Abstract
MHC class I proteins are cell-surface ligands that bind to T cell receptors and other immunoreceptors and act to regulate the activation state of immune cells. Recent work has shown that MHC class I genes and CD3ζ, an obligate component of T cell receptors, are expressed in neurons, are regulated by neuronal activity, and function in neuronal development and plasticity. A search for additional neuronally expressed T cell receptor components has revealed that the T cell antigen receptor β (TCRβ) locus is expressed in neurons of the murine central nervous system and that this expression is dynamically regulated over development. In neonates, expression is most abundant in various thalamic nuclei. At later ages and in adults, thalamic expression fades and cortical expression is robust, particularly in layer 6. In T cells, protein-encoding transcripts are produced only after recombination of the TCRβ genomic locus, which joins variable, diversity, and joining regions, a process that creates much of the diversity of the immune system. We detect no genomic recombination in neurons. Rather, transcripts begin in regions upstream of several joining regions, and are spliced to constant region segments. One of the transcripts encodes a hypothetical 207-aa, 23-kDa protein, which includes the TCRβ J2.7 region, and the entire C region. These observations suggest that TCRβ may function in neurons.
The MHC class I (MHCI) gene family encodes Ig superfamily cell-surface proteins that enable the immune system to distinguish normal cells from foreign, mutant, or infected cells. These proteins are expressed on the surface of cells of most tissues and have been studied intensively because of their crucial roles in initiating and regulating immune responses. They are most well known for their ability to bind intracellular-derived peptides and present them for inspection by cytotoxic T cells (1, 2).
The brain has long been considered an immune privileged organ (3), and neurons have been thought to express MHCI genes only in response to cytokines (4–7). Closer inspection by using highly sensitive in situ hybridization and RNase protection techniques has revealed that MHCI genes are expressed throughout the brains of healthy mammals, that different MHCI genes have overlapping but clearly distinct neuronal expression patterns within the brain, and that they can by dynamically regulated, both during normal development and by levels of neural activity (8, 9).
Recent experiments have demonstrated that proper surface expression of MHCI proteins is required for normal formation of neuronal connections and for normal synaptic plasticity in the mature hippocampus (9). Mutant mice with deletions of genes encoding β2-microglobulin and tobacco acid pyrophosphatase 1 have dramatically reduced levels of MHCI proteins on the cell surface (10). Neuronal projections from the retina to the visual area of the thalamus in these mutant mice are abnormal, and they have abnormal hippocampal long-term potentiation and long-term depression as well. Together, these phenotypes suggest that neuronal MHCI proteins are involved in regression and/or elimination of inappropriate synaptic connections. In addition to synaptic plasticity and development, neuronal MHCI proteins may play a role in pheromone detection. Two groups have recently shown that several M family MHCI genes are expressed in the mouse vomeronasal organ (11, 12) and are needed for normal mating behavior (12). These observations suggest that these genes, long studied as mediators of immune function, can also play distinct, nonimmune roles in neurons.
CD3ζ is a critical transmembrane-signaling component of T cell receptor complexes (13), which bind MHCI ligands. The CD3ζ gene is also expressed in the brain and regulated over development (8). Mice with deleted CD3ζ genes display nearly identical phenotypes as the mutant mice with reduced MHCI surface expression; retinal ganglion axon arbors in the lateral geniculate nucleus do not to fully remodel, and hippocampal long-term potentiation is abnormal (9). These observations suggest that, as in the immune system, some neurons may use CD3ζ-bearing MHCI receptors. These findings compelled us to search for MHCI receptors in the CNS.
Neuronal receptors for MHCI proteins have not been identified. Known receptors for MHCI ligands in the immune system include the T cell receptors, NKG2/CD94 receptors, human Kir and Lir/Ilt proteins, and mouse Ly49 proteins (14–16). Although none of these genes are known to be expressed in the CNS, we investigated the possibility that some of these receptors serve neuronal functions. Here, we report the expression of the T cell antigen receptor β (TCRβ) locus in the murine brain.
Methods
In Situ Hybridization. All experiments were performed by using C57BL6 mice. Wild-type mice were acquired from Charles River Breeding Laboratories, and TCRβ-/- mice were acquired from The Jackson Laboratory. All procedures were performed according to institutional guidelines and approved protocols. Mice were anesthetized with Halothane (Halocarbon Products, River Edge, NJ) and killed by injection of 0.1 ml of Euthasol (Delmarva Laboratories, Midlothian, VA). Brains were removed, placed in M1 embedding matrix (Shandon, Pittsburgh), and frozen in a dry ice/ethanol bath. In situ hybridization was performed as described (17). Cryostat sections (12 μm) of brain were cut, air-dried, fixed for 30 min in sodium phosphate-buffered 4% paraformaldehyde, dehydrated with ethanol, and stored at -80°C. Sections were thawed, permeabilized by proteinase K treatment, acetylated, dehydrated with ethanol, and hybridized at 62°C for 12–18 h with a riboprobe labeled with α-35S-UTP (1,250 Ci/mmol). The sections were then incubated with 50 μg/ml RNase A for 30 min at 37°C and washed with a series of SSC solutions, with a high-stringency wash of 0.1× SSC (0.15 M sodium chloride/0.015 M sodium citrate, pH 7) at 60°C for 30 min. After exposure to Kodak XAR-5 film at room temperature, sections were coated with NTB-2 emulsion and developed after 2–4 weeks. TCRβ constant (C) region probe was transcribed from a cDNA encoding the TCRβ C region and Jb2.3 (GenBank accession no. BE864116). Other probes were transcribed from templates generated by PCR or RT-PCR by using primers encoding either T3 or T7 RNA polymerase binding sites. Variable (V) region probes were used as individual probes, or as pools of three: pool A, 18S1, 15S1, and 7S1; pool B, 9S1, 12S1, and 13S1; pool C, 5S1, 8S2, and 2S1; and pool D, 1S1, 4S1, and 13S1.
Retrograde Tracing. A P25 C57BL6 mouse was anesthetized with isoflurane (Abbott) and a craniotomy was performed. Injected into the striatum by using a 5-μl Hamilton syringe was 0.5 μl of 4% Fluoro-Gold (Fluorochrome, Denver) dissolved in sterile 0.9% saline. After 4 days, to permit retrograde axonal transport, the mouse was anesthetized with Halothane (Halocarbon Products) and killed by injection of 0.1 ml of Euthasol (Delmarva Laboratories). The brain was removed and prepared for in situ hybridization as described above. After in situ hybridization, sections were stained with Hoechst 33258 (Sigma) to visualize nuclei. Sections were imaged under a ×100 oil objective on a Nikon Eclipse Fluoresence microscope. Silver grains were visualized by darkfield or brightfield microscopy.
Anterograde Tracing of Retinogeniculate Projections. P12 mouse pups (10 wild-type and 10 TCRβ-/-) were anesthetized with isoflurane, and 0.5–1 μl of a 0.2% solution of cholera toxin B subunit (List Biological Laboratories, Campbell, CA) conjugated to either rhodamine (product 107) or FITC (product 106) was injected into each eye (FITC, left eye; rhodamine, right eye) by using a glass micropipette. After 2 days, animals were killed with Euthasol (Delmarva Laboratories). Brains were fixed by cardiac perfusion with 0.9% saline and 4% paraformaldehyde. Brains were removed and postfixed overnight in 4% paraformaldehyde at 4°C, then bathed in 20% sucrose overnight. Coronal sections (50 μM) were cut on a freezing microtome, mounted on glass slides by using Vectashield (Vector Laboratories), and imaged on a Nikon Eclipse fluorescence microscope.
RT-PCR and 5′ RACE. Blood was removed from brains by cardiac perfusion with 0.9% saline. RNA was extracted with TRIzol (GIBCO/BRL), and reverse transcription was performed by using Retroscript (Ambion, Austin, TX). PCR was performed by using SuperTaq Plus (Ambion).
5′ RACE was performed by using the RNA ligase-mediated RACE system (Ambion). Ten micrograms of total RNA from saline-perfused cerebral cortex of a P40 mouse was treated with calf intestinal phosphatase for 1 h at 37°C, extracted with phenol/chloroform, precipitated in isopropyl alcohol, and resuspended in H2O. Sample was then treated with tobacco acid pyrophosphatase for 1 h at 37°C. The 5′ RACE adapter (GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUGAA) was then ligated to the RNA for 1 h at 37°C by using T4 RNA ligase. Ligated RNA was then used in a reverse transcription reaction with Moloney murine leukemia virus reverse transcriptase and random decamers for1hat42°C. PCR was performed in two steps by using nested primers. In the first step the primers were 5′-GCTGATGGCGATGAATGAACACTG and 3′-CCAAGCACACGAGGGTAGCCTTTTGTTTG. Thirty-five cycles of PCR were performed by using Taq polymerase, 1 μl of reverse transcriptase reaction, an annealing temperature of 64°C, and a 2-min extension time in a 50-μl reaction. One microliter of the first reaction was then used as a template in a second round of amplification using nested primers. The 5′ nested primer sequence is CGCGGATCCGAACACTGCGTTTGCTGGCTTTGATG, and the 3′ primer sequence is GGCTCAAACAAGGAGACCTTGG. PCR products were separated on agarose gels and sequenced.
Primers for in Situ Hybridization and Northern Blot Probes. V regions. T3-V1S, 3′-AGAATTAACCCTCACTAAAGGGACTCTGGATCCACGGCAGATA; T7-V1S1, 5′-GATTAATACGACTCACTATAGGGATGTCTTGTGGAAACAGCACTC; T3-V2S1, 3′-AGAATTAACCCTCACTAAAGGGACCATGTTGGCCACTTGCAG; T7-V2S1, 5′-GATTAATACGACTCACTATAGGGAGTGGCTACAGACCCCACAGT; T3-V3S1, 3′-AGAATTAACCCTCACTAAAGGGACCCTGCCTCAGAGGACTGAAT; T7-V3S1, 5′-GATTAATACGACTCACTATAGGGATCTCCTGGGTGCAAGAATTT; T3-V4S1, 3′-AGAATTAACCCTCACTAAAGGGACGCCGAGTCATCAGGCTTTAG; T7-V4S1, 5′-GATTAATACGACTCACTATAGGGATGTCTCCTGGTGGCAGGT; T3-V5S1, 3′AGAATTAACCCTCACTAAAGGGACCTCCAAGGCACTCATGTTCA; T7-V5S1, 5′-GATTAATACGACTCACTATAGGGACCTGCTATCTTGGGTTGCTC; T3-V5S2, 3′-AGAATTAACCCTCACTAAAGGGACTTCCAAGGCACTCATGTTCA; T7-V5S2, 5′-GATTAATACGACTCACTATAGGGAGCATCACCCTGCTATCTTGG; T3-V7S1, 3′-AGAATTAACCCTCACTAAAGGGACCCAGAATCAGGGAGAAATGC; T7-V7S1, 5′-GATTAATACGACTCACTATAGGGAAGGCTCATCTCTGCTGTGGT; T3-V8S2, 3′-AGAATTAACCCTCACTAAAGGGACGGGGTAGCCAACTCCAGAAT; T7-V8S2, 5′-GATTAATACGACTCACTATAGGGAGGCTGCAGTCACCCAAAG; T3-V8S3, 3′-AGAATTAACCCTCACTAAAGGGACTCCAGCAGGAGGAAGAAGTC; T7-V8S3, 5′-GATTAATACGACTCACTATAGGGACAGTCACCCAAAGCCCTAGA; T3-V9S1, 3′-AGAATTAACCCTCACTAAAGGGACCCTGCAGAGCCAATGTAGAG; T7-V9S1, 5′-GATTAATACGACTCACTATAGGGATGCAGCCACTTTTGTGGATA; T3-V12S1, 3′-AGAATTAACCCTCACTAAAGGGACCAGCTGAGTCCTTGGGTTCT; T7-V12S1, 5′-GATTAATACGACTCACTATAGGGATGCTGGAGTTACCCAGACAC; T3-V13S1, 3′-AGAATTAACCCTCACTAAAGGGACTGCTGGCACAGAGATAGGTG; T7-V13S1, 5′-GATTAATACGACTCACTATAGGGAGGGCAGTGTTCTGTCTCCTT; T3-V15S1, 3′-AGAATTAACCCTCACTAAAGGGACTCCCTAGCACCACAGAGATA; T7-V15S1, 5′-GATTAATACGACTCACTATAGGGAGGCTTGGAGCACTCGTCTAT; T3-V18S1, 3′-AGAATTAACCCTCACTAAAGGGACCGGCTGATTGGAAACTTGTC; T7-V18S1, 5′-GATTAATACGACTCACTATAGGGACTTTGGAGCCAAGGTTCTGT. J regions. Jb1.1, 5′-AAACACAGAAGTCTTCTTTGGTAAAG; Jb2.1, 5′-TATGCTGAGCAGTTCTTCGGAC; Jβ1, 3′-T7 GATTAATACGACTCACTATAGGGACTAAATTTCCCAGTCCCTTCCACC; Jβ2, 3′-T7 GATTAATACGACTCACTATAGGGATTTCCCTCCCGGAGATTCCCTAA.
Primers for Jβ C Region RT-PCR. Jb1.1, 5′-CTCTTCACCCCTTAAGATTATT; Jb1.2, 5′-AAGCATGTCCTCCGTGTCCATA; Jb1.3, 5′-GGAGAAAGGCTCACAGAGAG; Jb1.4, 5′-GGCAGGAAGCATGAGGAAGT; Jb1.5, 5′-CATCAGCTCTTTGGGAGTT; Jb1.6, 5′-GCTTTGGGACCGCTGGTAGGT; Jb2.1, 5′-AAGGTGAGGAAAGAGGAAGAA; Jb2.2, 5′-CCTGTATCAGGAAAGAGGAG; Jb2.3, 5′-GGAAGCACTGAGTTTTTGTC; Jb2.4, 5′-AGGATAGCCAGAGCCAGTTT; Jb2.5, 5′-AGGTAAGCTGGGGTATAGTT; Jb2.7–5′ GAGAATACCTCGCTGAACC; Constant, 3′-CGAGGGTAGCCTTTTGTTTG.
Northern Blots. Northern blots were performed by using the NorthernMax Glyoxal-based system (Ambion). Blood was removed from brains of animals of various ages (see Fig. 3) by cardiac perfusion with 0.9% saline. Brains were dissected and RNA was extracted with TRIzol (GIBCO/BRL). Twenty micrograms of brain-derived RNA and 3 μg of spleen-derived RNA were separated on a 1% agarose gel, transferred to a positively charged nylon membrane, UV-crosslinked, and probed by using a biotinylated RNA probe. The probes were detected by using streptavidin alkaline phosphatase and the BrightStar BioDetect system (Ambion), and imaged on Kodak XAR-5 film. The C region probe was transcribed from the same template used for in situ hybridization. The V region probe was a pool of probes for 12 V regions (Vβ1.1, 2.1, 3.1, 4.1, 5.1, 7.1, 8.2, 9.1, 12.1, 13.1, 15.1, and 18.1). Probes were transcribed from templates derived from PCR products.
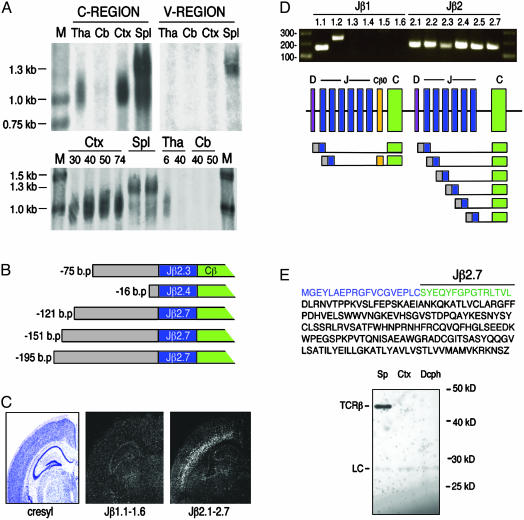
Fig. 3.
Neuronal TCRβ is transcribed from unrearranged genomic loci. (A Top) Northern blot analysis of RNA derived from P9 thalamus (Tha), P29 cerebellum (Cb), P30 cortex (Ctx), and adult spleen (Spl), probed with TCRβ C region or mix of V region probes. (Bottom) RNA derived from P30, P40, P50, and P74 cortex, adult spleen, P6 and P40 thalamus, and P40 and P50 cerebellum, probed for TCRβ C region. M, molecular weight markers. (B) RACE-derived sequences indicate that the 5′ ends of neuronal TCRβ are identical with genomic sequences 5′ to several of the TCRβ J regions. Numbers indicate bases from the beginning of transcript to Jβ exon junction. (C) In situ hybridization of adult mouse brain sections by using probes for genomic sequence spanning Jβ1.1–1.6 or Jβ2.1–2.7 indicates that the majority of neuronal TCRβ mRNA includes Jβ2 regions. (D) RT-PCR by using primers ≈20 bp 5′ to the start of each Jβ coding region indicated, plus a common C region 3′ primer, indicate that neuronal TCRβ transcripts include at least 8 of the 12 Jβ regions and include noncoding intergenic sequences upstream of Jβ regions. One sequence that included Jβ1.2 also included the Cβ0 exon. (E) Hypothetical truncated TCRβ protein sequence. (Top) Amino acid sequence of hypothetical protein encoded by genomic region 5′ to Jβ2.7 (blue), Jβ2.7 (green), and C region (black). (Bottom) Immunoprecipitation/Western blot analysis of saline-perfused brain tissue using an antibody to the C region of TCRβ. LC, light chain; Sp, spleen; Ctx, cortex; Dcph, diencephalon. Analyzed were 100 μg of spleen tissue and 5 mg of cortex and diencephalon.
Western Blots. Saline-perfused brains from C57 BL6 mice at P28 were dissected and homogenized in ice-cold buffer containing 1% Nonidet P-40, 150 mM NaCl, 50 mM Tris (pH 7.4), 1 mM EDTA, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride. Samples were separated by SDS/PAGE, transferred to poly(vinylidene difluoride) membranes, probed with the H57-587 monoclonal antibody (Pharmingen), or the A-19 polyclonal antibody (Santa Cruz Biotechnology), and visualized with enhanced chemiluminescence (Perkin–Elmer) and Kodak XAR-5 film.
Results and Discussion
The canonical receptor for MHCI proteins is the αβ T cell receptor. The α- and β-subunits are encoded by genes composed of separate genomic segments that are assembled by genomic recombination during T cell development (18–20). In the mouse, the locus for the β-subunit encodes approximately 20 variable (V) regions, 2 diversity (D) regions, 12 joining (J) regions, and two constant (C) regions. The V, D, and J segments are joined by genomic recombination during development, which requires the coordinate expression of RAG-1 and RAG-2 proteins. The many combinations of V, D, and J regions provide much of the diversity characteristic of the immune system.
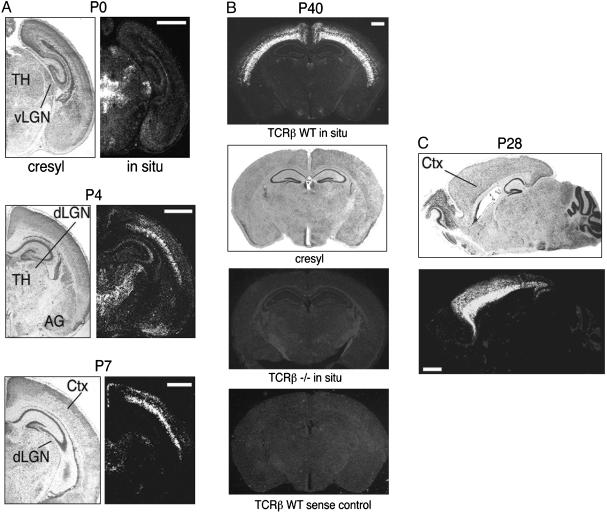
To determine whether TCRβ is expressed in neurons, in situ hybridization experiments were performed on mouse brain sections of various ages by using a probe specific for the C regions of mouse TCRβ. Expression is seen clearly in the thalamus and cortex of newborn mice (Fig. 1A). Regions of expression include the cerebral cortex, medial habenular nucleus, and the ventral lateral geniculate nucleus. By postnatal day 4, expression appears in the dorsal lateral geniculate nucleus, midline thalamic nuclei, and amygdala. Thalamic expression persists through P7 and is high throughout the cortex, including the retrosplenial granular cortex. By postnatal day 21, thalamic expression cannot be detected (data not shown), but robust expression persists in the cortex into adulthood, with the heaviest expression in layer 6 (Fig. 1 B and C). In addition to standard sense strand controls, sections from TCRβ knockout mice were hybridized to verify that our probe detects exclusively TCRβ mRNA. The knockout animals displayed no signal from our TCRβ probe (Fig. 1B), indicating that TCRβ is indeed being detected in the wild-type brain.
Fig. 1.
TCRβ mRNA is expressed in the mouse brain. Cryostat sections from postnatal days 0, 4, 7, 28, or 40 were either stained with cresyl violet or hybridized with a probe for TCRβ C regions. (A) Coronal sections. (B) P40 TCRβ-/- and TCRβ wild-type sense control sections are similar to the P40 section hybridized with TCRβ antisense probe. (C) Sagittal sections, anterior to the left. TH, thalamus; AG, amygdala; dLGN, dorsal lateral geniculate nucleus; vLGN, ventral lateral geniculate nucleus; CTX, cortex. (Scale bars, 1 mm.)
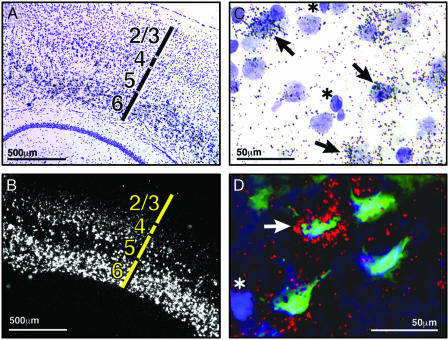
Detailed examination of sections that have undergone in situ hybridization for TCRβ expression reveals that distinct populations of neurons express TCRβ (Fig. 2 A and B). For example, expression is robust in layer 6 of the cerebral cortex, whereas other cortical layers express TCRβ at lower levels; no signal can be detected in hippocampus at this age (P40). This observation suggests that neurons express TCRβ, because glial cells are not known to be segregated in similar laminar patterns.
Fig. 2.
TCRβ is expressed in a subset of cortical neurons. (A and B) P40 coronal section showing cortical layers 1–6 (A) and TCRβ C region in situ hybridization shown in darkfield optics (B) (silver grains appear white). (C) High-power image of layer 6 cortical cells double-stained with cresyl violet (purple) and TCRβ in situ hybridization (black silver grains). Arrows point to large pale nuclei with associated silver grains; asterisks indicate smaller dark nuclei. (D) Triple labeling of cortical layer 6 neurons from posterior cortical region. Hoechst-stained nuclei (blue), retrograde label from Fluoro-Gold injection in striatum (green), and TCRβ in situ hybridization (red) are shown. Arrow indicates triple-labeled cell; asterisk indicates nucleus of a cell with no retrograde label and no TCRβ expression.
Inspection of cresyl violet-stained brain sections that have undergone the TCRβ in situ hybridization procedure reveals that within layer 6, most nuclei that are surrounded by silver grains are relatively large and pale, indicative of neuronal expression; the smaller, darkly stained nuclei, typical of glia, do not colocalize with silver grains and thus, do not express TCRβ mRNA (Fig. 2C).
To determine definitively whether neurons express TCRβ, we performed double-retrograde tracing/in situ hybridization experiments. The tracer Fluoro-Gold was injected into the striatum to retrogradely label neuronal cell bodies in cortical layers 5 and 6, which are known to project descending axons there (21). Four days later, the brain was sectioned and analyzed by in situ hybridization. Fig. 2D shows several layer 6 cortical neurons that have been retrogradely labeled (green) from the distant injection site. These pyramidal neurons also clearly express the TCRβ mRNA (red and yellow), indicating that at least some, if not all, TCRβ expression is neuronal. The nuclei of all cells in the section are also stained (blue), which reveals that not all cells in the region are stained with Fluoro-Gold; only those that project axons to the striatum are labeled retrogradely.
To determine whether TCRβ transcripts are derived from rearranged genomic loci, we attempted to detect V region transcripts in the brain. In situ hybridization experiments were performed by using pools of three probes representing 12 V regions. No V region genes were detected (data not shown), suggesting that the neuronal transcripts we detect are transcribed from unrearranged genomic loci.
To investigate this possibility further, Northern blot analysis was performed on total RNA derived from saline-perfused thalamus, cortex, and cerebellum from animals of various ages (Fig. 3A). It has been reported that two TCRβ mRNA species occur in normal lymphocyte populations in both humans and mice: a 1.3-kb mRNA representing mature rearranged transcripts, and a 1.0-kb species representing immature transcripts that do not contain V regions and are derived from either partially rearranged or unrearranged genomic loci (22–25). Using a probe specific for the TCRβ C regions, we found that transcripts from the brain were consistently smaller than mature 1.3-kb transcripts derived from the spleen and were instead similar in size to the immature 1.0-kb transcripts seen in lymphocytes. With pooled probes specific for several V region segments, no signal was detected from brain-derived RNA, yet the 1.3-kb mature band from the spleen was indeed detected (Fig. 3A). These results suggest that brain-derived transcripts do not include V regions at their 5′ ends and thus may be transcribed from unrearranged loci. In addition, Southern blot analysis of cortical tissue and single-cell RT-PCR from layer 6 neurons with primers specific for all 20 V regions did not detect recombined genomic DNA or recombined TCRβ mRNA, respectively (data not shown).
To determine the nature of the 5′ ends of neuronal TCRβ transcripts, we performed 5′ RACE analysis of cerebral cortical mRNA taken at P40. This procedure only amplifies RNA with a 5′ cap and thus should only identify 5′ ends of mRNA. These experiments revealed several transcripts with 5′ ends identical with genomic regions located just upstream of several Jβ2 regions (Fig. 3B). This finding indicates that transcription likely starts 5′ to unrearranged J regions. The J regions are spliced onto C regions, resulting in the transcripts described in Fig. 3B. A similar transcript has been described in the human T cell line Jurkat. This transcript is 1.0 kb and begins just upstream of an unrearranged Jβ2.3 segment, is spliced to the C region, and is polyadenylated (25).
To determine whether the majority of brain-derived transcripts do indeed include sequences from Jβ2 as opposed to Jβ1, in situ hybridization experiments were performed by using probes specific for either the entire Jβ1 genomic region or the entire Jβ2 region. The majority of transcripts contained sequences from the Jβ2 region (Fig. 3C). To confirm this observation, PCR primers were designed to detect genomic intergenic sequences ≈20 bases upstream from the beginning of each of the 12 J region coding sequences and used in combination with 3′ primers specific for the C regions in RT-PCR reactions with RNA derived from saline-perfused adult cerebral cortex. Transcript sequences included genomic regions upstream of each of the six Jβ2 regions and two of the six Jβ1 regions (Fig. 3D). All the RACE-derived sequences begin in regions just upstream of a Jβ segment, and all neuronal TCRβ transcripts appear to be of roughly one size on Northern blots. Hence, we believe it is likely that most, if not all, neuronal TCRβ transcripts begin 5′ to unrearranged Jβ regions.
Of the transcripts detected, only Jβ2.7 encodes an in-frame ATG start codon downstream of all stop codons. The hypothetical protein encoded by this transcript is 23 kDa (Fig. 3E) and may contain a minimal consensus signal sequence (26) and much of the TCRβ extracellular domain, the transmembrane and cytoplasmic regions. Immunoprecipitation and Western blot experiments were performed by using antibodies specific for the TCRβ C region. Tissue was examined from P28 cortex, where we detected TCRβ mRNA, and from P28 thalamus, a major target of axonal projections from layer 6 neurons. No TCRβ protein of any size was detected in these experiments (Fig. 3E). Brain tissue from other ages, including P0, P14, and P38, were also examined, and no protein was detected (data not shown). In contrast, TCRβ was detectable in spleen lysate (Fig. 3E). Although we detected no protein, the hypothetical 23-kDa truncated TCRβ may be expressed at low levels or may resist detection by our reagents. The protein may only be stable in the presence of other components of a receptor complex, such as T cell receptor α or CD3ζ (27–29). We detect no expression of T cell receptor α in the brain (data not shown), and we have not detected expression of CD3ζ in the cortex (8, 9). However, these proteins may be transiently expressed at times that were not tested in these studies. Alternatively, the RNA itself may have a function; numerous examples exist of RNA regulating the expression of other genes and even entire chromosomes (30, 31).
TCRβ mRNA is expressed at high levels in the lateral geniculate nucleus (LGN) of the thalamus at P4 and P7, when retinal ganglion cell axonal arbors remodel into eye-specific domains. TCRβ requires CD3ζ to signal in the immune system, and CD3ζ is also expressed in LGN neurons at this time. Mice deficient for CD3ζ and mice deficient for MHCI have abnormal retinogeniculate projections (9). Thus, if TCRβ is a component of a CD3ζ-bearing receptor complex in LGN dendrites, one might expect TCRβ knockout mice to display a similar connectivity phenotype. To test this hypothesis, retinogeniculate projections were labeled with fluorescent cholera toxin injected into the eyes of P12 mice. Two days later, the brains were sectioned and the dorsal LGN was analyzed. No apparent defects in the patterning of retinogeniculate projections in TCRβ mutant mice (n = 10) were detected when compared with wild-type controls (n = 10) (data not shown). In addition, the gross appearance of the brains of TCRβ-/- mice is normal; cortical layers and thalamic nuclei are histologically indistinguishable from wild-type when viewed in cresyl violet-stained sections (data not shown). The organization of somatosensory cortex, including barrels, is also normal (n = 2, A. Datwani, unpublished data). Thus, our results suggest that it is unlikely that TCRβ is a neuronal receptor for MHCI in developing LGN dendrites under normal physiological conditions.
These experiments, however, do not rule out a role for TCRβ in axonal remodeling; TCRβ may be required at axon terminals instead of dendrites, in which case no retinogeniculate phenotype would be expected. Instead, synapses from LGN axons projecting to cortical neurons, or synapses of layer 6 cortical axons projecting to thalamic, striatal, or other neuronal populations, could be affected. Aside from colocalization in the neonatal thalamus, TCRβ and CD3ζ have very different expression patterns. TCRβ is expressed in the neonatal thalamus and in the cerebral cortex and is absent from the hippocampus (Fig. 1), whereas CD3ζ is expressed primarily in neonatal thalamus and the adult hippocampus (9). The mismatch in timing and location of mRNA of TCRβ and CD3ζ in these structures suggests that, if neuronal TCRβ has a function, it does not require CD3ζ.
A major finding of this study is that transcripts from the TCRβ locus are expressed in selected subsets of neurons in the murine CNS and that these transcripts are derived from unrearranged genomic loci. Transcription of unrearranged TCRβ has been reported in early T cell development (32–35), where it is driven by a promoter near Dβ1. This activation of transcription is important for initiation of VDJ recombination, possibly by allowing recombination machinery greater access to chromatin. VDJ recombination requires the expression of RAG-1 and RAG-2 proteins, which function to cut and paste genomic regions, forming virtually unique, cell-specific genes (20). The many combinations of V, D, and J regions and the imprecise recombination process itself produce much of the diversity required by the immune system to distinguish between the myriad of antigens it encounters. The brain, with its plethora of precise synaptic connections and circuits, is also characterized by enormous diversity. The VDJ recombination system is attractive as a potential source for cell- or lineage-specific molecular markers, and the subject has been the topic of study (36, 37). RAG-1 is expressed in the brain (38), but RAG-2 has not been detected to date in mammals. Our finding that TCRβ is transcribed in neurons, an event that normally precedes VDJ recombination in lymphocytes, suggests that the TCRβ locus may be poised to undergo recombination in neurons, with the limiting factor being expression of RAG-2. Whether the RAG-2 gene can be induced in neurons and, under circumstances not tested in this study, elicit neuronal VDJ recombination is not known.
An alternative possibility is that the transcripts we have observed in the CNS are regulated by the remnants of a primordial gene expression program that may have once driven an ancestral receptor gene. This gene is thought to have been split into multiple segments by insertion of a transposable element, ultimately giving rise to modern Ig and T cell receptor genes (39, 40). Whatever the case, the finding of these unusual transcripts from the tightly regulated TCRβ gene implies an unanticipated use of the TCRβ locus by CNS neurons.
Acknowledgments
We thank Lisa Boulanger for help with single-cell RT-PCR, Akash Datwani for examination of TCRβ mutant barrels, Bella Printseva and Michael Marcotrigiano for excellent technical assistance, and David Schatz, Karl Münger, Dietmar Schmucker, and Nicola Tolliday for critical comments. This work was supported by National Research Service Award 5F32NS41890-02.
Abbreviations: MHCI, MHC class I; TCRβ, T cell antigen receptor β; LGN, lateral geniculate nucleus; D, diversity.
References
- 1.Ploegh, H. L., Orr, H. T. & Strominger, J. L. (1981) Cell 24 287-299. [DOI] [PubMed] [Google Scholar]
- 2.Rammensee, H. G., Falk, K. & Rotzschke, O. (1993) Curr. Opin. Immunol. 5 35-44. [DOI] [PubMed] [Google Scholar]
- 3.Lampson, L. A. (1987) Trends Neurosci. 10 211-216. [Google Scholar]
- 4.Wong, G. H., Bartlett, P. F., Clark-Lewis, I., Battye, F. & Schrader, J. W. (1984) Nature 310 688-691. [DOI] [PubMed] [Google Scholar]
- 5.Wong, G. H., Bartlett, P. F., Clark-Lewis, I., McKimm-Breschkin, J. L. & Schrader, J. W. (1985) J. Neuroimmunol. 7 255-278. [DOI] [PubMed] [Google Scholar]
- 6.Neumann, H., Cavalie, A., Jenne, D. E. & Wekerle, H. (1995) Science 269 549-552. [DOI] [PubMed] [Google Scholar]
- 7.Neumann, H., Schmidt, H., Cavalie, A., Jenne, D. & Wekerle, H. (1997) J. Exp. Med. 185 305-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corriveau, R. A., Huh, G. S. & Shatz, C. J. (1998) Neuron 21 505-520. [DOI] [PubMed] [Google Scholar]
- 9.Huh, G. S., Boulanger, L. M., Du, H., Riquelme, P. A., Brotz, T. M. & Shatz, C. J. (2000) Science 290 2155-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorfman, J. R., Zerrahn, J., Coles, M. C. & Raulet, D. H. (1997) J. Immunol. 159 5219-5225. [PubMed] [Google Scholar]
- 11.Ishii, T., Hirota, J. & Mombaerts, P. (2003) Curr. Biol. 13 394-400. [DOI] [PubMed] [Google Scholar]
- 12.Loconto, J., Papes, F., Chang, E., Stowers, L., Jones, E. P., Takada, T., Kumanovics, A., Fischer Lindahl, K. & Dulac, C. (2003) Cell 112 607-618. [DOI] [PubMed] [Google Scholar]
- 13.Howe, L. R. & Weiss, A. (1995) Trends Biochem. Sci. 20 59-64. [DOI] [PubMed] [Google Scholar]
- 14.Barten, R., Torkar, M., Haude, A., Trowsdale, J. & Wilson, M. J. (2001) Trends Immunol. 22 52-57. [DOI] [PubMed] [Google Scholar]
- 15.Lanier, L. L. (1998) Annu. Rev. Immunol. 16 359-393. [DOI] [PubMed] [Google Scholar]
- 16.Trowsdale, J. (2001) Immunity 15 363-374. [DOI] [PubMed] [Google Scholar]
- 17.Simmons, D. M., Arriza, J. L. & Swanson, L. W. (1989) J. Histotechnol. 12 169-181. [Google Scholar]
- 18.Schatz, D. G., Oettinger, M. A. & Schlissel, M. S. (1992) Annu. Rev. Immunol. 10 359-383. [DOI] [PubMed] [Google Scholar]
- 19.Tonegawa, S. (1983) Nature 302 575-581. [DOI] [PubMed] [Google Scholar]
- 20.Bassing, C. H., Swat, W. & Alt, F. W. (2002) Cell 109 Suppl., S45-S55. [DOI] [PubMed] [Google Scholar]
- 21.Selemon, L. D. & Goldman-Rakic, P. S. (1985) J. Neurosci. 5 776-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siu, G., Kronenberg, M., Strauss, E., Haars, R., Mak, T. W. & Hood, L. (1984) Nature 13 344-350. [DOI] [PubMed] [Google Scholar]
- 23.Clark, S. P., Yoshikai, Y., Taylor, S., Sui, G., Hood, L. & Mak, T. W. (1984) Nature 311 387-389. [DOI] [PubMed] [Google Scholar]
- 24.Hurwitz, J. L., Samaridis, J. & Pelkonen, J. (1988) Cell 52 821-829. [DOI] [PubMed] [Google Scholar]
- 25.Yasunobu, Y., Anatoniou, D., Clark, S. P., Yanagi, Y., Sangster, R., Van den Elsen, P., Terhorst, C. & Mak, T. W. (1984) Nature 312 521-524. [DOI] [PubMed] [Google Scholar]
- 26.Martoglio, B. & Dobberstein, B. (1998) Trends Cell Biol. 8 410-415. [DOI] [PubMed] [Google Scholar]
- 27.Minami, Y., Weissman, A. M., Samelson, L. E. & Klausner, R. D. (1987) Proc. Natl. Acad. Sci. USA 84 2688-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sussman, J. J., Bonifacino, J. S., Lippincott-Schwartz, J., Weissman, A. M., Saito, T., Klausner, R. D. & Ashwell, J. D. (1988) Cell 52 85-95. [DOI] [PubMed] [Google Scholar]
- 29.Bonifacino, J. S., Suzuki, C. K., Lippincott-Schwartz, J., Weissman, A. M. & Klausner, R. D. (1989) J. Cell Biol. 109 73-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eddy, S. R. (2001) Nat. Rev. Genet. 2 919-929. [DOI] [PubMed] [Google Scholar]
- 31.Brockdorff, N. (2002) Trends Genet. 18 352-358. [DOI] [PubMed] [Google Scholar]
- 32.Sikes, M. L., Gomez, R. J., Song, J. & Oltz, E. M. (1998) J. Immunol. 161 1399-1405. [PubMed] [Google Scholar]
- 33.Whitehurst, C. E., Chattopadhyay, S. & Chen, J. (1999) Immunity 10 313-322. [DOI] [PubMed] [Google Scholar]
- 34.Doty, R. T., Xia, D., Nguyen, S. P., Hathaway, T. R. & Willerford, D. M. (1999) Blood 93 3017-3025. [PubMed] [Google Scholar]
- 35.Pardoll, D. M., Fowlkes, B. J., Lechler, R. I., Germain, R. N. & Schwartz, R. H. (1987) J. Exp. Med. 165 1624-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abeliovich, A., Gerber, D., Tanaka, O., Katsuki, M., Graybiel, A. M. & Tonegawa, S. (1992) Science 257 404-410. [DOI] [PubMed] [Google Scholar]
- 37.Matsuoka, M., Nagawa, F., Okazaki, K., Kingsbury, L., Yoshida, K., Muller, U., Larue, D. T., Winer, J. A. & Sakano, H. (1991) Science 254 81-86. [DOI] [PubMed] [Google Scholar]
- 38.Chun, J. J., Schatz, D. G., Oettinger, M. A., Jaenisch, R. & Baltimore, D. (1991) Cell 64 189-200. [DOI] [PubMed] [Google Scholar]
- 39.Cannon, J. P., Haire, R. N. & Litman, G. W. (2002) Nat. Immunol. 3 1200-1207. [DOI] [PubMed] [Google Scholar]
- 40.Agrawal, A., Eastman, Q. M. & Schatz, D. G. (1998) Nature 394 744-751. [DOI] [PubMed] [Google Scholar]