection proceeds. A surprisingly large fraction of these genes are unique to mycobacteria and closely related species, indicating that many of the strategies used by this unusual group of organisms are fundamentally different from other pathogens.
Tuberculosis has been a major killer throughout history, and is still the cause of millions of deaths every year. The prevalence of this infection is largely due to the ability of Mycobacterium tuberculosis to persist in the host for long periods of time and cause disease even in the face of a highly orchestrated host immune response (1). This unusual ability, as well as the phylogenetic distance between M. tuberculosis and other common bacterial pathogens, suggests that mycobacteria may use unique pathogenic mechanisms. Furthermore, the chronic subacute nature of this disease is likely to require the bacterium to adapt to a continually changing host environment, and recent evidence suggests that different virulence determinants may be used at distinct stages of the disease (2). To define the molecular mechanisms used by this pathogen at each stage of the infection, we used transposon site hybridization (TraSH) to identify M. tuberculosis genes that are specifically required for survival during infection in a mouse model of tuberculosis. TraSH is a microarray-based technique designed to analyze large pools of transposon mutants to comprehensively identify genes that are essential for growth under different conditions (3). The ability to simultaneously monitor the growth of thousands of mutant strains throughout the infection has allowed the identification of both specific stresses to which the bacterium is exposed as well as mechanisms used by the bacterium to resist these insults.
Methods
TraSH to Identify Genes That Are Required for Infection. A library of transposon mutants was made in M. tuberculosis strain H37Rv by using the MycoMarT7 phage, as described (3). C57BL/6J mice were infected intravenously with 106 colony-forming units of the mutant library. At the indicated times after infection, surviving bacteria (≈200,000 clones) were recovered from spleen homogenates by plating on 7H10 agar medium (in vivo pools). Four mice were killed at the 1 week time point, and five mice were used at each other time point. The in vitro pool was generated by replating the library. Genomic DNA was isolated from each pool, and TraSH probes were generated for each pool and hybridized to microarrays as described (4). Probes were generated from each pool twice and analyzed on duplicate microarrays. Microarray data were collected by using GENEPIX software (Axon, Union City, CA) and analyzed by using GENESPRING software (Silicon Genetics, Redwood City, CA). To define mutations that produce in vivo growth defects, the data for each time point (8–10 microarray hybridizations) was averaged and filtered to include only those features whose ratios were significantly different from 1 (P < 0.05 by t test). We then excluded mutants with subtle defects (ratios that differed from the median by <2.5-fold).
Determination of in Vitro Growth Rates Using TraSH. Relative in vitro growth rates were determined previously (4). Briefly, the M. tuberculosis transposon library was allowed to grow as separated colonies on 7H10 agar, collected, and replated. The resulting colonies were collected and TraSH probe was generated. This probe was mixed with differentially labeled chromosomal DNA and hybridized to the microarray. Ratios <0.2 (TraSH probe/ chromosomal probe) indicated mutants with in vitro growth defects.
In Vivo and in Vitro Competition Experiments. The ΔbioF and ΔyrbE4A mutants were generated by allelic exchange, using specialized transduction, as described (5). In the ΔbioF strain, nucleotides 1776702–1777776 (encoding bioF) in the genome of H37Rv (GenBank accession no. AL123456) were replaced with the hygromycin resistance gene of Streptomyces hygroscopicus.In the ΔyrbE4A strain, nucleotides 3,920,862–3,920,096 were replaced. The other strains represent transposon mutants that were isolated by sequencing the insertion sites in random strains. To determine the relative in vivo growth rates of individual mutants, each mutant strain was mixed with wild-type H37Rv (≈1:1 ratio), and 2 × 105 colony-forming units were inoculated into the tail vein of C57BL/6J mice. Groups of three mice were killed at the indicated times after infection and the ratio of wild-type to mutant was determined by plating lung or spleen homogenates on 7H10 agar with and without antibiotic. To determine relative in vitro growth rates, each mutant was mixed with wild-type H37Rv and inoculated into 7H9 broth containing OADC enrichment and 0.05% Tween 80. The ratio of mutant to wild-type bacteria was determined by plating either immediately after inoculation or after 10 days of growth at 37°C.
Results
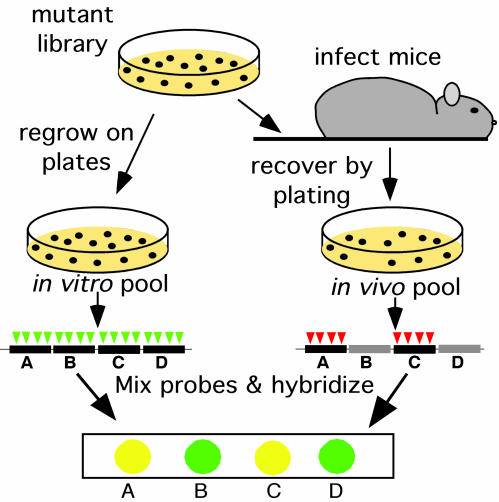
The strategy used to identify M. tuberculosis genes that are required for bacterial survival during infection is depicted in Fig. 1. A library of 100,000 transposon mutants in M. tuberculosis strain H37Rv, in which virtually every nonessential gene was mutated (4), was used to infect mice intravenously. The surviving bacteria were recovered from the spleen after 1, 2, 4, and 8 weeks (in vivo pools), and mutants that were underrepresented relative to an in vitro-grown pool were identified by using TraSH. Because 97% of the predicted genes in the genome are represented on the microarray, we were able to monitor the relative in vivo growth rate of nearly every viable mutant.
Fig. 1.
TraSH to identify attenuated mutants. C57BL/6J mice were infected intravenously with 106 colony-forming units of the mutant library. At the indicated times after infection, surviving bacteria were recovered by plating on agar medium (in vivo pools). The in vitro pool was generated by replating the library. The in vitro pool contained mutants with insertions (represented by triangles) in each nonessential gene (black bars). Mutants harboring insertions in genes that are specifically required for survival in the mouse spleen (gray bars) were lost from the in vivo pool. Genomic DNA was isolated from each pool, and TraSH probe was generated that was complementary to the chromosomal sequence flanking each insertion in the pool (4). The probes from the two pools were labeled with different colored fluorophores and mixed. Probes were then hybridized to a microarray onto which DNA fragments (features) were immobilized that were complementary to each gene in the genome of M. tuberculosis. Features that hybridized to the probe from the in vitro, but not the in vivo, pool represented genes that were specifically required for growth in the mouse.
For each time point, the data from 8–10 independent TraSH experiments were averaged (see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org, for the complete data set). Mutations that resulted in substantial in vivo growth attenuation were defined as those producing significantly reduced ratios of in vivo/in vitro-grown probes (defined in Methods). By applying these criteria to the data from each time point, 194 growth-attenuating mutations were defined (Table 3, which is published as supporting information on the PNAS web site). This represents ≈5% of the genome, which is similar to the number of attenuating mutations found in several large signature-tagged mutagenesis screens of other bacterial pathogens (6).
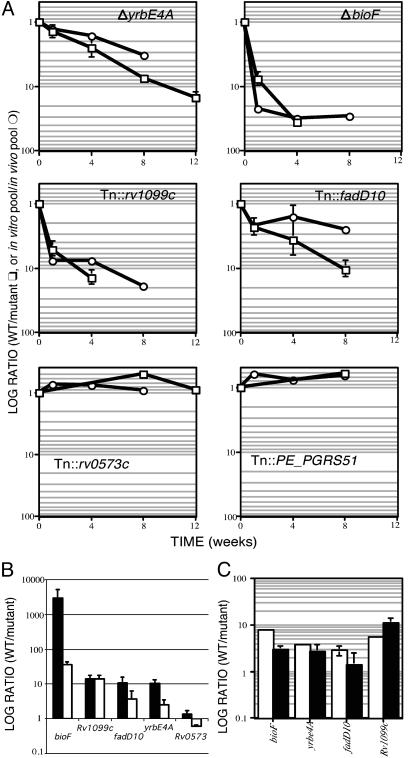
To assess the predictive value of the TraSH data, we isolated several mutant strains that we predicted to have varying in vivo-growth phenotypes. Mutants were isolated either by allelic exchange (ΔbioF and ΔyrbE4A) or by identifying strains with transposon insertions in appropriate genes (Tn::fadD10 and Tn::Rv1099c). The in vivo growth rate of each strain was then determined relative to wild-type bacteria in mixed infections (Fig. 2 A and B). Each of the strains that was predicted to be attenuated for growth did indeed show a significant defect compared with control strains (Tn::Rv0573c and Tn::PE_PGRS51). In addition, the TraSH data accurately predicted the time at which attenuation can first be detected (Fig. 2 A) and, therefore, these data can be used to distinguish between mutants with different in vivo growth kinetics. Although the TraSH ratio was correlated with the degree of attenuation, in some cases it underestimated the ratio of WT to mutant bacteria. It appears that the maximal attainable microarray ratio varies for each gene and, thus, although changes in the ratio for a particular gene over time are very informative, caution must be used when comparing the absolute ratios for different genes.
Fig. 2.
TraSH accurately predicts the in vivo growth characteristics of individual mutants. (A) Average TraSH ratios for the indicated genes (in vitro pool/in vivo pool) are plotted on a log scale as a function of time (circles). Individual strains carrying mutations in these genes were mixed with wild-type bacteria and inoculated into mice. The ratio (wild type/mutant) at each time point after infection is plotted (squares) on the same scale as the TraSH data. Data were normalized so the ratio of the inoculum equaled 1. Error bars represent standard deviations. (B) Growth of individual mutants in lung (filled bars) and spleen (open bars). Data were collected as in A from organs harvested after 4 weeks of infection except for Tn::Rv0573 (8 weeks). (C) Attenuated strains do not have in vitro growth defects. Each mutant strain was mixed with wild-type H37Rv and grown in triplicate broth cultures. The ratio of wild type to mutant was determined after inoculation (open bars) and after 10 days of growth (filled bars). Only the ratio for the ΔbioF mutant changed significantly (P = 0.0005; mutant grew faster than wild type).
In humans, where M. tuberculosis is generally transmitted via aerosol, the lung is the primary focus of infection. However, pathogenic mycobacteria can disseminate and grow in virtually any organ in the body; in the mouse model, both lung and spleen are infected. In this study, we analyzed the ability of mutants to survive in mouse spleens, because this allowed us to colonize a single organ with the entire library of 105 mutants. This model appears to be generally applicable to other sites of infection, because the four attenuated mutant strains that we have isolated based on TraSH data from spleen were also defective for growth in the lung (Fig. 2B).
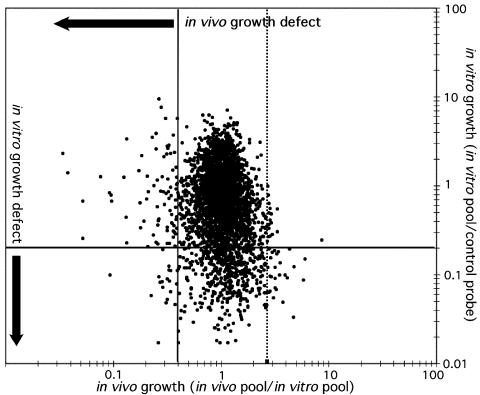
It is possible this analysis might identify mutants with generalized growth defects, because the in vivo pool has undergone several more rounds of division than the in vitro pool. To minimize this effect, we compared predicted in vivo growth rates of each mutant with previous data (4) on in vitro growth rates (Fig. 3). Only 22 of the 194 mutants that were predicted to have in vivo growth defects also grow poorly in vitro (annotated in Table 3). Furthermore, none of the four attenuated mutants that we isolated grew significantly slower than wild type in broth culture (Fig. 2C). Thus, the vast majority of the genes that we have identified are selectively required for growth in the mouse.
Fig. 3.
Comparison of in vivo and in vitro growth rates. Predicted in vivo growth rates are represented as TraSH ratios (8-week in vivo pool/in vitro pool) for each gene on the x axis. Solid vertical line represents cutoff value of 0.4×, which defines attenuating mutations. Dotted line indicates cutoff for the definition of mutants with increased in vivo growth rates. Relative in vitro growth rates were determined previously (4) and are represented on the y axis as the ratio of TraSH probe from the in vitro pool divided by a control probe of labeled chromosomal DNA. Horizontal line represents 0.2× cutoff value, identifying mutants with in vitro growth defects.
Genes of Known Function. Forty-five percent of the genes that we identified can be classified based on predicted function (Table 1). The distribution of functions for the genes that are necessary for in vivo growth differs greatly from those that were defined previously as important for growth on agar medium. Lipid metabolic genes and those involved in the transport or metabolism of inorganic ions and carbohydrates are prominently represented in the genes required in vivo. Genes with essential in vitro functions, such as those involved in translation and nucleotide and amino acid metabolism, are also likely to be important in vivo but, because strains with these mutations do not survive, they are not represented in the pools used for infection. Although functional predictions are lacking for only 29% of the genes that are required for growth on agar medium, the majority of those that are important for survival during infection have no annotated function. This observation high-lights our relative lack of knowledge regarding bacterial adaptation to the environment in the host.
Table 1. Predicted functional classifications of genes in this study.
|
In vivo
|
In vitro
|
|||
|---|---|---|---|---|
| Functional classification | No. of genes | Percent of category | No. of genes | Percent of category |
| Lipid metabolism | 15 | 7.5 | 30 | 14.9 |
| Carbohydrate transport and metabolism | 9 | 8.4 | 24 | 22.4 |
| Inorganic ion transport and metabolism | 8 | 8.0 | 8 | 8.0 |
| Cell envelope biogenesis, outer membrane | 8 | 7.3 | 32 | 29.4 |
| Amino acid transport and metabolism | 8 | 4.3 | 80 | 43.0 |
| Transcription | 7 | 5.4 | 15 | 11.6 |
| Coenzyme metabolism | 7 | 6.0 | 38 | 32.8 |
| DNA replication, recombination and repair | 5 | 4.6 | 19 | 17.4 |
| Translation, ribosomal structure | 5 | 3.9 | 76 | 59.4 |
| Signal transduction mechanisms | 4 | 5.2 | 12 | 15.6 |
| Secretion | 3 | 13.6 | 8 | 36.4 |
| Energy production and conversion | 3 | 1.6 | 31 | 16.3 |
| Cell division and chromosome partitioning | 2 | 8.7 | 8 | 34.8 |
| Posttranslational modification, chaperones | 2 | 2.5 | 27 | 34.2 |
| Nucleotide transport and metabolism | 1 | 1.5 | 25 | 38.5 |
| Unknown | 107 | 4.7 | 181 | 8.0 |
| Total | 194 | 5.0 | 614 | 15.7 |
In vivo and in vitro refer to genes that are required for optimal growth under either condition. In vitro genes were identified previously (4). “Percent of category” refers to the fraction of genes of particular functional class that are important for growth under each condition.
The functions that are predicted to be important during growth in the host suggest specific stresses to which the bacterium is exposed at different points during infection. In the mouse model, the first week after infection is a period of unrestrained growth during which the bacteria multiply at approximately the same rate as in broth culture. Despite this rapid growth, it appears that the in vivo environment is still quite challenging, because we identified 80 mutants that were already underrepresented at the 1-week time point. Clearly, the nutrients available to the bacteria are limiting during infection, as several auxotrophic mutants are known to be unable to survive in the mouse even at very early time points (7–10). Similarly, the set of genes that we identify at this time contains several that are involved in nutrient acquisition and cofactor biosynthesis. For example, mutations in each of the genes in the biotin biosynthetic pathway (except bioD, for which no data were available) result in some of the most dramatic in vivo growth defects in our study (Fig. 2 A and Table 3). In addition, four genes (sugA-C and lpqY) that are highly homologous to four-subunit disaccharide importers were predicted to be required at 1 week after infection. At later time points after infection, mycobacteria are thought to mainly use a C2 carbon source, such as fatty acid (11). Our data suggest that, at early time points, the nutrients available to the bacterium are quite different, and the inability to metabolize carbohydrates results in a severe growth defect. Other genes required at one week include several predicted to be involved in lipid metabolism or cell wall synthesis (Table 3). Mutations affecting the lipid-rich cell envelope of M. tuberculosis are known to result in reduced bacterial survival several weeks after infection (12–14). The TraSH data indicate that the integrity of this structure may also be critical at early time points, perhaps to resist elements of the innate immune system such as complement or antimicrobial peptides.
At ≈10 days after infection, the host environment changes dramatically, as do the genetic requirements for bacterial survival. The adaptive immune response is responsible for arresting the growth of M. tuberculosis at this time, and bacterial numbers in the spleen decrease by 10-fold between 2 and 4 weeks. This control of bacterial growth is largely dependent on increased production of reactive nitrogen intermediates (RNI) by activated macrophages (15). A potential mechanism to minimize the damage caused by these oxidizing agents is encoded by the kefB gene (Rv3236c). The Escherichia coli KefB protein is a K+ channel that protects the cell from the toxic effects of electrophilic species by reducing intracellular pH (16). Because the mutation of this gene in M. tuberculosis results in a significant growth defect at 4 weeks after infection, it is possible that the mycobacterial KefB is acting in an analogous fashion to limit the damage caused by host-derived radicals. Despite this system, RNI are still toxic to mycobacteria. These species have the ability to cause several different types of DNA damage, including deamination of cytosine (17), and the base excision repair system (BER) is important for mycobacterial resistance to RNI in vitro (18). We found that members of the BER system, uracil glycosylase and exonuclease III (encoded by ung and xthA, respectively), are required for in vivo growth at the 2-week time point. Uracil glycosylase specifically removes deaminated cytosine (uracil) residues from DNA. Thus, although RNI have the capacity to damage many different cellular constituents, a significant effect of RNI exposure in vivo appears be this specific DNA lesion. Interestingly, another BER enzyme, endonuclease IV (encoded by end, the mycobacterial ortholog of nfo), is required earlier, at 1 week after infection. In E. coli, exonuclease III and endonuclease IV have differing levels of activity toward different DNA lesions (19). Therefore, our data suggest that mycobacteria are exposed to different DNA-damaging agents as infection proceeds. As reactive oxygen intermediates are responsible for much of the DNA damage sustained by Salmonella typhimurium during the first week of infection (20), it is likely that these compounds are at least partly responsible for mycobacterial DNA damage at this time.
Genes of Unknown Function. No functional predictions were available for 55% of the genes that we identified as important for in vivo growth, and even more strikingly, 25% of these genes have no obvious homologs outside of mycobacteria and closely related species (Table 3). In part, this is a result of the unusual biochemistry and cellular architecture of these organisms, but it also undoubtedly reflects the presence of virulence mechanisms that are unique to mycobacteria. Many of these genes belong to three relatively large gene families, discussed below, that appear to have arisen by duplication and the TraSH data suggest that several members of these families have acquired specialized functions.
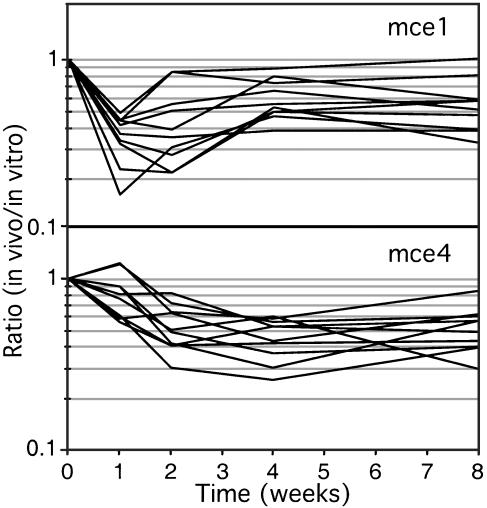
The first member of one such family (mce) was identified as a factor that increased the uptake of bacteria into nonphagocytic cells (21). This gene (mce1A) is located in a 12 gene operon (22) that has been duplicated four times around the chromosome (mce1-mce4). We find that two of these gene clusters are required for infection and, surprisingly, show kinetically distinguishable phenotypes. Mutations in the mce1 locus produce a growth defect one and two weeks after infection, and a partial recovery thereafter (Fig. 4). In contrast, mce4 mutants show a progressive growth defect that is not detectable until 2–4 weeks after infection. The microarray data are corroborated by the behavior of the ΔyrbE4A mutant (Fig. 2 A), in which the first gene of the mce4 operon was deleted. Thus, these structurally similar gene clusters are specialized to perform distinct functions and appear to be required at different times during infection. Notably, several of the genes in the mce1 and mce4 loci do not meet our strict criteria for attenuation. However, the similar phenotypes of the individual mutants (Fig. 4) strongly suggests that they are involved in a coordinate pathway and indicates that more extensive analyses that incorporate operon structure can reveal these subtle phenotypes.
Fig. 4.
In vivo behavior of mce mutants. TraSH ratios for each gene in the mce1 (Rv0169-Rv0178) or mce4 (Rv3492c-Rv3501c) loci are plotted as a function of time (yrbE1A and yrbE1B of mce1 locus were omitted because of lack of data). TraSH data compare mutants grown in vitro with those isolated from mouse spleens.
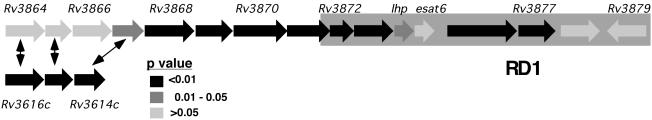
During the serial in vitro passage used to create the attenuated vaccine strain, M. bovis bacillus Calmette–Guérin, several large deletions occurred. One of these, termed RD1, removed the locus surrounding the secreted antigen ESAT-6 (23), and this event appears to be responsible for much of the attenuation seen in this strain (24, 25). Because this deletion removed all or part of nine genes, it remained uncertain which of these was responsible for the attenuated phenotype. We find that nearly all of the genes that surround esat-6 are required for infection (Fig. 5). In part, this could be caused by polar effects of transposon insertions on the transcription of downstream genes. However, the presence of a promoter in the transposon mitigates the attenuation of downstream transcription, and polar effects have not been observed in TraSH data (4). Furthermore, the transcription of downstream genes is unaffected in individual strains harbouring transposon insertions in two of the genes in this locus, Rv3870 and Rv3876 (D. Sherman, personal communication). Once again, the organization of these genes into an apparent transcriptional unit and the similarity of their mutant phenotypes suggest that the genes of this locus are performing a coordinated function, perhaps as steps in a biochemical pathway or subunits in a larger structure. This model is supported by recent work demonstrating that the secretion of the Esat-6 protein depends on at least two other genes in this locus (26).
Fig. 5.
Genes surrounding esat6 are required for in vivo survival. The esat-6 locus is depicted and shaded according to the significance level (P value) that the ratio for each gene differs from 1 (at the 4-week time point). Homology between Rv3614c-Rv3616c and the esat-6 locus is indicated by arrows. “RD1” refers to the region deleted in M. bovis bacillus Calmette–Guérin.
The esat-6 locus is a member of another gene family that appears to be specific to mycobacteria and close relatives (27). The genome of M. tuberculosis contains five imperfect copies of this gene cluster and one of these, Rv0282-Rv0291, was predicted to be essential for optimal growth on laboratory medium (4). Thus, like the mce genes, the different loci belonging to this family appear to have been retained because they have become specialized to function under different conditions. Although we cannot predict the importance of the Rv0282-Rv0291 locus in vivo, we suspect that it may play a role because only this cluster and the esat-6 locus are conserved in Mycobacterium leprae, an obligate pathogen. In addition to these large duplications, at least 10 smaller loci in the M. tuberculosis genome share some homology with these genes. One such cluster, Rv3614c-Rv3616c, is also predicted by our data to be essential for in vivo growth. Mutation of any of the genes in this locus results in severe attenuation at all time points (Fig. 5).
The largest and most distinctive class of mycobacteria-specific genes encode a group of 167 proteins of repetitive sequence belonging to the PE and PPE families, which have been implicated in the pathogenesis of several mycobacterial species (28–30). In our experiments, only three of these genes met the criteria for defining growth-attenuating mutations, Rv1807, Rv3872, and Rv3873, two of which are encoded in the RD1 region. Although mutations in several other PE and PPE genes appeared to have subtle defects, the fact that such a small fraction are detected in our system suggests either that most of these genes are able to functionally complement each other, or that they are required under conditions that we are unable to test.
Discussion
The set of attenuating mutations described here includes several genes that were previously found to be necessary for bacterial survival in the spleen. These include the alternative secA gene, secA2 (31); pirG, encoding a secreted protein of unknown function (32); and the RD1 locus (24). In addition, we identified the locus responsible for the synthesis and export of a cell wall-associated lipid, PDIM, which was identified in two different signature-tagged mutagenesis screens for mutants with growth defects in the lung (13, 29). Several of the genes in this locus were identified, such as drrA-C and mmpl7 (Table 3). Other attenuated mutants may not have been identified for several reasons. Mutations in some genes, such as icl, the gene that encodes the glyoxylate shunt enzyme isocitrate lyase (11), were not detected because they cause in vitro growth defects in the M. tuberculosis strain that we studied (data not shown). Other genes, such as relA, which is required during stationary phase growth (33), did not produce sufficient signal in all experiments to meet our criteria for attenuation. A closer analysis of gene clusters, such as the mce loci, suggests that the use of these strict criteria excluded several mutants that are truly attenuated, and likely resulted in an underestimation of the actual number of attenuating mutations. The specific animal model that was used for these experiments also influenced which genes were identified. For example, genes that are necessary for the initial colonization of the lung during an infection with aerosolized bacteria were likely missed in our study because of the route of infection that was used. Disease in mice differs markedly from human illness (34). Additionally, the phenotypes of individual mutants could have been influenced by the presence of the fully virulent bacteria that predominate the pool. This could have lead to an accentuation of mutant phenotypes caused by competition, or alternatively, specific mutants could have been complemented in trans by neighboring bacteria.
In many other bacterial pathogens, a surprisingly large number of mutations cause increased virulence in animal models of infection and the corresponding genes have been described as “anti-virulence” genes (35). It is thought that these genes are maintained either because they favor host survival and thereby increase transmission or alternatively, they may promote bacterial survival in a nonhost environment. When we used similar criteria as those that were used to identify attenuated mutants, we found 19 mutants that were overrepresented in the in vivo pool at the 8-week time point (Fig. 3; genes are annotated in Table 2). Interestingly, most of these mutants were predicted to grow poorly in vitro, suggesting that the increase in bacterial growth in vivo is balanced by a decrease in growth rate under other conditions. Although growth on laboratory medium is clearly not directly relevant to the evolution of M. tuberculosis, it is likely that the decrease in growth rate under this condition is indicative of a general deficit in the fitness of these mutants.
Several parallels can be drawn between the set of genes that we have defined as critical for mycobacterial infection and those that have been identified in other pathogens. Mutations that affect nutrient uptake systems or result in auxotrophy are attenuating in M. tuberculosis, as well as a variety of other intracellular and extracellular bacteria (9, 36, 37). This suggests that one of the common barriers to in vivo growth is the acquisition of essential nutrients. Similarly, as the production of reactive oxygen intermediates and RNI is a common host response to bacterial infection, most pathogens are able to resist the damage done by these toxic species. DNA repair mechanisms appear to be critical for surviving these insults in diverse species of bacteria, but the primary mechanisms that are used differ. Recombinational repair is of primary importance in Gram-negative organisms (20, 38). In contrast, the BER system is required for resistance to RNI in high G+C Gram-positive organisms, such as mycobacteria (18). We found that genes of the BER system, but not the recA gene were essential for mycobacterial growth in vivo, implying that this system, and not recombinational repair, is predominantly used by M. tuberculosis to resist oxidative damage during the first weeks of infection. At very late time points, an additional mechanism involving an error-prone polymerase is important for mycobacterial survival (39). We have also identified the KefB potassium channel as a potential mechanism of resistance to oxidative stress. Interestingly, expression of the kefB gene of Salmonella typhimurium is induced during intracellular growth in macrophages (40), indicating that the activity of this gene may represent a common virulence mechanism.
A surprisingly large fraction of the genes that we identified are unique to mycobacteria and this has profound implications for understanding the evolution of M. tuberculosis. For many bacteria, the acquisition of a pathogenic phenotype is associated with the horizontal transfer of DNA from another organism and this has resulted in many pathogens using conserved virulence mechanisms (41). However, recent acquisition of foreign DNA is not apparent in the genome of M. tuberculosis, and most of these common virulence systems are not used by this organism. These observations suggest a model in which mycobacterial evolution from saprophyte to pathogen has occurred largely through the adaptation of ancestral genes to function in the host environment. This model provides a rationale for the large fraction of the genome that appears to have arisen through duplication. Members of the resulting gene families have evolved different functions, and this specialization is critical to bacterial survival in the continually changing environment of the host.
Supplementary Material
Acknowledgments
We thank Ilona Breiterene for expert technical assistance, David Sherman for sharing unpublished data, Bruce Demple for helpful conversations, and Lalita Ramakrishnan, Andries Steyn, and members of the Rubin laboratory for critical review of this manuscript. This work was supported by grants from the National Institutes of Health (to E.J.R.). C.M.S. is a Damon Runyon fellow supported by Damon Runyon Cancer Research Foundation Grant 1647.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TraSH, transposon site hybridization; RNI, reactive nitrogen intermediates; ROI, reactive oxygen intermediates; BER, base excision repair.
References
- 1.Flynn, J. L. & Chan, J. (2001) Annu. Rev. Immunol. 19, 93-129. [DOI] [PubMed] [Google Scholar]
- 2.Glickman, M. S. & Jacobs, W. R., Jr. (2001) Cell 104, 477-485. [DOI] [PubMed] [Google Scholar]
- 3.Sassetti, C. M., Boyd, D. H. & Rubin, E. J. (2001) Proc. Natl. Acad. Sci. USA 98, 12712-12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sassetti, C. M., Boyd, D. H. & Rubin, E. J. (2002) Mol. Microbiol. 48, 77-84. [DOI] [PubMed] [Google Scholar]
- 5.Bardarov, S., Bardarov, S., Jr., Pavelka, M. S., Jr., Sambandamurthy, V., Larsen, M., Tufariello, J., Chan, J., Hatfull, G. & Jacobs, W. R., Jr. (2002) Microbiology 148, 3007-3017. [DOI] [PubMed] [Google Scholar]
- 6.Mecsas, J. (2002) Curr. Opin. Microbiol. 5, 33-37. [DOI] [PubMed] [Google Scholar]
- 7.Smith, D. A., Parish, T., Stoker, N. G. & Bancroft, G. J. (2001) Infect. Immun. 69, 1142-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordhan, B. G., Smith, D. A., Alderton, H., McAdam, R. A., Bancroft, G. J. & Mizrahi, V. (2002) Infect. Immun. 70, 3080-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hondalus, M. K., Bardarov, S., Russell, R., Chan, J., Jacobs, W. R., Jr., & Bloom, B. R. (2000) Infect. Immun. 68, 2888-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson, M., Phalen, S. W., Lagranderie, M., Ensergueix, D., Chavarot, P., Marchal, G., McMurray, D. N., Gicquel, B. & Guilhot, C. (1999) Infect. Immun. 67, 2867-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKinney, J. D., Honer zu Bentrup, K., Munoz-Elias, E. J., Miczak, A., Chen, B., Chan, W. T., Swenson, D., Sacchettini, J. C., Jacobs, W. R., Jr., & Russell, D. G. (2000) Nature 406, 735-738. [DOI] [PubMed] [Google Scholar]
- 12.Dubnau, E., Chan, J., Raynaud, C., Mohan, V. P., Laneelle, M. A., Yu, K., Quemard, A., Smith, I. & Daffe, M. (2000) Mol. Microbiol. 36, 630-637. [DOI] [PubMed] [Google Scholar]
- 13.Cox, J. S., Chen, B., McNeil, M. & Jacobs, W. R., Jr. (1999) Nature 402, 79-83. [DOI] [PubMed] [Google Scholar]
- 14.Glickman, M. S., Cox, J. S. & Jacobs, W. R., Jr. (2000) Mol. Cell 5, 717-727. [DOI] [PubMed] [Google Scholar]
- 15.Chan, J., Tanaka, K., Carroll, D., Flynn, J. & Bloom, B. R. (1995) Infect. Immun. 63, 736-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferguson, G. P., Nikolaev, Y., McLaggan, D., Maclean, M. & Booth, I. R. (1997) J. Bacteriol. 179, 1007-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wink, D. A., Kasprzak, K. S., Maragos, C. M., Elespuru, R. K., Misra, M., Dunams, T. M., Cebula, T. A., Koch, W. H., Andrews, A. W., Allen, J. S., et al. (1991) Science 254, 1001-1003. [DOI] [PubMed] [Google Scholar]
- 18.Venkatesh, J., Kumar, P., MuraliKrishna, P. S., Manjunath, R. & Varshney, U. (2003) J. Biol. Chem. 278, 24350-24358. [DOI] [PubMed] [Google Scholar]
- 19.Levin, J. D. & Demple, B. (1996) Nucleic Acids Res. 24, 885-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiloh, M. U., MacMicking, J. D., Nicholson, S., Brause, J. E., Potter, S., Marino, M., Fang, F., Dinauer, M. & Nathan, C. (1999) Immunity 10, 29-38. [DOI] [PubMed] [Google Scholar]
- 21.Arruda, S., Bomfim, G., Knights, R., Huima-Byron, T. & Riley, L. W. (1993) Science 261, 1454-1457. [DOI] [PubMed] [Google Scholar]
- 22.Tekaia, F., Gordon, S. V., Garnier, T., Brosch, R., Barrell, B. G. & Cole, S. T. (1999) Tuber. Lung Dis. 79, 329-342. [DOI] [PubMed] [Google Scholar]
- 23.Mahairas, G. G., Sabo, P. J., Hickey, M. J., Singh, D. C. & Stover, C. K. (1996) J. Bacteriol. 178, 1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis, K. N., Liao, R., Guinn, K. M., Hickey, M. J., Smith, S., Behr, M. A. & Sherman, D. R. (2003) J. Infect. Dis. 187, 117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pym, A. S., Brodin, P., Brosch, R., Huerre, M. & Cole, S. T. (2002) Mol. Microbiol. 46, 709-717. [DOI] [PubMed] [Google Scholar]
- 26.Pym, A. S., Brodin, P., Majlessi, L., Brosch, R., Demangel, C., Williams, A., Griffiths, K. E., Marchal, G., Leclerc, C. & Cole, S. T. (2003) Nat. Med. 9, 533-539. [DOI] [PubMed] [Google Scholar]
- 27.Gey Van Pittius, N. C., Gamieldien, J., Hide, W., Brown, G. D., Siezen, R. J. & Beyers, A. D. (2001) Genome Biol. 2, 0044.1-0044.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brennan, M. J., Delogu, G., Chen, Y., Bardarov, S., Kriakov, J., Alavi, M. & Jacobs, W. R., Jr. (2001) Infect. Immun. 69, 7326-7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Camacho, L. R., Ensergueix, D., Perez, E., Gicquel, B. & Guilhot, C. (1999) Mol. Microbiol. 34, 257-267. [DOI] [PubMed] [Google Scholar]
- 30.Ramakrishnan, L., Federspiel, N. A. & Falkow, S. (2000) Science 288, 1436-1439. [DOI] [PubMed] [Google Scholar]
- 31.Braunstein, M., Espinosa, B. J., Chan, J., Belisle, J. T. & Jacobs, W. R. (2003) Mol. Microbiol. 48, 453-464. [DOI] [PubMed] [Google Scholar]
- 32.Berthet, F. X., Lagranderie, M., Gounon, P., Laurent-Winter, C., Ensergueix, D., Chavarot, P., Thouron, F., Maranghi, E., Pelicic, V., Portnoi, D., et al. (1998) Science 282, 759-762. [DOI] [PubMed] [Google Scholar]
- 33.Primm, T. P., Andersen, S. J., Mizrahi, V., Avarbock, D., Rubin, H. & Barry, C. E., III (2000) J. Bacteriol. 182, 4889-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKinney, J. D., Jacobs, W. R. & Bloom, B. R. (1998) in Emerging Infections, eds. Krause, R., Gallin, J. I. & Fauci, A. S. (Academic, New York), pp. 51-146.
- 35.Foreman-Wykert, A. K. & Miller, J. F. (2003) Trends Microbiol. 11, 105-108. [DOI] [PubMed] [Google Scholar]
- 36.Leung, K. Y. & Finlay, B. B. (1991) Proc. Natl. Acad. Sci. USA 88, 11470-11474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Attridge, S. R. (1995) Microb. Pathog. 19, 11-18. [DOI] [PubMed] [Google Scholar]
- 38.Spek, E. J., Wright, T. L., Stitt, M. S., Taghizadeh, N. R., Tannenbaum, S. R., Marinus, M. G. & Engelward, B. P. (2001) J. Bacteriol. 183, 131-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boshoff, H. I., Reed, M. B., Barry, C. E. & Mizrahi, V. (2003) Cell 113, 183-193. [DOI] [PubMed] [Google Scholar]
- 40.Eriksson, S., Lucchini, S., Thompson, A., Rhen, M. & Hinton, J. C. (2003) Mol. Microbiol. 47, 103-118. [DOI] [PubMed] [Google Scholar]
- 41.Finlay, B. B. & Falkow, S. (1997) Microbiol. Mol. Biol. Rev. 61, 136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.