Abstract
The variant antigen Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1), present on the surface of P. falciparum-parasitized erythrocytes (PE), plays a central role in naturally acquired immunity, although antibodies to PfEMP1 are predominantly variant specific. To overcome this major limitation for vaccine development, we immunized mice with three cysteine-rich interdomain 1 (CIDR1) domains of PfEMP1 that have the critical function of binding the PE to CD36 on endothelium and thus preventing spleen-dependent killing of the parasite. The immunizations consisted of different combinations of three CIDR1 encoded by DNA followed by recombinant protein boost. Immunizations with a single variant in a prime-boost regimen induced no or low cross-reactivity toward heterologous CIDR1; however, a broad range of crossreactivity was detected in mice that were immunized with all three variants simultaneously. The induced crossreactivity suggests that an anti-PfEMP1 vaccine may be possible.
It is accepted that an effective malaria bloodstage vaccine would greatly reduce deaths in infants and children in Africa (1). John Robbins, one of the fathers of glycoconjugate vaccines for infants, always asks, “What is the best target antigen for vaccine development against malaria?” As the antigens on merozoites are only exposed for seconds to minutes after release from a parasitized erythrocyte (PE), high antibody titers are probably required. Alternatively, the parasite protein expressed on the surface of PE, Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1), is exposed for 2/3 of the 48 h that the parasite is within the erythrocyte. PfEMP1 is critical for parasite survival because it binds endothelium and placenta to evade spleen-dependent killing of PE. Study of immune mechanisms in natural infection suggests that immunity in African children relates primarily to antibodies to multiple PfEMP1 (2). In endemic areas, women who were immune before pregnancy from hundreds to thousands of exposures again become susceptible to malaria during pregnancy, in part because of the novel nature of PfEMP1 that bind in placenta (3–6). If other important mechanisms of immunity, such as those against invasion of erythrocytes, were sufficient to prevent disease, then placental malaria would not be a complication of pregnancy.
Why is PfEMP1 not an ideal target for malaria vaccine development? The major problem is that the parasite has evolved to have ≈60 copies of this gene in each genome (7). Each copy differs, so that the parasite undergoes antigenic variation to escape antibody-dependent killing (8). With some exceptions, it has been found that copies differ from each other and that induced antibodies are variant specific (9). Immunity is believed to develop as a child experiences multiple infections that eventually induce immunity to a variety of PfEMP1. This is not encouraging for vaccine development.
One approach that may circumvent these problems is a vaccine oriented toward a subdomain of PfEMP1 involved in the critical function of binding endothelium. One such domain is the cyteinerich interdomain region 1 (CIDR1) that binds CD36 (10), a receptor universally found on PE from children. Although it is variant, it was hoped that its variation would be more limited than the potentially more immunodominant and more variable regions of PfEMP1. In a vaccine trial with CIDR1 of the Malayan Camp (MC) strain of P. falciparum, vaccination with one copy of MC CIDR1 protected Aotus monkeys against an otherwise lethal challenge with MC parasites (11). The monkeys were protected despite antigenic variation and persistence of parasites. Unfortunately, they were not protected against another P. falciparum strain, FVO. The immunization induced antibodies that reacted with PE of MC but not with those of FVO, indicating that immunovariability exists for CIDR1 as well as for the full-length PfEMP1. In addition, antibody was not induced to FVO CIDR1 by continuous infection with MC parasites after immunization with MC CIDR1. In an effort to increase the reactivity with a multitude of CIDR1, a DNA-based vaccine to three PfEMP1 was tested (12). Reactivity with other CIDR1 was minimal after vaccination. In the present study, we have boosted the immunity from DNA vaccination with recombinant proteins to the three immunogens used in the DNA vaccine to determine its effect on reactivity to other CIDR1. The present study demonstrates greatly increased crossreactivity after the mixed recombinant protein boost.
Materials and Methods
Design of CIDR1 Synthetic Genes. Because of P. falciparum genome adenosine/thiamine (AT) richness, synthetic CIDR1 genes (Bionexus, Oakland, CA) were designed to optimize codon usage for mammalian and Pichia expression, removing transcription stop signals and N-linked glycosylation by converting asparagine to glutamine or lysine. The GenBank accession numbers for the synthetic genes are: MC CIDR1, AY338479; FVO CIDR1, AY338480; and A4tres CIDR1, AY338481.
Construction and Preparation of VR1020/CIDR1 DNA Vaccine Plasmid. MC CIDR1 (residues 1–267), FVO CIDR1 (residues 1–260), and A4tres CIDR1 (residues 1–262) were cloned in VR1020 vector (Vical Incorporated, San Diego, CA). The synthetic genes coding for the three CIDR1 were amplified by PCR using the High Fidelity PCR Master (Roche Applied Science) and specific primers carrying BamHI and BglII restriction sites (MC forward, 5′-GCACAGGTAGGATCCAATGGTGGTGGTTGGAAGGCTAAGG-3′, reverse 5′-GCACAGGTAAGATCTTCATTAAGAACGAGCAACGGAACGATCCTCTTG-3′; FVO forward, 5′-GCACAGGTAGGATCCGAAGATGGTACTTGGGAACGTAAAG-3′, reverse 5′-GCACAGGTAAGATCTTCATTAGGAACGAGCTGGAGAACGATCAGTTGG-3′; A4tres forward, 5′-GCACAGGTAGGATCCTCTGACGGTTCCTTTCGTGTTCGTG-3′, reverse 5′-GCACAGGTAAGATCTTCATTAATTGTAGTGAGTTTCGTCTTCATCGTC-3′). The PCR-amplified products were cleaned by GeneClean Spin kit (Qbiogene, Carlsbad, CA) before BamHI/BglII digestion and ligation in the predigested and dephosphorylated VR1020 DNA vector. This vector contains a human cytomegalovirus promotor, a kanamycin-resistance gene, and an upstream tissue plasminogen activator fusion leader sequence. After transformation in Escherichia coli,the recombinant clones were selected on LB agar plates containing 50 μg/ml kanamycin. Four clones of each construct were completely sequenced, and the one showing no mutations and the correct reading frame was used. Plasmid DNA was prepared by using the endofree plasmid DNA purification kit (Qiagen, Valencia, CA) as described in the manufacturer's protocol.
Recombinant Protein Expression in Pichia pastoris (Pp). Recombinant PpMC-179 (residues 88–267; GenBank accession no. AY338479), PpFVO CIDR1 (residues 1–260; GenBank accession no. AY338480), and PpA4tres CIDR1 (residues 1–262; GenBank accession no. AY338481) proteins were expressed in Pichia pastoris (Pp). PpMC-179 production and purification have been described (13). The synthetic genes encoding FVO and A4tres CIDR1s (optimized for codon usage in yeast and containing a His6-tag on the C terminus) were cloned into pPIC9K vector (Invitrogen) by standard methods. The pPIC9K plasmid contains the α-factor secretion signal that directs the recombinant protein into the secretory pathway. The constructs were digested with SacI and used to transform Pichia pastoris strain GS115 by electroporation. This resulted in insertion of the construct at the AOX1 locus of Pichia pastoris, generating a His+ Mut+ phenotype. Transformants were selected for the His+ phenotype on 2% agar containing regeneration dextrose biotin (1 M sorbitol, 2% dextrose, 1.34% yeast nitrogen base, 4 × 10-5 percent biotin, and 0.005% of l-glutamic acid, l-methionine, l-lysine, l-leucine, and l-isoleucine) medium and then further selected for high copy number by their ability to grow on 2% agar containing 1% yeast extract, 2% peptone, 2% dextrose medium, and the antibiotic G418 at various concentrations (0.5–4 mg/ml) (Invitrogen).
A4tres protein was expressed in a shaker flask and harvested at 96 h after induction by methanol. The protein was purified by using nickel-nitrilotriacetic acid-agarose (Ni-NTA; Qiagen) followed by size-exclusion chromatography on a Superdex 75 column (Amersham Pharmacia). FVO CIDR1 was expressed in a 2.5-liter fermenter (Bioflo 3000; New Brunswick Scientific, Edison, NJ) containing 1.25 liters of rich medium (1% yeast extract/2% peptone/ 100 mM potassium phosphate, pH 6.0/1.34% yeast nitrogen base/ 4 × 10-5 percent biotin/4% glycerol) supplemented with 4 ml/liter of trace salt solution (PTM4) (14). Set points for fermentation were: pH 6.5; air flow rate, 2.5 liter/min; temperature, 30°C; dissolved oxygen, 30%; agitation, 360–1,000 rpm (cascaded with the dissolved oxygen). The culture was induced with methanol and harvested at 8 h after induction. Cells were separated from the supernatant by centrifugation at 13,000 × g for 15 min at 4°C and discarded. The protein was purified from the supernatant by using Ni-NTA agarose followed by an anion-exchange column (Hiload 16/10, Q Sepharose HP, Amersham Pharmacia) and by a cation-exchange column (Hiload 16/10, SP Sepharose HP).
Protein Characterization. Proteins were separated by SDS/PAGE with (R) or without (NR) DTT at a final concentration of 50 mM (Invitrogen) on 4–20% gradient gels (Invitrogen) per the manufacturer's instructions. Gels were either stained with Coomassie brilliant blue or prepared for electrophoretic transfer to poly(vinylidene difluoride) (PVDF) membranes (Invitrogen). Samples bound to PVDF were subjected to amino acid sequencing by automated Edman degradation (performed at the Biological Resources Branch, National Institute of Allergy and Infectious Diseases, National Institutes of Health). Protein concentrations were determined by using the bicinchoninic acid protein assay (Pierce).
CD36 Binding Assay. The three recombinant proteins were assayed for their ability to bind CD36 by using a protocol similar to that described (10). Recombinant CIDR1 (1 μg) were bound to 50 μl of Ni-NTA magnetic agarose beads (Qiagen) in PBS at 4°C overnight. After extensive washing with PBS, soluble recombinant CD36 (15) was added to the magnetic beads and incubated with shaking at 25°C for 2 h. Beads were washed extensively with PBS, and bound proteins were eluted by boiling in SDS/PAGE sample buffer. Proteins were separated on SDS/PAGE and transferred to a PVDF membrane. Western blot was performed by standard methods. CD36 bound on the membrane was detected by incubating blots with the MAb-179 monoclonal antibody (Affymax Research Institute, Santa Clara, CA) that recognized an epitope tag on the C terminus of CD36. The membranes were incubated with horseradish peroxidase-conjugated goat anti-mouse IgG (Kirkegaard and Perry Laboratories, Gaithersburg, MD). Blots were developed by using the BM chemiluminescence detection system (Roche Diagnostics) according to the manufacturer's instructions.
Vaccination of Mice. For DNA vaccination, six groups (four mice per group) of 6- to 8-week-old female BALB/c mice were immunized by intradermal injection. Each mouse was vaccinated with plasmid DNA (1 mg/ml) in 0.9% sodium chloride (Abbott Laboratories, Chicago) at two sites at the tail base (50 μl per site for a total of 100 μl). The mice were immunized with DNA three times at 3-week intervals. Groups 1, 2, and 3 were immunized with MC CIDR1-α, FVO CIDR1-α, and A4tres CIDR1-α, respectively. Mice in group 4 (mixed) were immunized with equal amounts (33 μg each) of MC, FVO, and A4tres CIDR1 for a total of 100 μg. Mice in group 5 (sequential) were immunized sequentially with MC CIDR1-α (week 0) followed by FVO CIDR1-α (week 3) and then A4tres CIDR1-α (week 6). Mice in group 6 were immunized with control VR1020 plasmid DNA. All animals except in group 6 were boosted intramuscularly with 10 μg of recombinant CIDR1 formulated with Montanide ISA 720 (Seppic, France) in a final volume of 100 μl (50 μl in each anterior tibialis muscle). Groups 1, 2, and 3 were immunized with PpMC-179, PpFVO CIDR1, and PpA4tres CIDR1 recombinant proteins, respectively. Groups 4 and 5 were immunized with equal amounts (3.3 μg each) of the three recombinant proteins. Sera were collected from the retro-orbital sinus (see Fig. 3 for times of the bleeds) and stored at -80°C.
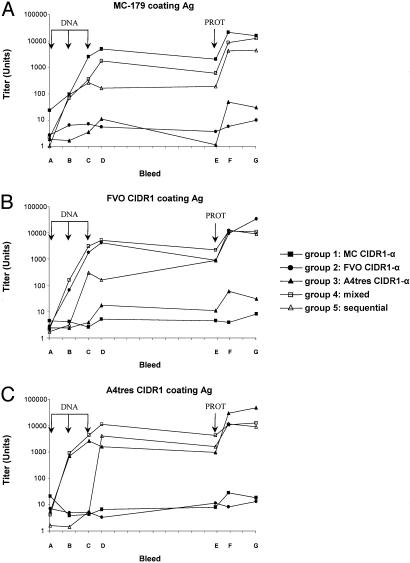
Fig. 3.
Antibody titers in the sera of the mice immunized three times with DNA vaccine (DNA) and once with recombinant protein vaccine (PROT) as measured by ELISA. Pooled sera from each of the five groups was tested for reactivity with MC-179 recombinant protein (A), FVO CIDR1-α recombinant protein (B), and A4tres CIDR1-α recombinant protein (C). Sera collected at seven collection dates from group 1 (MC CIDR1 only), 2 (FVO CIDR1 only), 3 (A4tres CIDR1 only), 4 (three constructs mixed together), 5 (sequential immunization), and 6 (VR1020 only, data not shown) were tested for antibody activity to the three CIDR1 in ELISA. The x axis corresponds to the sera collected at day 0 (bleed A), day 21 (bleed B), day 42 (bleed C), day 56 (bleed D), day 180 (bleed E), day 194 (bleed F), and day 224 (bleed G). The group 6 serum immunized with the DNA vector VR1020 alone did not react with any recombinant proteins (data not shown). Arrows represent the immunization days.
Enzyme-Linked Immunosorbent Assay (ELISA). Serum antibodies to PpMC-179, PpFVO, and PpA4tres were assayed by ELISA using an internal standard operating procedure. Briefly, flat-bottom 96-well ELISA plates were coated at 4°C overnight with 100 ng of antigen per well diluted in 15 mM sodium carbonate/35 mM sodium bicarbonate (pH 9.6). Plates were washed with 0.1% Tween 20 in Tris-buffered saline (TBS) (Biosource International, Camarillo, CA) and then blocked with 5% skim milk (Difco) in TBS for 2 h at room temperature. After the plates were washed with 0.1% Tween 20 in TBS, sera from each group were pooled and diluted in 0.1% BSA (Sigma) and 0.05% Tween 20 in TBS, added to antigen-coated wells in triplicate, and incubated for 2 h at room temperature. A duplicate dilution series of a standard mouse antiserum to the plate antigen were added to each plate. After extensive washing with 0.1% Tween 20 in TBS, plates were incubated with alkaline phosphatase-linked goat anti-mouse IgG (Kirkegaard and Perry Laboratories) diluted at 1:1,000 in 0.1% BSA and 0.05% Tween 20 in TBS for 2 h at room temperature. Bound antibodies were visualized by addition of 1 mg/ml of p-nitrophenyl phosphate (Sigma Diagnostics, St. Louis, MO) in 15 mM sodium carbonate/35 mM sodium bicarbonate (pH 9.6). Absorbance at 405 nm was read at 20 min with an ELISA plate reader (Spectromax 340PC; Molecular Devices, Sunnyvale, CA). ELISA results of the sera are expressed in arbitrary antibody units relative to the reference mouse serum that has been assigned a value of 1,500, 1,200, and 2,000 units for MC-179, FVO, and A4tres, respectively, based on the fact that a 1:1,500, 1:1,200, and 1:2,000 dilution, respectively, of the sera gave an absorbance of 1.0 at 405 nm.
Construction of Recombinant Plasmids for Surface Expression in Chinese Hamster Ovary (CHO) Cells. A total of 16 CIDR1 domains was expressed in this study, including nine CD36 binding CIDR1-α and three nonbinding CIDR1-α1 from the 3D7 genome. In addition, four CIDR1 from other genomic background were expressed: MC, FVO, A4, and A4tres CIDR1. Constructs were amplified from genomic DNA by PCR and cloned into either the pSRα5 or pSRα5(12CA5) vector (Affymax Research Institute) (16). Both vectors supply a signal sequence and a glycosylphosphatidylinositol (GPI) anchor for cell surface expression as well as a selectable marker for stable integration. The vectors differ in that pSRα5 uses an epitope tag recognized by the 179 monoclonal antibody to monitor surface expression, whereas the pSRα5(12CA5) vector flanks the insert with an upstream hemagglutinin epitope tag recognized by monoclonal antibody 12CA5 and a downstream 179 epitope tag. The following 3D7 CIDR1 sequences were cloned in pSRα5(12CA5) (GenBank accession numbers and amino acid boundaries of each clone are given): 3D7var2T.2 CIDR1-α (NP_472931, 400–843); PFD0005w_CIDR1-α (NP_702661, 422–877); 3D7var3T.1_CIDR1-α (NP_473136, 363–807); PF07_0049_CIDR1-α (NP_704061, 420–929); PFL1970w_CIDR1-α (AL035475, 412–933); PF08_0106_CIDR1-α (NP_704467, 405–889); PFD0995c/1000c_CIDR1-α (NP_702855, 417–895); PF13_0364_CIDR1-α,(NP_705581, 413–939); PF07_0050_CIDR1-α (NP_704063, 384–819); PFE1640w_CIDR1-α1, (NP_703663, 395–809); PF08_0140_CIDR1-α1, (NP_704542, 403–812); and PFD1235w_CIDR1-α1, (NP_702903, 399–814). In addition, four other CIDR1-α from four different P. falciparum strains were cloned in pSRα5: MC_CIDR1 (AAB60251, 395–852); A4_CIDR1 (L42244, 401–846); A4tres_CIDR1 (AF193424, 375–724); and FVO_CIDR1 (AF286005, 1–480).
Surface Expression and Cloning of Various Domains in CHO Cells. The K1 line of CHO cells was obtained from the American Type Culture Collection (Manassas, VA). The cells were grown in RPMI medium 1640 (Life Technologies, Gaithersburg, MD) supplemented with 10% heat-inactivated FCS (Life Technologies), 20 mM Hepes (pH 7.2; Life Technologies), 4 mM l-glutamine (Biosource International), and penicillin/streptomycin (Biosource International). Cells were transfected with 2.5 μg of plasmid DNA by using the Superfect transfection reagent (Qiagen) according to the manufacturer's recommendations and selected with 1 mg/ml Geneticin (Life Technologies). Stable transfectants expressing the various domains on the surface of CHO K1 cells were selected by single-cell cloning for high expression by using a FACS sorter as described (17). To facilitate the annotation, we gave different names (bold) to the CHO cell lines expressing various CIDR1 domains. 6F9: 3D7 var2T2_CIDR1-α, 4D10: PFD0005w_CIDR1-α, 5D3: 3D7var3T.1_CIDR1-α, 3G4: PF07_0049_CIDR1-α, 11G10: PFL1970w _CIDR1-α, 10F9: PF08_0106_CIDR1-α, 2B5: PFD0995c/1000c_CIDR1-α, 1G4: PF13_0364_CIDR1-α, 8H11: PF07_0050_CIDR1-α, 13D2: PFE1640w_CIDR1-α1, 12H9: PF08_0140_CIDR1-α1, 14F8: PFD1235w_CIDR1-α1.
Flow Cytometry Assays. The binding of antibodies in the sera of vaccinated mice to the surface of transfected CHO cell lines was measured by flow cytometry as described (18). Sera for each group were pooled and diluted to 1:50 in RIA buffer (PBS1X, BSA 0.5%, azide 0.1%). MAb-179, which recognizes an epitope tag incorporated to the C terminus of each clone, was used at 8 μg/ml to determine the expression level of each clone. Flow cytometry was performed by using a Becton Dickinson FACScan (Franklin Lakes, NJ) and FLOWJO 3.4 analysis software (Tree Star, Inc., San Carlos, CA).
Sequence Analysis. Phylogenetic analyses were done by using CLUSTALX for multiple alignments and PAUP*4.0b10 to generate neighbor-joining trees with 1,000 bootstrap replicates. Percent sequence identities were calculated by using the algorithm in DNAStar MEGALIGN, version 5.0.
Results
Expression and Purification of MC-179, FVO CIDR1, and A4tres CIDR1. In an attempt to elicit crossreactive antibodies, we expressed and purified three different CIDR1-α (MC, FVO, and A4tres) in Pichia pastoris. The CIDR1-α region has been divided into three domains: M1, M2, and M3 (19). The minimal sequence of MC CIDR1-α for binding to CD36 was a 179-aa sequence that defined the M2 region (10). A 5′ extension into the M1 domain was necessary to obtain high binding to CD36 for the FVO clone (19) and to obtain any binding to A4tres clone of P. falciparum (20); thus, we produced the M2 179-amino acid segment for MC and the longer domains for A4tres and FVO. Synthetic genes of the three domains, containing a His-tag on the C terminus sequence, were cloned into pPIC9K expression vector in frame with the α-factor secretion signal. The expression and purification of MC-179 are described elsewhere (13).
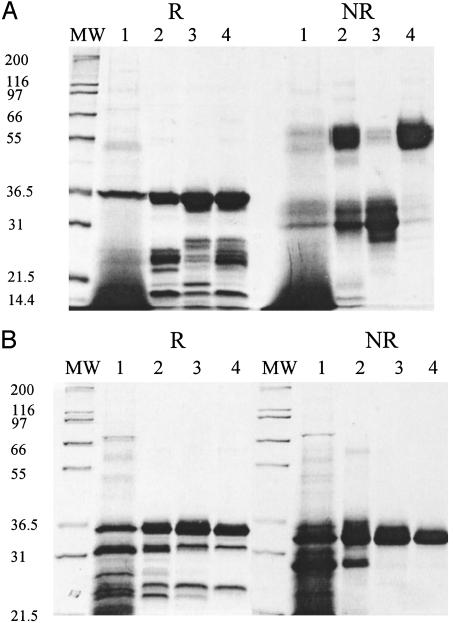
The A4tres CIDR1-α protein was purified from yeast proteins in the supernatant by using a batch nickel-chelate chromatography step. The multiple bands at 32 and 55 kDa on nonreduced (NR) SDS/PAGE shifted to a single band in reduced (R) gels (Fig. 1A; R, lane 2; NR, lane 2), indicating that the protein had multiple conformers in its native state, including a dimer. Sequencing of the two major bands on the NR SDS/PAGE had the same N terminus as expected. Multiple bands at lower molecular weight on the R SDS/PAGE indicated that the protein was partially nicked but held together by disulfide bonds in the NR gel. The monomer and dimer were separated from each other by size exclusion chromatography, and the monomer (Fig. 1A; R, lane 3; NR, lane 3) was used for immunization of mice and on ELISA plates.
Fig. 1.
A4tres CIDR1-α (5 μg) or FVO CIDR1-α (3 μg) proteins were loaded on a SDS/4–20% PAGE gel under reduced (R) or nonreduced (NR) conditions and stained with Coomassie blue. (A) A4tres CIDR1-α purification steps. MW, molecular mass markers. Lane 1, shake flask supernatant; lane 2, Ni-NTA fraction; lane 3, S75 monomer fraction; lane 4, S75 dimer fraction. (B) FVO CIDR1-α purification steps. Lane 1, fermentation supernatant; lane 2, Ni-NTA fraction; lane 3, anion exchange fraction (Q column); lane 4, cation exchange fraction (SP column). Molecular mass standard sizes (in kDa) are indicated.
FVO CIDR1-α was purified from the supernatant by using affinity Ni-NTA column chromatography. R SDS/PAGE analysis indicated a major band running at the apparent molecular size of 36 kDa and several minor bands running faster (Fig. 1B; R, lane 2). The terminal sequence analysis and a Western blot with an anti-his-tag antibody demonstrated that all of the bands were FVO CIDR1 (data not shown). The NR SDS/PAGE showed multiple conformers at the point of the major band and lower bands (Fig. 1B; NR, lane 2). Further purification steps (anion- and cation-exchange chromatography) were performed to eliminate the different conformers observed on NR SDS/PAGE (Fig. 1B; NR, lanes 3 and 4). The final purified product in NR SDS/PAGE showed one major band at 32.5 kDa, indicating the presence of a single conformer (Fig. 1B; NR, lane 4). The presence of two small lower-molecular-mass bands in the R SDS/PAGE indicated proteolytic cleavage of a small amount of the protein (Fig. 1B; R, lane 4).
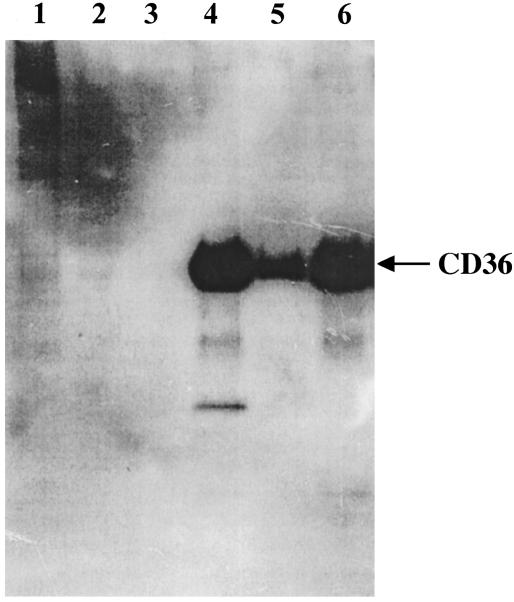
To determine whether the three recombinant proteins had the correct conformation, they were tested for their ability to bind CD36. Results of the assay demonstrated that the three CIDR1 bound to CD36 (Fig. 2, lanes 4–6). Nevertheless, A4tres CIDR1 bound more weakly to CD36 than did FVO and MC-179 CIDR1-α. The lower binding of A4tres to CD36 may result from the fact that only one of the multiple conformers was folded correctly to bind CD36 or that its binding to CD36 was of lower affinity. No binding to CD36 was detected for two control proteins, PpPfs25 and PpPvs25 (recombinant forms of the 25-kDa P. falciparum and P. vivax surface antigen of ookinetes) (Fig. 2, lanes 2 and 3), validating the specificity of the binding assay.
Fig. 2.
The three CIDR1 recombinant proteins were assayed for binding to CD36. CIDR1 recombinant protein (1 μg) was incubated with Ni-NTA magnetic agarose beads (50 μl) overnight at 4°C; this complex was then incubated with a soluble recombinant CD36 protein. After several washings, CD36 was eluted with SDS/PAGE sample buffer. The CD36 protein was detected by Western blotting using MAb 179, which recognizes an epitope tag in the CD36 recombinant protein. Lane 1, no recombinant CIDR1 added to the beads; lane 2, PpPvs25 recombinant protein; lane 3, PpPfs25 recombinant protein; lane 4, PpFVO CIDR1 recombinant protein; lane 5, PpA4tres CIDR1 recombinant protein; lane 6, PpMC-179 recombinant protein.
Antibody to MC-179, FVO, and A4tres Measured by ELISA Against the Recombinant Proteins. BALB/c mice were immunized three times with 100 μg of DNA by intradermal injection. Six months later, a protein boost was performed to the first five groups as follows: PpMC-179 to group 1, PpFVO CIDR1 to group 2, PpA4tres CIDR1 to group 3, and the three proteins together for groups 4 and 5. The vaccination protocol is summarized in Table 1. Sera collected at seven collection dates were tested for antibody activity to the three CIDR1 in ELISA. The results are displayed in Fig. 3.
Table 1. Vaccination protocol.
| Number of vaccinations |
Vaccination doses, μg |
|||
|---|---|---|---|---|
| Experimental groups | DNA | Protein boost | DNA | Protein boost |
| MC CIDR1 | 3 | 1 | 100 | 10 |
| FVO CIDR1 | 3 | 1 | 100 | 10 |
| A4tres CIDR1 | 3 | 1 | 100 | 10 |
| Three antigens mixed | 3 | 1 | 33 per antigen | 3.3 per antigen |
| Sequential | ||||
| MC CIDR1 | 1st | 100 | ||
| FVI CIDR1 | 2nd | 1 | 100 | 3.3 per antigen |
| A4tres | 3rd | 100 | ||
| VR1020 alone | 3 | No | 100 | No |
Two weeks after the third immunization with DNA (Fig. 3, bleed D), sera from groups 1, 2, and 3 immunized with DNA constructs of MC CIDR1, FVO CIDR1, or A4tres CIDR1, respectively, showed an antibody titer above 1:1000 against the homologous antigen (Fig. 3). Two and six weeks after the recombinant protein boost, the antibody titer from these sera showed a marked rise (>1:10,000) against the homologous protein (Fig. 3, bleeds F and G); however, these sera did not crossreact with the heterologous proteins to a major degree, even after protein boost (Fig. 3, bleeds F and G). No antibody against the three different antigens was detected in mice immunized with the control VR1020 vector (data not shown).
In an attempt to raise crossreactive antibodies, mice were immunized with the three DNA vaccine constructs simultaneously (group 4). All sera (except the prebleed) from this group reacted against the three CIDR1 by ELISA (Fig. 3) and gave a similar titer to the homologous immunization, indicating no immune competition among the immunogens. Intriguingly, mice from this group were immunized with only 33 μg of each DNA vaccine construct compared with the 100 μg of DNA that mice from groups 1, 2, and 3 received. It is possible that the injection of these three DNA together had a synergystic effect or that 33 μg of DNA vaccine construct was sufficient to reach the same antibody level as 100 μg.
Another way to overcome the restricted antibody response to one specific CIDR1 was to immunize the mice sequentially with the different DNA vaccine constructs (group 5). The animals immunized sequentially gave a lower titer response to all three CIDR1 but gave a similar titer to the homologous immunogen after boosting with the three-protein mixture (Fig. 3, bleeds F and G). Of interest, in the group of animals sequentially immunized (group 5), there was evidence that earlier immunizations boosted the heterologous challenge. For example, after a single DNA immunization with A4tres, the titer against the A4tres antigen increased by a factor of ≈125 (Fig. 3C; group 3, bleeds A and B). By comparison, in mice sequentially challenged with first (MC), second (FVO), and third (A4tres), the A4tres titer rose by a factor of ≈800 after injection of the A4tres DNA construct (Fig. 3C; group 5, bleeds C and D). The same effect but to a lower extent could be seen for the FVO CIDR1 coating antigen (Fig. 3; compare group 2, bleeds A and B with group 5, bleeds B and C). Thus, sequential DNA immunizations primed the immune system against other variants.
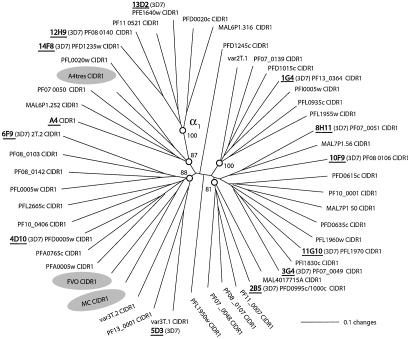
FACS Analysis of Sera Using CHO Cell Lines Expressing Different CIDR1 on Their Surface. To get more information on the specificity or crossreactivity of all these sera, we performed a FACS assay using 16 CHO cell lines expressing different CIDR1 domains on their surface (Table 2). The full-length CIDR1 used in this assay included 12 from the 3D7 genome and 4 from other genomes (see Materials and Methods). Among the 16 CIDR1 that we chose, 13 are CIDR1-α type (bind to CD36) and 3 are CIDR1-α1 type (do not bind to CD36) (21). These CIDR1 were chosen to represent the diversity of CIDR1-α and -α1 sequences (Fig. 4) and had ≈18–63% amino acid identity, with most sequences being 25–40% identical.
Table 2.
Sera reactivity from immunized mice with CHO K1 cells expressing different CIDR1 domains of the surface measured by flow cytometry
| 1G4 | 2B5 | 3G4 | 4D10 | 5D3 | 6F9 | 8H11 | 10F9 | 11G10 | 12H9 | 13D2 | 14F8 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MC | A4tres | FVO | A4 | (3D7) | (3D7) | (3D7) | (3D7) | (3D7) | (3D7) | (3D7) | (3D7) | (3D7) | (3D7) | (3D7) | (3D7) | |||
| CHO K1 | CIDR1-α | CIDR1-α | CIDR1-α | CIDR1-α | CIDR1-α | CIDR1-α | CIDR1-α | CIDR1-α | CIDR1-α | CIDR1-α | CIDR1-α | CIDR1-α | CIDR1-α | CIDR1-α | CIDR1-α | CIDR1-α | ||
| MC CIDR1 (G1) | Bleed D | 9.4 | 543 (2.08) | 10.2 (0.02) | 28.4 (0.01) | 13.1 (0.08) | 10.9 (0) | 19.2 (0.09) | 12.1 (0.01) | 21.7 (0.02) | 12.0 (0.05) | 10.8 (0.01) | 21.5 (0.21) | 20.4 (0.05) | 17.1 (0.04) | 12.9 (0.01) | 22.5 (0.02) | 19.9 (0.02) |
| Bleed E | 9.6 | 537 (2.06) | 11.2 (0.04) | 23.3 (0.01) | 11.6 (0.05) | 15.2 (0.02) | 19.0 (0.09) | 12.5 (0.01) | 23.0 (0.02) | 12.2 (0.05) | 10.9 (0.01) | 16.9 (0.13) | 19.7 (0.04) | 16.9 (0.04) | 13.0 (0.01) | 28.6 (0.02) | 20.9 (0.02) | |
| Bleed F | 11.0 | 604 (2.31) | 12.7 (0.04) | 26.9 (0.01) | 12.1 (0.02) | 13.1 (0.01) | 19.9 (0.09) | 13.1 (0.01) | 19.3 (0.01) | 11.0 (0) | 10.3 (0) | 15.0 (0.07) | 17.6 (0.03) | 18.0 (0.04) | 12.1 (0) | 29.3 (0.02) | 17.2 (0.01) | |
| FVO CIDR1 (G2) | Bleed D | 7.8 | 9.9 (0.01) | 8.2 (0.01) | 2,173 (1.2) | 7.8 (0) | 27.5 (0.05) | 14.4 (0.06) | 9.3 (0) | 12.1 (0.01) | 7.5 (0) | 10.9 (0.02) | 13.9 (0.11) | 13.2 (0.02) | 12.2 (0.02) | 11.0 (0.01) | 13.7 (0.01) | 11.8 (0.01) |
| Bleed E | 8.2 | 11.3 (0.01) | 15.6 (0.18) | 1,201 (0.65) | 11.1 (0.07) | 22.5 (0.04) | 17.5 (0.09) | 11.5 (0.01) | 14.4 (0.01) | 11.5 (0.06) | 10.0 (0.01) | 18.6 (0.18) | 19.0 (0.05) | 17.4 (0.05) | 10.6 (0.01) | 18.6 (0.01) | 18.3 (0.02) | |
| Bleed F | 9.6 | 15.6 (0.02) | 12.1 (0.06) | 2,783 (1.5) | 12.3 (0.06) | 11.7 (0.01) | 19.6 (0.10) | 12.2 (0.01) | 15.2 (0.01) | 13.0 (0.06) | 28.2 (0.13) | 17.1 (0.13) | 18.6 (0.04) | 17.5 (0.04) | 13.9 (0.01) | 16.3 (0.01) | 17.7 (0.02) | |
| A4tress CIDR1 (G3) | Bleed D | 8.1 | 8.8 (0) | 195 (4.63) | 18.3 (0.01) | 8.9 (0.02) | 10.5 (0.01) | 14.3 (0.06) | 7.9 (0) | 13.4 (0.01) | 8.6 (0.01) | 8.4 (0) | 15.2 (0.12) | 15.9 (0.03) | 12.9 (0.03) | 10.1 (0) | 14.1 (0.01) | 15.4 (0.01) |
| Bleed E | 14.0 | 15.0 (0) | 178 (4.06) | 28.0 (0.01) | 16.0 (0.05) | 15.4 (0) | 23.9 (0.09) | 15.2 (0) | 19.0 (0.01) | 15.1 (0.02) | 13.7 (0) | 21.6 (0.13) | 22.8 (0.04) | 23.1 (0.05) | 15.0 (0) | 22.0 (0.01) | 24.1 (0.02) | |
| Bleed F | 14.1 | 20.4 (0.02) | 253 (5.91) | 32.4 (0.01) | 15.8 (0.04) | 15.4 (0) | 25.0 (0.10) | 14.9 (0) | 26.7 (0.02) | 13.8 (0) | 15.8 (0.01) | 18.6 (0.08) | 21.5 (0.03) | 19.7 (0.03) | 13.3 (0) | 16.0 (0) | 18.9 (0.01) | |
| Mixed (G4) | Bleed D | 6.0 | 370 (1.42) | 230 (5.54) | 2,779 (1.5) | 8.4 (0.05) | 59.0 (0.15) | 16.2 (0.10) | 8.2 (0.01) | 11.7 (0.01) | 7.9 (0.03) | 8.0 (0.01) | 13.1 (0.12) | 13.9 (0.03) | 12.9 (0.04) | 8.0 (0) | 39.3 (0.04) | 12.8 (0.01) |
| Bleed E | 8.1 | 327 (1.24) | 239 (5.72) | 1,910 (1.0) | 9.1 (0.02) | 18.0 (0.03) | 16.4 (0.08) | 11.1 (0.01) | 12.2 (0.01) | 8.7 (0.01) | 9.9 (0.01) | 12.9 (0.08) | 15.8 (0.03) | 15.1 (0.04) | 9.2 (0) | 41.3 (0.04) | 16.5 (0.02) | |
| Bleed F | 14.4 | 575 (2.19) | 264 (6.18) | 3,369 (1.8) | 19.8 (0.12) | 32.7 (0.05) | 80.3 (0.63) | 9.8 (0) | 34.2 (0.02) | 19.1 (0.09) | 36.7 (0.16) | 46.9 (0.56) | 93.0 (0.34) | 71.6 (0.31) | 23.3 (0.02) | 108.0 (0.11) | 71.9 (0.11) | |
| Sequential (G5) | Bleed D | 7.6 | 172 (0.64) | 109 (2.51) | 485 (0.26) | 8.9 (0.03) | 9.8 (0.01) | 15.3 (0.07) | 12.9 (0.02) | 12.0 (0.01) | 8.4 (0.01) | 8.1 (0) | 15.2 (0.13) | 14.5 (0.03) | 14.7 (0.04) | 10.0 (0.01) | 16.4 (0.01) | 16.2 (0.02) |
| Bleed E | 15.8 | 123 (0.42) | 166 (3.72) | 761 (0.40) | 18.7 (0.07) | 34.7 (0.05) | 30.8 (0.14) | 17.6 (0.01) | 22.5 (0.01) | 15.9 (0) | 20.3 (0.03) | 26.0 (0.18) | 27.5 (0.05) | 29.3 (0.07) | 18.3 (0.01) | 35.4 (0.01) | 23.4 (0.01) | |
| Bleed F | 22.0 | 362 (1.33) | 240 (5.40) | 3,095 (1.7) | 20.9 (0) | 23.4 (0) | 24.0 (0.02) | 25.7 (0.01) | 22.1 (0) | 17.2 (0) | 26.0 (0.03) | 22.2 (0) | 25.0 (0.01) | 23.5 (0.01) | 21.9 (0) | 35.7 (0.02) | 22.3 (0) | |
| VR1020 (G6) | Bleed D | 5.1 | 4.5 | 4.6 | 11.3 | 5.8 | 6.7 | 10.6 | 5.9 | 9.2 | 5.3 | 5.2 | 6.1 | 8.5 | 8.3 | 5.9 | 10.5 | 7.5 |
| MAb 179‡ | 5.5 | 261.0 | 45.0 | 1,856.0 | 50.0 | 372.0 | 115.0 | 313.0 | 803.0 | 60.3 | 143.0 | 63.8 | 239.0 | 192.0 | 471.0 | 833.0 | 543.0 |
Reactivity is given as the median fluorescence activity (arbitrary units)
Normalized data: (FI intensity for each data point — CHO K1)/(FI intensity for MAb 179 for each parasite — VR1020 for each parasite
MAb 179 recognizes an epitope tag incorporated in the extracellular C-terminal region of each construct
Fig. 4.
Sequence comparison of CIDR domains used for immunization and serologic characterization: a neighbor-joining tree comparing the CIDR sequences of recombinant proteins expressed in stable CHO cells lines. Statistically distinct branches are indicated by circles with percent bootstrap support. Six of the α1 subtype that do not bind CD36 come off a single node at 12 o'clock; the node is marked on the figure (α1). Other CIDR sequences in the tree bound to CD36 (21). CHO cell-expressing lines are marked in underlined bold. The three CIDR sequences used for immunization are highlighted in gray.
After immunization with individual CIDR1 (MC, FVO, or A4tres), reactivity was specific for the immunizing antigen and showed little to no crossreactivity to the heterologous proteins, even though MC was closest in sequence to FVO. There was also no crossreactivity to other CIDR1 after DNA vaccination with the exception of 1G4, where DNA gave some weak crossreactivity. After protein boost with the homologous proteins (groups 1, 2, and 3), there was minimal to no crossreactivity to the CHO cells expressing heterologous CIDR1 despite the increase in the homologous reactivity.
The sera from mice after DNA immunization and protein boost with three different sequences (group 4, bleed F) were the only sera to consistently show crossreactivity (Table 2). The mean fluorescent intensity was normalized by subtracting background and dividing by the level of fluorescence with a monoclonal antibody 179 against an epitope tag on the extracelular C terminus of each CHO line. The significance was defined as 2 times or more above the value for any sera against that CHO clone. Six (2B5, 8H11, 10F9, 11G10, 13D2, and 14F8) of 12 CHO lines were positive crossreactive by this critera. The protein boost in this group also elicited crossreactive antibodies recognizing CHO cells expressing CIDR1-α1 (13D2 and 14F8) as well as CIDR1-α. This demonstrates that some epitopes are shared among the CD36-binding CIDR1-α and the non-CD36-binding CIDR1-α1. The sera from DNA-vaccinated mice gave no crossreactivity with the possible exception of 1G4. This general failure of DNA vaccination to induce crossreactivity was similar to the DNA vaccination studies of Baruch et al. (12). It is clear that priming with the mixed DNA vaccine followed by boost with the three recombinant proteins was critical in inducing crossreactivity in this study. Whether protein immunization alone will give equivalent or greater crossreactivity is unknown.
Discussion
DNA vaccination followed by recombinant protein boost with three CIDR1 recombinant proteins, but not with each individually, led to crossreactive antibodies to other CIDR1 than those used for vaccination. The crossreactivity was not seen after DNA vaccine alone. By ELISA, the titers markedly increased after recombinant protein boost, although there was no obvious increase by FACS analysis. The titers also rose by ELISA on immunization with individual proteins, although these did not lead to crossreactivity.
Why should immunization with three recombinant proteins lead to crossreactivity? What are the possible explanations for crossreactivity? The first is a trivial possibility. The apparent crossreactivity occurs as a result of the additive effects of each individual antigen alone. The second possibility is more interesting for such a strategy to protect against all variants by immunization with a limited number of CIDR1. Each antigen drives clonal expansion of the highest affinity B cells. It is possible that some B cells, during somatic mutation and maturation of the immune response, are stimulated by shared antigenic determinants favored by the simultaneous presentation of the three CIDR1, increasing or leading to unique antibody specificities different from those detected after immunization with a single recombinant protein. Such events may, in some cases, lead to crossreactivity.
The way forward is evident from these studies. First, will immunization with the three proteins be more protective in Aotus monkeys challenged with FVO PE than immunization with FVO CIDR1 alone? This can be tested for proof of principle. Second, the CIDR1 expressed on CHO cells that are nonresponsive after immunization with three recombinant proteins can be use as a recombinant protein to determine whether immunizing with four recombinant proteins will lead to still broader crossreactivity. Will this present an endless problem of chasing after parasites with different variants? Despite variation in the field, children do eventually become immune to disease, although never to infection. Can this state be induced in children in the field, lessening their disease so that CIDR1-based vaccines will be effective? In addition, will the low-titer but crossreactive antibodies be boosted rapidly enough to protect children from infection by heterologous strains? These questions can be answered only after animal trials and extensive field trials. In conclusion, prime-boost DNA vaccine immunization of multiple variants at the same time elicits specific but also crossreactive antibodies against CIDR1 domains. These very promising data encourage us to continue research on CIDR1 vaccine development. Moreover, the newly sequenced 3D7 genome (7) associated with phylogenetic analysis, combined with this study, will allow us to design better strategies in choosing a CIDR1 mixture able to raise a wider crossreactive antibody response against PfEMP1.
Abbreviations: CHO, Chinese hamster ovary; CIDR, cysteine-rich interdomain region; Ni-NTA, nickel-nitrilotriacetic acid-agarose; PE, parasitized erythrocytes; PfEMP1, Plasmodium falciparum erythrocyte membrane protein 1; PVDF, poly(vinylidene difluoride); R, reduced; NR, nonreduced; MC, Malayan camp.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY338479–AY338481).
References
- 1.Sachs, J. D. (2002) Science 298 122-124. [DOI] [PubMed] [Google Scholar]
- 2.Bull, P. C. & Marsh, K. (2002) Trends Microbiol. 10 55-58. [DOI] [PubMed] [Google Scholar]
- 3.Beeson, J. G., Brown, G. V., Molyneux, M. E., Mhango, C., Dzinjalamala, F. & Rogerson, S. J. (1999) J. Infect. Dis. 180 464-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fried, M., Nosten, F., Brockman, A., Brabin, B. J. & Duffy, P. E. (1998) Nature 395 851-852. [DOI] [PubMed] [Google Scholar]
- 5.Fried, M. & Duffy, P. E. (1996) Science 272 1502-1504. [DOI] [PubMed] [Google Scholar]
- 6.Gamain, B., Gratepanche, S., Miller, L. H. & Baruch, D. I. (2002) Proc. Natl. Acad. Sci. USA 99 10020-10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner, M. J., Hall, N., Fung, E., White, O., Berriman, M., Hyman, R. W., Carlton, J. M., Pain, A., Nelson, K. E., Bowman, S., et al. (2002) Nature 419 498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts, D. J., Craig, A. G., Berendt, A. R., Pinches, R., Nash, G., Marsh, K. & Newbold, C. I. (1992) Nature 357 689-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newbold, C. I., Pinches, R., Roberts, D. J. & Marsh, K. (1992) Exp. Parasitol. 75 281-292. [DOI] [PubMed] [Google Scholar]
- 10.Baruch, D. I., Ma, X. C., Singh, H. B., Bi, X., Pasloske, B. L. & Howard, R. J. (1997) Blood 90 3766-3775. [PubMed] [Google Scholar]
- 11.Baruch, D. I., Gamain, B., Barnwell, J. W., Sullivan, J. S., Stowers, A., Galland, G. G., Miller, L. H. & Collins, W. E. (2002) Proc. Natl. Acad. Sci. USA 99 3860-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baruch, D. I., Gamain, B. & Miller, L. H. (2003) Infect. Immun. 71 4536-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yipp, B. G., Baruch, D. I., Brady, C., Murray, A. G., Looareesuwan, S., Kubes, P. & Ho, M. (2003) Blood 101 331-337. [DOI] [PubMed] [Google Scholar]
- 14.Stratton, J., Chiruvolu, V. & Meagher, M. (1998) Methods Mol. Biol. 103 107-120. [DOI] [PubMed] [Google Scholar]
- 15.Baruch, D. I., Pasloske, B. L., Singh, H. B., Bi, X., Ma, X. C., Feldman, M., Taraschi, T. F. & Howard, R. J. (1995) Cell 82 77-87. [DOI] [PubMed] [Google Scholar]
- 16.Whitehorn, E. A., Tate, E., Yanofsky, S. D., Kochersperger, L., Davis, A., Mortensen, R. B., Yonkovich, S., Bell, K., Dower, W. J. & Barrett, R. W. (1995) Bio/Technology 13 1215-1219. [DOI] [PubMed] [Google Scholar]
- 17.Smith, J. D., Craig, A. G., Kriek, N., Hudson-Taylor, D., Kyes, S., Fagen, T., Pinches, R., Baruch, D. I., Newbold, C. I. & Miller, L. H. (2000) Proc. Natl. Acad. Sci. USA 97 1766-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamain, B., Miller, L. H. & Baruch, D. I. (2001) Proc. Natl. Acad. Sci. USA 98 2664-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gamain, B., Smith, J. D., Miller, L. H. & Baruch, D. I. (2001) Blood 97 3268-3274. [DOI] [PubMed] [Google Scholar]
- 20.Miller, L. H., Hudson-Taylor, D., Gamain, B. & Saul, A. J. (2002) Mol. Biochem. Parasitol. 120 321-323. [DOI] [PubMed] [Google Scholar]
- 21.Robinson, B. A., Welch, T. L. & Smith, J. D. (2003) Mol. Microbiol. 47 1265-1278. [DOI] [PubMed] [Google Scholar]