Abstract
Little is known about how the human brain differs from that of our closest relatives. To investigate the genetic basis of human specializations in brain organization and cognition, we compared gene expression profiles for the cerebral cortex of humans, chimpanzees, and rhesus macaques by using several independent techniques. We identified 169 genes that exhibited expression differences between human and chimpanzee cortex, and 91 were ascribed to the human lineage by using macaques as an outgroup. Surprisingly, most differences between the brains of humans and non-human primates involved up-regulation, with ≈90% of the genes being more highly expressed in humans. By contrast, in the comparison of human and chimpanzee heart and liver, the numbers of up- and down-regulated genes were nearly identical. Our results indicate that the human brain displays a distinctive pattern of gene expression relative to non-human primates, with higher expression levels for many genes belonging to a wide variety of functional classes. The increased expression of these genes could provide the basis for extensive modifications of cerebral physiology and function in humans and suggests that the human brain is characterized by elevated levels of neuronal activity.
The origin of humans was accompanied by the emergence of new behavioral and cognitive functions, including language and specialized forms of abstract representation (1, 2). However, the neural foundations of these human capabilities are poorly understood. Although the human brain is characterized by its unusually large size and disproportionate expansion of the neocortex (3, 4), the only differences in its internal organization that have been identified involve the number and size of spindle cells in the anterior cingulate cortex (5), the organization of the planum temporale minicolumns (6), and the compartmental organization of the primary visual cortex (7). Moreover, little is known about underlying changes at the molecular level. Because of the extensive similarity between human and chimpanzee DNA sequences, it has been suggested that many of the key phenotypic differences between species result primarily from alterations in the regulation of genes rather than in their sequences (8). Current genomic techniques allow us to examine the expression of thousands of genes at the same time and to address these questions (9, 10). A recent comparison of expression patterns in brain, liver, and leukocytes from humans, chimpanzees, and an orangutan reported that species-specific differences in overall gene expression patterns were particularly pronounced in the human brain relative to other organs (11), but the specific genes that underwent expression changes during human evolution were not described.
In the present study, we used high-density oligonucleotide arrays to identify genes differentially expressed in the brain of humans or chimpanzees by using macaques as an outgroup and validated many of the observed differences by quantitative RT-PCR, cDNA arrays, and in situ hybridization. Our results indicate that gene expression changes in the human cortex involved predominantly increased expression, and that many of the genes up-regulated in humans could be related to higher levels of neuronal activity. Identifying the specific genes that underwent expression changes during human brain evolution could provide important clues to the biochemical, anatomical, and functional specializations of the human brain and help us understand why humans are more vulnerable to certain neurodegenerative diseases, such as Alzheimer's dementia (12), that are rare in other primates.
Materials and Methods
Samples. Human cortex samples were collected from two females and three males during autopsy [Homo sapiens (Hs)1, -2, and –3] or surgical procedures (Hs4 and -5) and were obtained from the Brain and Tissue Bank for Developmental Disorders at the University of Maryland (BTBUM; Baltimore) or the University of California at San Diego Medical Center (approved and monitored by the Institutional Review Board). Cortex samples of non-human primates were provided by the University of Louisiana at Lafayette New Iberia Research Center and the Salk Institute for Biological Studies (in accordance with Institutional Animal Care and Use Committees guidelines). Common chimpanzee [Pan troglodytes (Pt)] samples were removed during postmortem dissections of three females and one male that died of natural causes (Pt1–4). Rhesus macaque [Macaca mulatta (Mm)] samples were dissected from four females and three males killed with a lethal dose of barbiturate (Mm1–7). Individuals were mostly adults, with an average age of 43.4 years for humans, 18.5 years for chimpanzees, and 6.1 years for rhesus macaques. Tissue was derived from several regions of frontal, parietal, and temporal cortex of the left hemisphere of all species (see Fig. 1). Heart samples of one female and two male adult humans and two neonate male pygmy chimpanzees (Pan paniscus), also known as bonobos, were obtained from the BTBUM and the Zoological Society of San Diego. Additional details on materials and experimental procedures can be found in Supporting Materials and Methods and Table 1, which are published as supporting information on the PNAS web site, www.pnas.org, and at www.teragenomics.com.
Fig. 1.
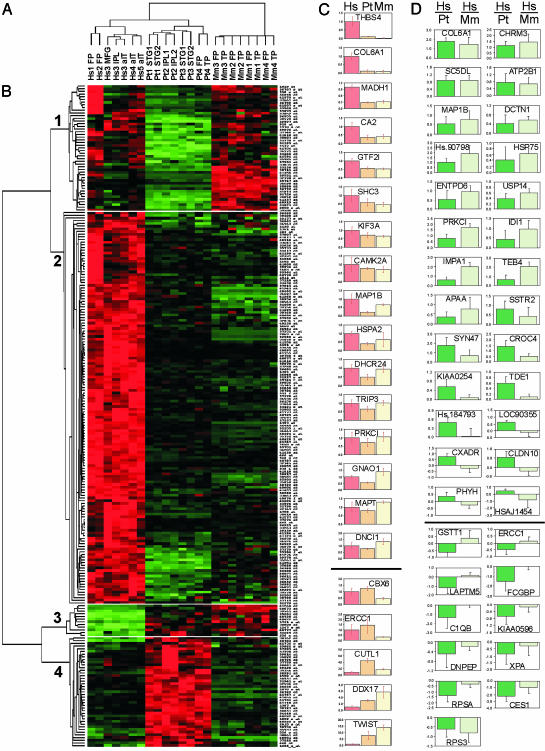
Gene expression analysis of human, chimpanzee, and rhesus macaque cerebral cortex. (A) Dendrogram showing the hierarchical clustering of the different cortex samples according to the hybridization signals of 9,733 probe sets detected in at least one of the samples from any species (Hs1–5, humans; Pt1–4, chimpanzees; Mm1–4, rhesus macaques). FP, frontal pole; MFG, medial frontal gyrus; IPL, inferior parietal lobule; aIT, anterior inferotemporal cortex; STG, superior temporal gyrus; TP, temporal pole. (B) Hybridization levels for 246 probe sets that show differences in signal intensity between human and chimpanzee cortex. Columns correspond to the different cortex samples in the same order as in the dendrogram. Rows represent the individual probe sets with their identifying number indicated (Right) (see Table 2). For each probe set, red, green, and black indicate increased, decreased, and equal hybridization levels relative to the median, respectively. Hierarchical cluster analysis classified the probe sets into four different groups related to their species-specific hybridization pattern. (C) Expression levels of 21 genes showing differences between humans and chimpanzees by real-time RT-PCR. The y axis corresponds to the average expression level in three chimpanzees (Pt, orange) and three rhesus macaques (Mm, yellow) relative to three humans (Hs, red). Standard deviation within each species is indicated by error bars. (D) Relative hybridization levels in human and non-human primate cortex samples of 37 genes interrogated by using cDNA arrays. The y axis represents the base-2 logarithm of the average hybridization signal ratio of four human–chimpanzee comparisons (Hs/Pt, dark green) and four human–rhesus comparisons (Hs/Mm, light green). Positive and negative values denote higher and lower hybridization levels, respectively, in humans than in non-human primates. In C and D, horizontal lines separate the genes that show increased expression in humans and in chimpanzees. The gene symbol or UniGene accession number is indicated inside each graph.
Oligonucleotide Arrays. Gene expression levels were measured by using human oligonucleotide arrays (Affymetrix GENECHIP Human Genome U95Av2 arrays, Affymetrix, Santa Clara, CA), which contain 12,625 probe sets for ≈10,000 different genes. RNA extraction, labeling, and hybridization were performed as described (9, 13), with the exception that hybridization was done at 50°C. Each tissue sample was processed independently and hybridized to a different array, except for the cortex samples of Pt1, -2, and -3, where the specimen was cut into two different pieces and processed independently. Array results were analyzed using several methods, including the MICROARRAY SUITE (MAS) software, Ver. 4.0 (Affymetrix) and Teragenomics (Information Management Consultants); see supporting information for details. All arrays were normalized separately to the same average intensity on the basis of probe sets corresponding to the 60–90th percentile of hybridization signals. To identify genes with signal intensity differences between primate species, we used the BULLFROG 4.5 (14) and DCHIP 1.0 programs (15). In the BULLFROG analysis, all pair-wise comparisons between the arrays of each species were generated by using MAS 4.0, and only those probe sets that showed consistent differences among all of the samples compared were selected. The criteria used were a call of increase/marginal increase or decrease/marginal decrease, fold change >1.8, and absolute difference change >50 in at least 75% of the comparisons, a fold change of 1.3 in at least 90% of the comparisons, and a present call in at least one of the arrays. In the DCHIP analysis, the expression values for each probe set were calculated by using the average difference. The criteria used to identify probe sets with signal differences between species were a fold-change >1.8 by using the lower bound of the 90% confidence interval, absolute difference between means >100, t test P value <0.001, and a present call in >25% of the samples involved. For each tissue, the probe sets identified with both analyses were then combined to generate the final list. Cluster analysis was carried out with the CLUSTER and TREEVIEW programs (16).
Sequence Difference Detection. Because the oligonucleotide arrays are designed on the basis of human sequences, sequence differences between the mRNAs measured and the array probes could result in an underestimation of expression levels in non-human primates. Thus, we used an algorithm developed in the Barlow laboratory (J. A. Greenhall, M.A.Z., C.B., and D.J.L., unpublished results) to identify probe sets that may contain one or more probes that could be affected by sequence differences between humans and chimpanzees and then reanalyzed the data after excluding those probes. Briefly, the algorithm analyzes the hybridization patterns of all of the oligonucleotide probes for each probe set, after normalizing for expression level differences, and determines the probability that a probe has different hybridization behavior between two sets of samples. Four thousand five hundred and seventy-seven probes (corresponding to 2,285 probe sets) potentially compromised by sequence variation between humans and chimpanzees were found. The reanalysis of the array data without these probes detected 65 probe sets with sequence differences that might contribute to higher signal intensities in humans compared with non-human primates [Table 2 (genes indicated by asterisk), which is published as supporting information on the PNAS web site].
Real-Time RT-PCR. Real time RT-PCR was performed by using the DNA-binding dye SYBR green (Applied Biosystems) with total RNA from the cortex samples of three humans, three chimpanzees, and three rhesus macaques. To ensure that interspecific sequence differences did not affect the amplification, ≈1 kb of the region covered by the array probes for each of the genes was sequenced in non-human primates, and PCR primers were designed in areas conserved among the three species. Amplification of the gene of interest and the housekeeping control gene β-actin was done in triplicate from each sample. Results were analyzed with the DISSOCIATION CURVE 1.0 and SEQUENCE DETECTOR 1.7 programs (Applied Biosystems). The gene amplification levels were normalized by dividing by β-actin levels, and the three samples for each species were combined in a single expression value. Expression changes were identified by a 1.3-fold difference between the average expression levels of each species.
cDNA Arrays. Arrays containing ≈7,500 different human cDNAs obtained from Incyte Genomics (Palo Alto, CA) were spotted in duplicate at the Salk Institute microarray facility. For hybridization to the arrays, 1 μg of total RNA was labeled with Cy5 or -3 by an aminoallyl indirect labeling procedure. Four comparisons of human and chimpanzee cortex and human and rhesus cortex were done. After scanning the hybridized slides, background-subtracted data of all spots in the Cy5 and -3 channels were scaled to a common value in each slide, and only spots with a signal greater than background in at least 25% of the hybridizations were considered. The criteria for detecting expression differences between humans and non-human primates were a probability of <0.05 by a paired t test of the hybridization signals in each spot and an average relative change between the two species >1.3-fold.
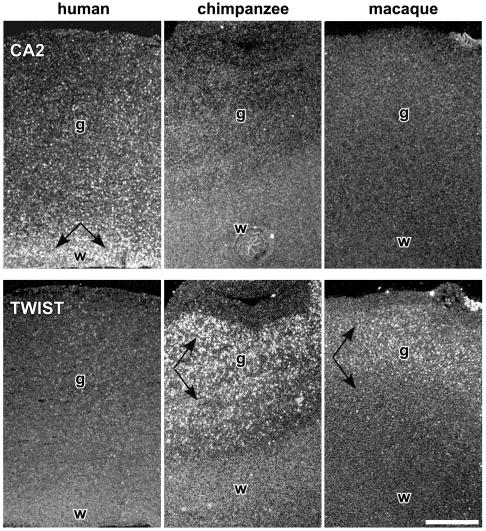
In Situ Hybridization. In situ hybridization of two genes, carbonic anhydrase II (CA2) and TWIST, was performed with multiple 20-μm coronal or sagittal sections derived from the inferior parietal lobule and the cerebellum of one individual of each species and following a previously described protocol (17) by using radiolabeled riboprobes. Films and emulsion-dipped slides were analyzed by visual inspection and light microscopy by using bright- and dark-field illumination. Control sections incubated with sense RNA showed no specific hybridization. CA2 (1,055 bp) and TWIST (906 bp) probes were derived from the 3′ region of the human and chimpanzee cDNAs, respectively. Sequence divergence between the three species for both regions is <3%.
Results
Hierarchical clustering of oligonucleotide array hybridization signals (Fig. 1A) segregated the human, chimpanzee, and rhesus samples into discrete groups, with humans and chimpanzees more similar to each other (r = 0.900, SD = 0.017) than either is to rhesus (human–rhesus: r = 0.785, SD = 0.053; chimpanzee–rhesus: r = 0.817, SD = 0.016). These relationships agree well with the known phylogeny and reflect both the divergence of gene expression profiles and nucleotide sequences between species, because sequence differences with respect to the array probes could affect the hybridization efficiency in non-human primates and produce low measures of some transcripts.
Genes differentially expressed between humans and chimpanzees were identified on the basis of pair-wise comparisons between the hybridization patterns of all cortical samples from each species, and 230 probe sets (212 genes) were found. A complementary analysis by using the program DCHIP (15) detected significant differences between the average signal intensity of humans and chimpanzees for 89 probe sets (86 genes). By combining the two methods, we detected 246 probe sets (227 genes) with consistent hybridization differences between humans and chimpanzees (Table 3, which is published as supporting information on the PNAS web site). When we repeated the analysis excluding one of the human or one of the chimpanzee samples each time, very similar results were obtained, with 75% of the genes identified in the initial analysis present in all cases. A striking observation was that the vast majority of the genes had higher hybridization signals in the human samples. Hierarchical clustering of the signal intensities for the 246 probe sets in human, chimpanzee, and rhesus revealed four distinct clusters (Fig. 1B). Clusters 1 and 4 contained 88 probe sets (84 genes) with a chimpanzee-specific hybridization pattern, and they include a similar number of genes with increased (cluster 4, 38 genes) and decreased (cluster 1, 46 genes) signal in chimpanzees compared with humans and rhesus. By contrast, clusters 2 and 3 represent 158 probe sets (143 genes) with human-specific hybridization patterns, and most of these genes showed increased signal intensity in humans compared with chimpanzees and rhesus: cluster 2, containing 132 genes, increased in humans, vs. cluster 3, containing 11 genes, decreased in humans.
To verify this unexpected finding, we applied the same analysis to previously published gene expression data derived from human, chimpanzee, and orangutan cerebral cortex (11). Hybridization signals for 311 probe sets (290 genes) differed between humans and chimpanzees (Table 3). Of the 221 genes that showed a human-specific pattern of hybridization, 211 (95%) had higher signal intensities in humans, consistent with the predominance of gene expression up-regulation in the human brain recently reported from the same data (18). Therefore, two independent data sets provide evidence for an asymmetry of gene-expression changes in human brain evolution, with the great majority of changes involving expression increases.
To investigate whether the preponderance of up-regulation of gene expression in humans was unique to the brain, we analyzed samples from human and chimpanzee heart (this work) and liver (data from ref. 11). In clear contrast to the brain, in heart and liver, the number of genes differentially expressed between humans and chimpanzees that were up- and down-regulated was nearly equal. For the heart samples, the hybridization signals for 841 probe sets (778 genes) were significantly different between species, and just over half of them had higher signals in humans (54%). Analysis of the liver data identified 521 probe sets (485 genes) with signal differences between humans and chimpanzees, and 55% showed greater abundance in humans. Similar results were also obtained by using several additional normalization and analytical methods (Table 3). Furthermore, the distribution of the relative differences in hybridization signal between humans and chimpanzees for all of the genes in the arrays showed a significant skewing toward higher signals in humans in the cortex, but not in heart or liver (Fig. 3, which is published as supporting information on the PNAS web site). Thus, the up-regulation of a large set of genes in humans compared with chimpanzees is cortex- or brain-specific and does not depend on how the array data are analyzed.
The use of oligonucleotide arrays based on human sequences is not expected to affect the quantification of most chimpanzee mRNAs, because divergence between human and chimpanzee genomic and mRNA sequences is <1.3% (19, 20) and 0.7% (21), respectively, and only sequence mismatches located near the central region of the probes are expected to have a significant effect on hybridization (22). In addition, sequence differences are not likely to account for the cortex-specific directional bias of gene-expression changes between humans and non-human primates, because this bias was not seen in heart or liver. To be conservative, however, we investigated the extent to which sequence differences might affect measurements of mRNA abundance by sequencing the region interrogated by the arrays in 36 genes (showing an average identity between humans and chimpanzees of 99.45%) and by using an algorithm to identify probes that do not behave consistently between species. The algorithm detected 65 probe sets that may be affected by sequence differences between humans and chimpanzees (Table 2). Elimination of these 65 probe sets did not significantly change the marked predominance of up-regulation of gene expression in the human cortex: 89% of the remaining genes with human-specific hybridization patterns showed higher levels in humans than in non-human primates.
To independently validate the results obtained with the oligonucleotide arrays, quantitative real-time RT-PCR was used to verify the expression differences for 26 genes of interest with a variety of gene expression patterns in humans, chimpanzees, and rhesus macaques. Of those genes, 21 showed at least a 1.3-fold difference between the average expression levels of humans and chimpanzees in the same direction as in the oligonucleotide arrays (Fig. 1C), and no expression difference was found for the other five (Table 2).
To further confirm our findings, we performed a limited gene expression analysis with cDNA arrays. The cDNA array results also indicated a bias toward increased gene expression levels in the human brain, with 64% of the genes that hybridized differentially in human and chimpanzee cortex showing a higher signal in humans. In addition, 62 of the genes with clear differences between humans and chimpanzees in the reanalysis of the oligonucleotide array data were detected on the cDNA arrays, and 33 showed significant hybridization differences consistent between the two methods (Fig. 1D). Most of the remaining genes showed similar trends in the cDNA and oligonucleotide arrays (three of which were confirmed by RT-PCR), and none had significant expression differences between humans and chimpanzees in opposite directions by using both arrays. Four additional genes showed differences with the cDNA and oligonucleotide arrays, although they were predicted by the algorithm to include probes affected by sequence variation between species (Fig. 1D and Table 2).
Finally, to put these observations into an anatomical context, we examined the spatial pattern of expression of two genes, CA2 and the transcription factor TWIST, which were found, respectively, to be up- and down-regulated in humans (Fig. 1C). Results of in situ hybridization for CA2 and TWIST in the cortex show the same pattern of differences across species observed with the previous methods. Differences for CA2 are apparent in the gray matter and especially the white matter of the cerebral cortex (Fig. 2) and cerebellum (data not shown). Expression differences in TWIST appear to be restricted to the cortical gray matter (Fig. 2) and are not as evident in the cerebellum (data not shown).
Fig. 2.
Histological study of gene-expression differences in the cortex between humans and non-human primates. In situ hybridization confirms that CA2 is expressed at higher levels in the cortex of humans than in chimpanzees or rhesus macaques (Upper). Note the particularly strong labeling for CA2 in the white matter (w) immediately below the cortical gray matter (g). TWIST is weakly expressed in human cortex compared with chimpanzees and macaques (Lower), where expression is strongest in cortical layers II–VI. Arrows point to the regions of expression of both genes. (Bar = 1 mm.)
Overall, several methods of gene expression analyses identified 169 genes with clear differential expression in human and chimpanzee cortex, 54 of which were confirmed using independent techniques (Table 2). Of those 169 genes, 167 show qualitatively similar expression differences between human and chimpanzee cortex in a previously published data set (11), and 77 show significant differences in the heart or liver comparison. The genes differentially expressed between human and chimpanzee cortex include 91 genes that are differentially expressed in humans relative to both chimpanzees and rhesus macaques, of which 83 show evidence of increased expression in humans, and only eight have decreased expression. Although the higher degree of divergence between human and rhesus sequences makes inferences of expression levels in rhesus based only on oligonucleotide array data somewhat less reliable (23), this group of 91 genes is the most readily interpretable as having undergone regulatory changes in the human lineage after the human–chimpanzee divergence. The classification of these genes into gene ontology categories using the MAPPFINDER program (24) identified a disproportionate number of expression changes in genes related to cell growth and maintenance (including many enzymes involved in metabolism, particularly of lipids and RNA) and chaperones (Table 4, which is published as supporting information on the PNAS web site), among other categories. However, the genes differentially expressed in the human cortex span a wide variety of functional classes and include many related to neural function, as discussed below.
Discussion
We used several experimental and analytical methods to identify 91 genes differentially expressed in human cortex compared with non-human primates and showed that there is a clear bias toward up-regulation in humans. Regardless of the analysis method, the directional asymmetry in expression changes is seen in the brain but not in other tissues. Moreover, the total number of genes scored as differentially expressed between humans and chimpanzees is higher in nonbrain tissues than in the cortex. These results contrast in part with those of Enard et al. (11), who indicated that gene-expression changes in the brain accumulated more rapidly during human evolution than during chimpanzee evolution, although divergence in gene expression between humans and chimpanzees was greater in liver than in brain. Reanalysis of the same data, however, suggested that gene-expression changes in the human brain involved more increases than decreases of expression (18). Thus, two different data sets support the conclusion that the human cortex is distinguished by a predominant up-regulation of gene expression, with elevated transcript levels for a variety of genes.
Two possible sources of error might confound these results. First, because human and chimpanzee samples are extremely difficult to obtain, it is not possible to match them perfectly in all pertinent parameters. However, we considered only gene-expression differences that were consistent across all our human and chimpanzee samples, and very similar gene lists were obtained when we reanalyzed the data eliminating one sample at a time. In addition, in our limited data set, variation in gene expression patterns related to gender, collection procedure, or cortex region is comparatively small (see Supporting Materials and Methods). Few expression differences are found in the human cortex in the comparison of males vs. females and postmortem vs. surgical samples, although the reduced number of individuals used makes it difficult to detect any subtle effects of these factors. Similarly, different cortical regions appear to be relatively homogeneous in terms of gene expression, as illustrated by the grouping of the cortex samples by individual and species in the hierarchical clustering analysis (Fig. 1 A) and the comparison of samples of the frontal and temporal lobes. Finally, most of the genes identified in this study show similar expression differences in a completely independent group of samples (11).
A second potential problem stems from the use of human arrays to measure expression levels in non-human primates and the importance of validating the array results with independent techniques (25, 26). In this study, we took several steps to deal with these potentially confounding effects. First, we used an algorithm to detect and exclude a high proportion of the oligonucleotide probes that are affected by sequence differences between species, eliminating many possible artifacts. Second, we confirmed one-third of the observed expression differences between human and chimpanzee cortex by using other quantitative RT-PCR, which would be less sensitive to small sequence differences and cDNA arrays.
The identification of the genes that exhibit regulatory changes in adult human cortex provides clues to the biochemical pathways and cell-biological processes that were modified during evolution. The apparent up-regulation of so many different genes suggests, among other things, that the general level of neuronal activity and the metabolic processes that support it may be unusually high in human cortex. Consistent with this is the up-regulation of genes involved in synaptic transmission, including the control of glutamatergic excitability (SYN47, also known as Homer 1b), plasticity at glutamatergic synapses [CAMK2A (27)], phosphatidylinositol signaling (IMPA1, CDS2), synaptic vesicle release [RAB3GAP, ATP2B1 (28)], axonal transport along microtubules (KIF3A, DCTN1), microtubule assembly (MAP1B), and targeting of proteins to postsynaptic densities [USP14 (29)].** We have also found expression changes related to energy metabolism. For example, CA2, which is expressed in glia, has been related to the generation and transport of lactate by astrocytes for use by neurons as an energy source (30, 31). To our knowledge, the possibility that the human brain has an unusually high metabolism has not been previously considered. Typically, larger brains have lower metabolic rates (per unit of tissue) than smaller brains (32). Nevertheless, recent studies with imaging techniques to measure cerebral glucose metabolism in the conscious state suggest that metabolic rates are as high or even higher in humans (33, 34) than in macaques (35, 36). Higher levels of neuronal activity are likely to have important consequences in cognitive and behavioral capacities, and of the genes up-regulated in humans, CAMK2A is involved in learning and memory (37), and mutations of GTF2I (Williams syndrome), CA2 (marble brain disease), and SC5DL (lathosterolosis) have been linked to mental retardation.
Increased activity would pose a serious challenge to the biochemical mechanisms that sustain normal cell function in the brain. Yet the greater life-span potential of humans (38) suggests that human neural cells could possess biochemical adaptations that enable them to function longer than those of other primates. It is thus noteworthy that human evolution was accompanied by up-regulation of genes with products linked to cytoprotection [CHRM3 (39) and SHC3 (40)] as well as protein chaperones (HSPA2, HSP75, ORP150, and BAG5). Abnormal processing, aggregation, and deposition of misfolded proteins are common features of diverse neurodegenerative syndromes (41), and chaperones could confer some protection from the above processes. Another group of genes with expression change in humans are related to lipid metabolism (ACADSB, CDS2, CES1, CYB5-M, IDI1, IMPA1, OSBPL8, PCCB, and SC5DL), which could have a role in membrane synthesis and turnover, steroid metabolism, and cell signaling. Several of these genes are involved in cholesterol metabolism, and cholesterol is thought to influence the accumulation of amyloid β protein, which is involved in the pathogenesis of Alzheimer's disease (42). In addition, the modification of genes related to lipid metabolism is interesting in view of the role attributed to increased meat consumption in human origins (43, 44), a point also pertinent to some of the gene expression differences observed between human and bonobo fibroblasts (26).
The distinctive pattern of up-regulation of gene expression that characterizes human cortical evolution casts the normal structure and function of human cortex in a new light. We suggest that these gene-regulation changes can be understood, at least in part, as adaptations for maintaining high levels of cerebral activity over a long life span. By investigating in detail the biological roles of the specific genes identified in this study, it should be possible to gain new insights into the modifications of structure and function that distinguish the human brain from that of other primates.
Supplementary Material
Acknowledgments
We thank Jo A. Del Rio for early work on the project; Dan Lockhart (The Salk Institute) for developing the bullfrog program; Information Management Consultants (McClean, VA) and Teradata for donating the Teradata database and programming of the teragenomics database; Robert Vigorito and the Brain and Tissue Bank for Developmental Disorders for human tissues; Peter Nakaji (The Salk Institute) and Chris Ames (The Salk Institute) for neurosurgical specimens; the Zoological Society of San Diego for bonobo heart samples; Ed Callaway (The Salk Institute) and Dan Pankratz (The Salk Institute) for rhesus cortical samples; New Iberia Research Center for their assistance in obtaining nonhuman primate tissue; and Joseph G. Hacia and Fred H. Gage for valuable comments. We also thank the members of the Barlow and Functional Genomics laboratories at the Salk Institute for technical assistance and help with array analysis. This work was supported by a National Institute of Mental Health grant and the Frederick B. Rentschler Developmental Chair (to C.B.), a European Molecular Biology Organization Fellowship, the Salk Institute President's Club Innovation Grant (to M.C.), NATO and Phillippe Foundation Fellowship (to J.L.), and a James S. McDonnell Foundation grant (to T.M.P.).
Abbreviations: Hs, Homo sapiens; Pt, Pan troglodytes; Mm, Macaca mulatta; CA2, carbonic anhydrase II.
Data deposition. The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY369785–AY369856).
Footnotes
For additional references to gene functions, see the LocusLink (www.ncbi.nlm.nih.gov/LocusLink), OMIM (www.ncbi.nlm.nih.gov/Omim), and SOURCE (http://source.stanford.edu/cgi-bin/sourceSearch) databases.
References
- 1.Tomasello, M. & Call, J. (1997) Primate Cognition (Oxford Univ. Press, New York).
- 2.Povinelli, D. J. (2000) Folk Physics for Apes (Oxford Univ. Press, Oxford).
- 3.Jerison, H. J. (1973) Evolution of the Brain and Intelligence (Academic, New York).
- 4.Stephan, H., Baron, G. & Frahm, H. D. (1988) in Comparative Primate Biology, eds. Steklis, H. D. & Erwin, J. (Liss, New York), Vol. 4, 1-38. [Google Scholar]
- 5.Nimchinsky, E. A., Gilissen, E., Allman, J. M., Perl, D. P., Erwin, J. M. & Hof, P. R. (1999) Proc. Natl. Acad. Sci. USA 96 5268-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buxhoeveden, D. P., Switala, A. E., Litaker, M., Roy, E. & Casanova, M. F. (2001) Brain Behav. Evol. 57 349-358. [DOI] [PubMed] [Google Scholar]
- 7.Preuss, T. M. & Coleman, G. Q. (2002) Cereb. Cortex 12 671-691. [DOI] [PubMed] [Google Scholar]
- 8.King, M. C. & Wilson, A. C. (1975) Science 188 107-116. [DOI] [PubMed] [Google Scholar]
- 9.Wodicka, L., Dong, H., Mittmann, M., Ho, M. H. & Lockhart, D. J. (1997) Nat. Biotechnol. 15 1359-1367. [DOI] [PubMed] [Google Scholar]
- 10.DeRisi, J., Penland, L., Brown, P. O., Bittner, M. L., Meltzer, P. S., Ray, M., Chen, Y., Su, Y. A. & Trent, J. M. (1996) Nat. Genet. 14 457-460. [DOI] [PubMed] [Google Scholar]
- 11.Enard, W., Khaitovich, P., Klose, J., Zollner, S., Heissig, F., Giavalisco, P., Nieselt-Struwe, K., Muchmore, E., Varki, A., et al. (2002) Science 296 340-343. [DOI] [PubMed] [Google Scholar]
- 12.Varki, A. (2000) Genome Res. 10 1065-1070. [DOI] [PubMed] [Google Scholar]
- 13.Sandberg, R., Yasuda, R., Pankratz, D. G., Carter, T. A., Del Rio, J. A., Wodicka, L., Mayford, M., Lockhart, D. J. & Barlow, C. (2000) Proc. Natl. Acad. Sci. USA 97 11038-11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zapala, M. A., Lockhart, D. J., Pankratz, D. G., Garcia, A. J., Barlow, C. & Lockhart, D. J. (2002) Genome Biol. 3 SOFTWARE0001–9; Epub 2002 May 23. [DOI] [PMC free article] [PubMed]
- 15.Li, C. & Wong, W. H. (2001) Proc. Natl. Acad. Sci. USA 98 31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geschwind, D. H., Ou, J., Easterday, M. C., Dougherty, J. D., Jackson, R. L., Chen, Z., Antoine, H., Terskikh, A., Weissman, I. L., Nelson, S. F., et al. (2001) Neuron 29 325-339. [DOI] [PubMed] [Google Scholar]
- 18.Gu, J. & Gu, X. (2003) Trends Genet. 19 63-65. [DOI] [PubMed] [Google Scholar]
- 19.Chen, F. C. & Li, W. H. (2001) Am. J. Hum. Genet. 68 444-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebersberger, I., Metzler, D., Schwarz, C. & Paabo, S. (2002) Am. J. Hum. Genet. 70 1490-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hellmann, I., Zollner, S., Enard, W., Ebersberger, I., Nickel, B. & Paabo, S. (2003) Genome Res. 13 831-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chee, M., Yang, R., Hubbell, E., Berno, A., Huang, X. C., Stern, D., Winkler, J., Lockhart, D. J., Morris, M. S. & Fodor, S. P. (1996) Science 274 610-614. [DOI] [PubMed] [Google Scholar]
- 23.Chismar, J. D., Mondala, T., Fox, H. S., Roberts, E., Langford, D., Masliah, E., Salomon, D. R. & Head, S. R. (2002) Biotechniques 33 516-522. [DOI] [PubMed] [Google Scholar]
- 24.Doniger, S. W., Salomonis, N., Dahlquist, K. D., Vranizan, K., Lawlor, S. C. & Conklin, B. R. (2003) Genome Biol. 4 R7; Epub 2003 Jan 06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chuaqui, R. F., Bonner, R. F., Best, C. J., Gillespie, J. W., Flaig, M. J., Hewitt, S. M., Phillips, J. L., Krizman, D. B., Tangrea, M. A., Ahram, M., et al. (2002) Nat. Genet. 32 Suppl., 509-514. [DOI] [PubMed] [Google Scholar]
- 26.Karaman, M. W., Houck, M. L., Chemnick, L. G., Nagpal, S., Chawannakul, D., Sudano, D., Pike, B. L., Ho, V. V., Ryder, O. A. & Hacia, J. G. (2003) Genome Res. 13 1619-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roche, K. W., Tu, J. C., Petralia, R. S., Xiao, B., Wenthold, R. J. & Worley, P. F. (1999) J. Biol. Chem. 274 25953-25957. [DOI] [PubMed] [Google Scholar]
- 28.Blaustein, M. P., Juhaszova, M., Golovina, V. A., Church, P. J. & Stanley, E. F. (2002) Ann. N.Y. Acad. Sci. 976 356-366. [DOI] [PubMed] [Google Scholar]
- 29.Ehlers, M. D. (2003) Trends Neurosci. 26 4-7. [DOI] [PubMed] [Google Scholar]
- 30.Ames, A., III (2000) Brain Res. Brain Res. Rev. 34 42-68. [DOI] [PubMed] [Google Scholar]
- 31.Deitmer, J. W. (2002) J. Neurochem. 80 721-726. [DOI] [PubMed] [Google Scholar]
- 32.Aiello, L. & Wheeler, P. (1995) Curr. Anthropol. 36 199-221. [Google Scholar]
- 33.Bentourkia, M., Bol, A., Ivanoiu, A., Labar, D., Sibomana, M., Coppens, A., Michel, C., Cosnard, G. & De Volder, A. G. (2000) J. Neurol. Sci. 181 19-28. [DOI] [PubMed] [Google Scholar]
- 34.Bohnen, N. I., Minoshima, S., Giordani, B., Frey, K. A. & Kuhl, D. E. (1999) Neurology 52 541-546. [DOI] [PubMed] [Google Scholar]
- 35.Cross, D. J., Minoshima, S., Nishimura, S., Noda, A., Tsukada, H. & Kuhl, D. E. (2000) J. Nucl. Med. 41 1879-1887. [PubMed] [Google Scholar]
- 36.Noda, A., Ohba, H., Kakiuchi, T., Futatsubashi, M., Tsukada, H. & Nishimura, S. (2002) Brain Res. 936 76-81. [DOI] [PubMed] [Google Scholar]
- 37.Miller, S., Yasuda, M., Coats, J. K., Jones, Y., Martone, M. E. & Mayford, M. (2002) Neuron 36 507-519. [DOI] [PubMed] [Google Scholar]
- 38.Hawkes, K., O'Connell, J. F., Jones, N. G., Alvarez, H. & Charnov, E. L. (1998) Proc. Natl. Acad. Sci. USA 95 1336-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Sarno, P., Shestopal, S. A., King, T. D., Zmijewska, A., Song, L. & Jope, R. S. (2003) J. Biol. Chem. 278 11086-11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelicci, G., Troglio, F., Bodini, A., Melillo, R. M., Pettirossi, V., Coda, L., De Giuseppe, A., Santoro, M. & Pelicci, P. G. (2002) Mol. Cell. Biol. 22 7351-7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor, J. P., Hardy, J. & Fischbeck, K. H. (2002) Science 296 1991-1995. [DOI] [PubMed] [Google Scholar]
- 42.Yanagisawa, K. & Matsuzaki, K. (2002) Ann. N.Y. Acad. Sci. 977 384-386. [DOI] [PubMed] [Google Scholar]
- 43.Aiello, L. & Wells, J. (2002) Annu. Rev. Anthropol. 31 323-328. [Google Scholar]
- 44.Finch, C. & Stanford, C. (2003) in Brain and Longevity, eds. Finch, C., Robine, J.-M., Christen, Y. & Schmidt-Atzert, L. (Springer, Heidelberg), 33-68.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.