Abstract
By comparing expression levels of MADS box transcription factor genes between near-isogenic winter and spring lines of bread wheat, Triticum aestivum, we have identified WAP1 as the probable candidate for the Vrn-1 gene, the major locus controlling the vernalization flowering response in wheat. WAP1 is strongly expressed in spring wheats and moderately expressed in semispring wheats, but is not expressed in winter wheat plants that have not been exposed to vernalization treatment. Vernalization promotes flowering in winter wheats and strongly induces expression of WAP1. WAP1 is located on chromosome 5 in wheat and, by synteny with other cereal genomes, is likely to be collocated with Vrn-1. These results in hexaploid bread wheat cultivars extend the conclusion made by Yan et al. [Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T. & Dubcovsky, J. (2003) Proc. Natl. Acad. Sci. USA 100, 6263–6268] in the diploid wheat progenitor Triticum monococcum that WAP1 (TmAP1) corresponds to the Vrn-1 gene. The barley homologue of WAP1, BM5, shows a similar pattern of expression to WAP1 and TmAP1. BM5 is not expressed in winter barleys that have not been vernalized, but as with WAP1, expression of BM5 is strongly induced by vernalization treatment. In spring barleys, the level of BM5 expression is determined by interactions between the Vrn-H1 locus and a second locus for spring habit, Vrn-H2. There is now evidence that AP1-like genes determine the time of flowering in a range of cereal and grass species.
Many plants from temperate regions are induced to flower by an extended exposure to low temperature: vernalization. In winter cereal crops, such as wheat and barley, plant breeders have selected for variation in vernalization responsiveness to produce cultivars suited to plantings in different climatic zones. Winter cultivars are sown in autumn, vernalized by the low temperatures of winter, and subsequently flower and develop grain in spring. Spring cultivars do not require vernalization and usually are planted in the late winter period (reviewed in refs. 1–3).
The genetics of the vernalization response have been studied in a number of cereals. In hexaploid bread wheat, where the effects of recessive traits are masked by the redundancy resulting from the three genomes, the dominant Vrn-1 gene has been found to be the major determinant of vernalization responsiveness. Vrn-1 is located on chromosome 5 in each of the A, B, and D genomes of wheat. Winter wheats carry only winter Vrn-1 alleles (vrn-A1/vrn-B1/vrn-D1), and without vernalization are late flowering. Spring alleles of Vrn-1 are dominant and reduce the requirement for vernalization. Spring alleles of the Vrn-1 gene on the A genome, Vrn-A1, have the strongest effect on flowering time, and plants with the Vrn-A1 spring allele do not require any vernalization (4). The effects of spring alleles of the Vrn-1 genes from the B and D genomes are weaker. Plants that carry Vrn-B1 or Vrn-D1 spring alleles (in the absence of a Vrn-A1 spring allele) flower earlier than winter wheats but still show some acceleration in flowering time when vernalized. Such plants are classed as semispring wheats.
The Vrn-1 locus has been mapped to the long arm of chromosome 5 (5–8). Orthologous genes have been identified in Triticum monococcum (Vrn-Am1) (9) and in barley (Vrn-H1) (10). In these diploid cereals recessive alleles of a second gene, Vrn-2 (vrn-2 for the recessive allele), can also confer the spring habit phenotype. The Vrn-H2 locus of barley maps to the long arm of chromosome 4(H) (10, 11) and the Vrn-Am2 locus maps to a syntenic region on the long arm of chromosome 5 in T. monococcum (9). vrn-Am2 is epistatic to Vrn-Am1, suggesting that these genes operate in a single genetic pathway controlling the vernalization response (12). Similar genetic interactions have been observed between Vrn-H1 and vrn-H2 in barley (11).
Recently, the Vrn-Am1 locus of T. monococcum was mapped to a small region of chromosome 5 that includes two MADS box genes (13). One of these MADS box genes, TmAP1, has an expression pattern consistent with a role in determining the vernalization response. In winter lines of T. monococcum, TmAP1 expression is low in plants that have not been vernalized, but increases after vernalization. In spring cultivars that have Vrn-Am1 spring alleles and flower early without vernalization, TmAP1 expression is high. TmAP1 is a strong candidate for the Vrn-1 gene (13). Sequencing of the promoter region of the TmAP1 gene from Vrn-Am1 spring cultivars revealed small deletions of a region that includes a putative MADS box transcription factor binding site. Yan et al. (13) suggested that the removal of this binding site renders the promoter of TmAP1 immune to the action of a proposed second MADS box protein that acts as a repressor to ensure that TmAP1 is expressed only after vernalization treatment. Spring alleles of the Vrn-Am1 locus would be caused by constitutively derepressed TmAP1 expression.
In this paper, we have compared the expression of MADS box transcription factor genes in near-isogenic winter and spring cultivars of hexaploid bread wheat, Triticum aestivum. We have identified 10 wheat MADS box genes that are expressed in vegetative tissues before the floral transition, including two vernalization-responsive MADS box genes. One of these is the hexaploid wheat orthologue of TmAP1, WAP1, and we suggest that this gene corresponds to the Vrn-1 locus of hexaploid wheat. We also suggest that the barley orthologue of WAP1, BM5, corresponds to the Vrn-H1 gene of barley.
Materials and Methods
Plant Growth. Seeds for the various wheat and barley cultivars were obtained from the Australian Winter Cereal Collection (www.dpi.qld.gov.au/extra/asp/auspgris). Plants were grown in pots of soil in glasshouse conditions under a 16-h light/8-h dark photoperiod. Average temperature was 19°C. Nonvernalized seedlings were grown for 14 days. Vernalized seedlings were imbibed overnight in water, then planted in pots and kept at 4°C for 60 days under long-day conditions. At the end of this treatment, vernalized seedlings were equivalent in size to 7-dayold nonvernalized seedlings and were placed under glasshouse conditions for 7 days before harvesting. Barley cultivars of known genotype are described in the Barley Germplasm Database (www.shigen.nig.ac.jp/barley/Barley.html). Summaries of information pertaining to the wheat and barley cultivars used in this study are included in Tables 2 and 3, which are published as supporting information on the PNAS web site, www.pnas.org.
Genomic DNA Extraction and Southern Blots. Genomic DNA was extracted by using cetyltrimethylammonium bromide and purified on a cesium chloride gradient (14). DNA digestion, electrophoresis, blotting, and hybridization were performed by using standard methods (14). The 3′ untranslated region of WAP1 was used for a gene specific probe.
RNA Extraction and Gel Blot Analysis. Total RNA was extracted by using the RNA Extraction protocol of Chang et al. (15). Twenty micrograms of total RNA was separated in formaldehyde gels and then blotted to Hybond-N membrane as described in Ausubel et al. (14). RNA blots were hybridized with riboprobes and washed by using the protocol of Dolferus et al. (16). A WAP1/BM5 specific riboprobe was amplified from the 3′ end of the WAP1 cDNA clone (BE431095) by using the sequences CTGAAACATATCAGATCCAGG and TATACACATCGCTGCAGCTTGC. Riboprobes were synthesized by using the Promega T7 in vitro transcription kit following the protocol provided. Blots were exposed to autoradiograph film (Fuji) for varying lengths of time, dependent on signal intensity.
RT-PCR Analysis. A oligo(T) primer (T18[G/C/A]) was used to prime first-strand cDNA synthesis from 5 μg of total RNA by using the SuperScript reverse transcriptase enzyme (Invitrogen) according to the manufacturer's instructions. For PCR, primers were designed against the untranslated regions of cDNA sequences of wheat MADS box genes (Table 4, which is published as supporting information on the PNAS web site). Negative control reactions that lacked template or reverse transcriptase enzyme and a positive control reaction using genomic DNA as template were run simultaneously for each primer and template combination. Amplification of a fragment from the wheat Ubiquitin gene (X56601) was used to compare loading for each sample. Reactions were run for 30 cycles of 20 s at 94°C, 20 s at 60°C, and 30 s at 72°C, by using Taq polymerase (Fisher Biotechnology) in the polymerase buffer supplied by the manufacturer with 2.5 mM magnesium chloride. Fragments were visualized by agarose gel electrophoresis. For the BM5 gene, the same reaction conditions were used with the primer pair TGAAGCTCAGAAATGGATTCG and TATGAGCGCTACTCTTATGC, which does not amplify a product from genomic DNA under these conditions but amplifies the predicted fragment size from first-strand cDNA. The resulting fragments were hybridized at high stringency with a BM5-specific DNA probe to demonstrate specificity of the PCR.
Results
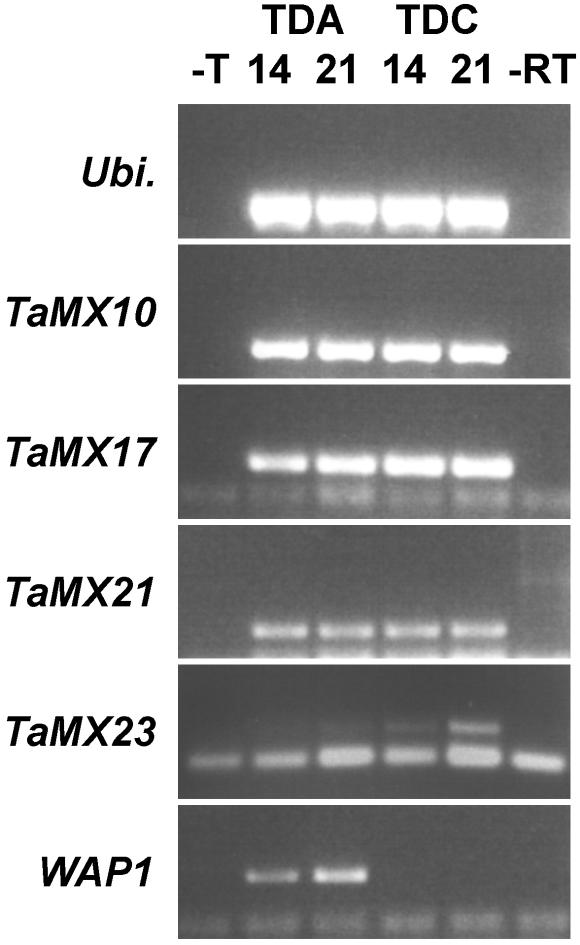
An RT-PCR assay was used to compare the expression levels of 25 different MADS box genes (Fig. 8, which is published as supporting information on the PNAS web site) between two near-isogenic wheat lines that differ for spring or winter habit (4). The Triple Dirk C line (TDC) is a winter wheat (vrn-A1/ vrn-B1/vrn-D1) that flowers late when not vernalized, whereas the Triple Dirk A line (TDA) has Vrn-A1 and Vrn-B1 spring alleles (Vrn-A1/Vrn-B1/vrn-D1) and flowers early without vernalization (17). Ten MADS box genes were expressed in vegetative (2-week-old) nonvernalized wheat plants before the floral transition. Differences in gene expression levels were observed between the spring and winter genotypes for two of these genes (Table 1). Strong expression of the WAP1 gene (18) was detected in the spring cultivar TDA, but no expression of WAP1 was detected in the winter line TDC (Fig. 1). The second MADS box sequence, TaMX23, was more strongly expressed in TDC (winter) than in TDA (spring) (Fig. 1).
Table 1. Summary of RT-PCR results.
| Sequence | EST | Winter | Spring |
|---|---|---|---|
| WAP1 | AB007504* | ++ | |
| TaMADS12 | AB007505* | ||
| TaMADS51 | AB007506* | ||
| TaMX1 | BJ316096 | ||
| TaMX2 | BE423660 | ||
| TaMX3 | BJ208073 | ||
| TaMX4 | BJ233536 | ++ | ++ |
| TaMX5 | BM137344 | ||
| TaMX6 | BF483385 | ||
| TaMX8 | BE445262 | ||
| TaMX9 | BE499520 | ++ | ++ |
| TaMX10 | BF484557 | ++ | ++ |
| TaMX11† | BJ245749 | ||
| TaMX12 | BJ245437 | ||
| TaMX13 | BJ224514 | ||
| TaMX14 | BJ314788 | ++ | ++ |
| TaMX15 | BJ247111 | ||
| TaMX16 | BQ806253 | ++ | ++ |
| TaMX17 | BF483481 | ++ | ++ |
| TaMX18 | BJ245890 | ||
| TaMX19 | BJ233463 | ||
| TaMX20 | AL823256 | ++ | ++ |
| TaMX21 | BJ258612 | ++ | ++ |
| TaMX22 | BE445505 | ||
| TaMX23 | BJ258117 | ++ | + |
The source ESTs for the MADS box sequences and whether the sequence was expressed in winter or spring plants. ++, strong amplification; +, weak amplification.
Previously characterized gene
Amplification was unsuccessful with TaMX11 primer set
Fig. 1.
RT-PCR analysis of wheat MADS box gene expression in 2-week-old plants. Shown are RT-PCR results for the positive control gene Ubiquitin and four of the MADS box genes expressed in wheat plants before floral transition, TaMX10, TaMX17, TaMX21, TaMX23, and WAP1. Control samples were water as template (-T) or without addition reverse transcriptase (-RT). Test samples included first-strand cDNA from 14- or 21-day old TDA (a strong spring wheat with the genotype Vrn-A1/Vrn-B1/vrn-d1) and 14- or 21-day-old TDC (a vernalization-responsive winter wheat, genotype vrn-a1/vrn-b1/vrn-d1). None of the plants were vernalized. The products were all ≈150 bp and match the sizes predicted on the basis of sequence.
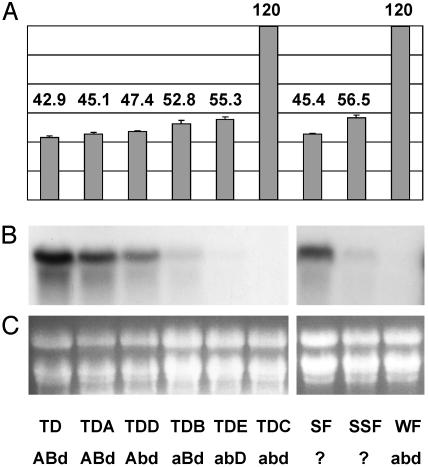
The near-isogenic Triple Dirk cultivar series generated by Pugsley (4, 17) includes winter, semispring, and spring lines, all of known genotype for the Vrn-1 locus on each of the three genomes. RNA gel blot analysis was used to examine the expression of WAP1 across the Triple Dirk series. In seedlings that were not vernalized, the level of WAP1 expression was found to be related to the effect of the Vrn-1 genotype on heading date (Fig. 2). High WAP1 expression was observed in the spring Triple Dirk lines TD, TDA, and TDD, which carry the strong Vrn-A1 spring allele and flower early. Moderate expression was detected in the semispring lines TDB and TDE, which carry either of the Vrn-B1 or Vrn-D1 spring alleles. No expression of WAP1 was detected in the winter line TDC, which flowers late.
Fig. 2.
Expression of WAP1 compared with head emergence time for plants from the Triple Dirk and Festival cultivar series. (A) Days until head emergence for nonvernalized plants from each of the following lines: Triple Dirk (TD), Triple Dirk A (TDA), Triple Dirk D (TDD), Triple Dirk B (TDB), Triple Dirk E (TDE), Triple Dirk C* (TDC), Spring Festival (SF), Semispring Festival (SSF), and Winter Festival* (WF). The winter lines (*) did not show head emergence within the 120-day time period assayed. (B) Expression of WAP1 in 2-week-old, nonvernalized plants from each line. WAP1 expression was assayed by high stringency hybridization of RNA gel blots with a WAP1-specific riboprobe. Ethidium bromide staining of ribosomal RNA is shown in C as a loading comparison. The genotype of each line is included underneath each line with uppercase letters indicating dominant spring Vrn-1 alleles. Lowercase letters represent recessive winter vrn-1 alleles.
Analysis of WAP1 gene expression was extended to other wheat cultivars. Spring, semispring, and winter lines of the cultivar Festival have been defined on the basis of heading dates. The exact Vrn-1 genotypes of these lines are unknown, although, on the basis of the winter habit phenotype, the winter Festival line must lack spring Vrn-1 alleles and therefore is likely to have the genotype vrn-A1/vrn-B1/vrn-D1. The pattern of WAP1 expression in the different Festival lines parallels that observed in the Triple Dirk series (Fig. 2). High expression of WAP1 was found in the spring Festival line, which flowers early without vernalization, moderate expression of WAP1 was detected in semispring Festival and no WAP1 expression was detected in winter Festival (Fig. 2). Furthermore, no expression of WAP1 was detected in the winter wheat Cheyenne (Fig. 3) or in three lines of the winter wheat Black Hull (Fig. 3).
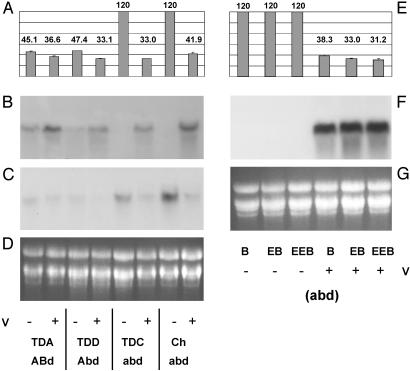
Fig. 3.
Expression of WAP1 compared with head emergence dates for vernalized and nonvernalized spring and winter wheat cultivars. (A–D) Data from nonvernalized and vernalized plants from the lines TDA, TDD, Triple Dirk C (TDC)*, and Cheyenne* (CH, a winter wheat). (E–G) Data from nonvernalized plants from three lines of the winter wheat Blackhull [Blackhull (B)*, Early Blackhull (EB)*, and Extra Early Blackhull (EEB)*], and then vernalized plants from each of the same three lines. Days until heading are shown for each combination of line and treatment in A and E. This does not include the 60-day vernalization period for the vernalized plants. Without vernalization, winter lines (*) showed no head emergence within the 120-day time period assayed. RNA gel blot analysis of WAP1 expression in 2-week-old plants from each combination of line and treatment is shown in B and F.(C) Expression of TaMX23 in 2-week-old vernalized or nonvernalized plants from the Triple Dirk series and winter wheat Cheyenne, analyzed by hybridization of RNA blots with a TaMX23 specific riboprobe. Ethidium bromide staining of ribosomal RNA is shown in D and G as a loading comparison. The genotype of each line is included (a capital letter indicates dominant spring Vrn-1 allele from the respective genome), as is vernalization status by + for vernalized and - for nonvernalized.
Expression of WAP1 was examined in spring, semispring, and winter wheats that had been vernalized for 60 days. WAP1 expression was strongly induced in winter lines by vernalization treatment (Fig. 3). Likewise, expression of WAP1 was induced in semispring lines and some induction was observed in the vernalized spring lines. After vernalization, the heading dates of the winter, semispring, and spring wheats examined were all similar (≈35 days after vernalization). Thus, the induction of WAP1 expression in winter and semispring wheats correlates with the promotion of flowering.
The second MADS box gene TaMX23, which was found by RT-PCR to be also differentially expressed between TDA and TDC, was repressed by vernalization in the TDC and Cheyenne winter wheats (Fig. 3). Whereas the expression pattern of WAP1 matches that which would be expected for a dominant promoter of flowering and is consistent across a number of wheat cultivars, the effect of vernalization on TaMX23 expression was found to vary depending on which cultivar was examined. Furthermore, the rice homologue of TaMX23 is located on chromosome 1 (marker S10304) in a region not known to be syntenic with any vernalization response genes in winter cereals. For these reasons, the role of TaMX23 in the vernalization response of cereals was not characterized further in this study.
The role of WAP1-like genes in controlling the vernalization response of cereals was further examined in the important (diploid) cereal crop barley. The barley MADS box gene BM5 shares a high degree of predicted amino acid similarity (95%) with WAP1 (19). Furthermore, during plant development, BM5 has a gene expression pattern similar to WAP1 and is likely to be the barley orthologue of WAP1 (18, 19). No expression of BM5 was detected in six nonvernalized winter barley cultivars. Vernalization treatment strongly induced BM5 expression in all six lines (Fig. 4, lanes 7–12 and 19–24). Thus, BM5 shows a response to vernalization similar to WAP1 and TmAP1.
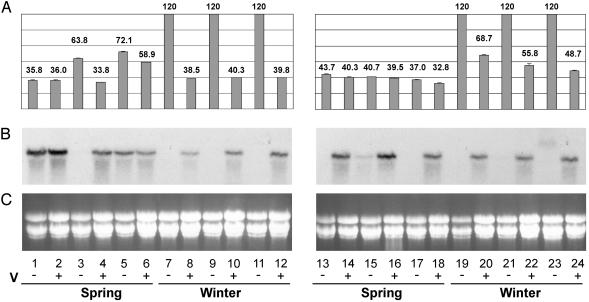
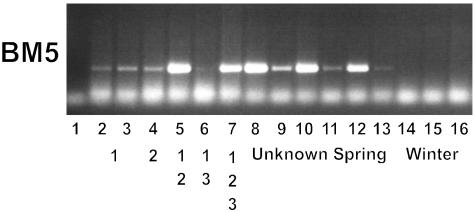
Fig. 4.
Expression of BM5 in vernalized and nonvernalized seedlings of 12 barley cultivars. Days until heading for nonvernalized or vernalized plants are shown in A. Lane 1, Morgenrot; lane 2, vernalized Morgenrot (+V); lane 3, Randolph; lane 4, Randolph +V; lane 5, Malta; lane 6, Malta +V; lane 7, Sonja*; lane 8, Sonja +V; lane 9, Hudson*; lane 10, Hudson +V; lane 11, Mirra*; lane 12, Mirra +V; lane 13, Perga; lane 14, Perga +V; lane 15, Dunja; lane 16, Dunja +V; lane 17, Will; lane 18, Will +V; lane 19, Hydra*; lane 20 Hydra +V; lane 21, Bollo*; lane 22, Bollo +V; lane 23 Igri*; lane 24, Igri +V. This does not include the 60-day vernalization period for the vernalized plants. Winter lines (*) did not show head emergence within the 120-day time period assayed when not vernalized. (B) Expression of BM5 determined by RNA gel blot analysis for 2-week-old plants from the same combinations of lines and treatments. Ethidium bromide staining of ribosomal RNA is shown in C as a loading comparison. Vernalization status (-, nonvernalized; +, vernalized) and spring habit phenotype are shown beneath each cultivar/treatment combination.
Spring barley lines of known genotype for the Vrn-H1, Vrn-H2, and Vrn-H3 genes were used to determine whether expression of BM5 correlates with the spring habit phenotype of barley. All five genotypic combinations for spring habit that occur naturally in spring barleys were assayed for BM5 expression (10). Expression of BM5 was detected in lines that carry spring habit alleles at both the Vrn-H1 and Vrn-H2 loci (Vrn-H1/-, vrn-H2/vrn-H2) (Fig. 5), but not in spring barleys that carry spring habit alleles at only one of Vrn-H1 or Vrn-H2. By using the more sensitive RT-PCR assay of BM5 gene expression, we were able to detect expression of BM5 in all spring barleys examined, including those lines that carry spring habit alleles at either of the Vrn-H1 or Vrn-H2 genes (Fig. 6). We found one winter barley line (Bollo) that also showed a low level of BM5 activity detectable by RT-PCR analysis. We are not able to explain why this winter line, in contrast to all of the other winter lines we have examined, has some expression of BM5.
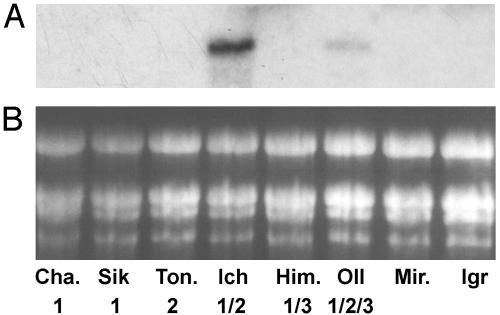
Fig. 5.
Expression of BM5 in seedlings of barleys of known spring habit genotype. Expression of BM5 in nonvernalized barley cultivars was determined by hybridizing RNA gel blots with a BM5-specific riboprobe. Cha., Chame 11 (Vrn-H1 spring); Sik, Sikangense Type 15 (Vrn-H1 spring); Ton., Tongyeong Covered (vrn-H2 spring); Ich, Icheon Naked (Vrn-H1/vrn-H2 spring); Him., Himalayense Type 5 (Vrn-H1/Vrn-H3 spring); Oll, Olli (Vrn-H1/ vrn-H2/Vrn-H3 spring); Mir., Mirra (winter); Igr, Igri (winter). Numbers below each lane indicate locus or loci where spring alleles are present: 1, Vrn-H1; 2, vrn-H2;or3, Vrn-H3. Ethidium bromide staining of ribosomal RNA is shown in B as a loading comparison.
Fig. 6.
RT-PCR assay of BM5 expression in spring and winter barley lines. RT-PCR was performed with 12 spring and 3 winter barley cultivars, and the resulting fragments were separated by gel electrophoresis. Lane 1, No template; lane 2, Chame 11; lane 3, Sikangense Type 15; lane 4, Tongyeong Covered; lane 5, Icheon Naked; lane 6, Himalayense Type 5; lane 7, Olli; lane 8, Morgenrot; lane 9, Randolph; lane 10, Malta; lane 11, Perga; lane 12, Dunja; lane 13, Will; lane 14, Hudson; lane 15, Mirra; lane 16, Hydra. For spring barleys of known genotype, the presence of spring alleles is shown beneath each sample as 1 (Vrn-H1), 2 (vrn-H2), or 3 (Vrn-H3).
In six spring barley lines of unknown genotype, Northern analyses showed two (presumably containing spring alleles at both Vrn-H1 and Vrn-H2) to have high expression levels of the BM5 gene, equivalent to vernalized winter lines, and one with a moderate level of expression of the gene (Fig. 4, lanes 1, 4, and 15). In the other three lines, the Northern analyses did not detect any expression of BM5 (Fig. 4, lanes 3, 13, and 17), but RT-PCR showed a low level of activity (Fig. 6). After vernalization treatment, BM5 transcript levels in all six lines were equivalent to those of vernalized winter lines (Fig. 4, lanes 4, 6, 8, 14, 16, and 18).
Our results suggest that Vrn-H1 and Vrn-H2 interact to regulate BM5 expression. A spring genotype at either the Vrn-H1 or Vrn-H2 locus leads to a low level of BM5 expression detectable by RT-PCR. This low level of BM5 expression is sufficient to cause an early flowering phenotype. The combination of spring alleles at both loci (Vrn-H1/-, vrn-H2/vrn-H2) has a synergistic effect on BM5 gene expression, leading to greatly increased BM5 transcript levels that are then detectable by both Northern analysis and by RT-PCR.
Discussion
Our results show that the WAP1 gene is likely to correspond to the Vrn-1 locus of bread wheat. The WAP1 genes are located on chromosomes 5A, 5B, and 5D in hexaploid wheats (Fig. 9, which is published as supporting information on the PNAS web site), the same chromosomes as Vrn-A1, Vrn-B1, and Vrn-D1, and by synteny with rice and T. monococcum, are most probably collocated with the Vrn-1 loci. In nonvernalized plants, WAP1 is strongly expressed in spring wheats, moderately expressed in semispring wheats, and is not expressed in winter wheats. Furthermore, the nearest rice homologue to WAP1, RAP1b (OsMADS14), causes an extreme early-flowering phenotype when overexpressed in rice (20), suggesting that WAP1 expression promotes flowering in this cereal. These data are consistent with the results of Yan et al. (13), which led them to suggest that the TmAP1 gene encodes the VrnAm1 gene of T. monococcum.
The Triple Dirk set of near-isogenic Vrn-1 genotypes in bread wheat enabled us to examine the levels of WAP1 expression associated with the Vrn-1 alleles of each of the three genomes of wheat. The A genome locus, Vrn-A1, generates the strongest spring phenotype, and plants carrying the Vrn-A1 allele show the strongest expression of WAP1 in the absence of vernalization. Spring alleles of Vrn-B1 and Vrn-D1, on the B and D genomes, respectively, produce semispring phenotypes and low levels of WAP1 gene expression. Comparison of WAP1 expression between spring and semispring lines shows that there is not a linear correlation between the level of WAP1 activity and flowering times; TDA has strong expression of WAP1 and a flowering time (heading date) of 45 days; the two semispring lines, TDB and TDE, have much lower WAP1 expression levels but a flowering time of ≈54 days. It seems that even a low level of WAP1 expression can elicit early flowering. Increases in WAP1 expression above the effective threshold level seem to have a limited effect on flowering time. This is shown in semispring wheats, where vernalization causes a large increase in WAP1 transcript level but has only a minor effect on heading date.
BM5, in barley, shows a similar expression pattern to TmAP1 and WAP1. BM5 is generally not expressed in nonvernalized winter barleys, but it is induced by vernalization. As is the case for TmAP1, BM5 is expressed in spring barleys that have not been exposed to vernalization. Barley lines carrying dominant spring Vrn-H1 alleles have a low level of BM5 expression. Likewise, barleys homozygous for the recessive vrn-H2 spring allele have a low level of BM5 expression. Cultivars with the spring genotype at both the Vrn-H1 and Vrn-H2 loci have a higher level of BM5 transcript, similar to that found in vernalized plants.
Small deletions occur in the promoter of the TmAP1 genes from spring lines of T. monococcum that carry the dominant Vrn-Am1 for spring habit (13). Spring wheats and Vrn-H1 spring barley lines may also carry alterations in the promoter of WAP1 and BM5. Yan et al. (13) suggested that the small deletions found in the promoter of TmAP1 from spring T. monococcum accessions render the promoter of TmAP1 immune to the action of the Vrn-Am2 repressor. This seems not to be the case in barley. The level of BM5 expression is much higher in plants with spring genotype at both the Vrn-H1 and Vrn-H2 loci than in those that carry the Vrn-H1 (or vrn-H2) spring genotype alone.
We suggest that BM5 expression may be controlled by both an activator and a repressor, which interact to determine the response to vernalization. In winter barleys that have not been vernalized, the BM5 promoter activity is low and is repressed by the Vrn-H2 protein. We propose that vernalization treatment removes repression by Vrn-H2 and further enhances the activity of the BM5 promoter through the induction of a transcription factor that promotes BM5 expression. A MADS box gene similar to SOC1 or AGL24, which are vernalization responsive and promote flowering in Arabidopsis (21, 22), could be the activator of the BM5 promoter in vernalized barley plants. The combined effect of these two regulatory systems would maintain a low level of BM5 expression in unvernalized plants, and ensure strong induction of BM5 by vernalization (Fig. 7). In Vrn-H1 spring barleys, the BM5 promoter would be active, but the action of the Vrn-H2 repressor would repress this activity and the result would be a low level of BM5 transcription. In a vrn-H2 spring barley, the Vrn-H2 repressor would not be active, but the activity of the BM5 promoter would remain low in the absence of vernalization, resulting in a low level of BM5 expression. The presence of spring alleles at both loci (Vrn-H1/-, vrn-H2/vrn-H2) would result in greatly enhanced expression of BM5, relative to barleys with Vrn-H1 or vrn-H2 genotype alone. In the Vrn-H1/vrn-H2 spring lines, the promoter of BM5 would be both active and derepressed. It has been suggested that, in T. monococcum, the Vrn-2 repressor is a MADS box gene (13).
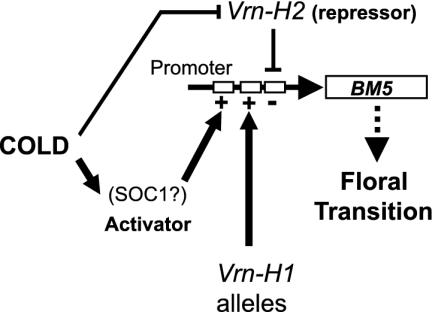
Fig. 7.
A model for the regulation of BM5 expression by vernalization and spring habit genotypes. Vernalization controls two genetic systems that regulate BM5 transcription through the BM5 promoter. One represses BM5 expression in plants that have not been vernalized. Extended cold treatment counteracts this repression and activates a second regulatory mechanism that activates BM5 transcription. Known flowering time genes such as the barley equivalent of SOC1 are likely to be involved in this activation pathway. Recessive spring habit alleles of the Vrn-H2 locus abolish the repression pathway and allow some expression of BM5. Dominant alleles of Vrn-H1 activate expression regardless of vernalization status, but do not completely counteract repression, leading to some BM5 activity. Combining both spring habit genotypes results in BM5 expression levels that approach those in vernalized plants, through simultaneous activation and de-repression of the BM5 promoter.
Experiments in Lolium temulentum offer further evidence for the involvement of AP1-like genes in the floral transition in all grasses. Long days promote flowering in L. temulentum, and an AP1-like MADS box gene, LtMADs1, closely related in sequence to BM5 and WAP, is one of the first genes to be induced after exposure to long day conditions (23). This raises the possibility that WAP1-like genes could also promote flowering in response to photoperiod in wheat and barley.
In cereals, two MADS box proteins with opposite effects appear to be involved in the regulation of the vernalization response. This pathway resembles the vernalization response pathway of Arabidopsis, where SOC1, a MADS box gene that promotes flowering, is repressed by FLC, also a MADS box gene, in plants that have not been vernalized (21, 24, 25). No FLC-like genes have been identified in cereals, making it unlikely that Vrn-2 is closely related to FLC. There is no evidence that AP1-like genes mediate the vernalization response in Arabidopsis, but overexpression of AP1 does result in early flowering (26). The vernalization response may have evolved separately in the ancestors of the cereals (monocots) and the Brassicaceae (dicots) through the recruitment of different MADS box transcription factor genes into a cold regulated switch that promotes flowering in vernalized plants.
Supplementary Material
Acknowledgments
We thank Janice Norman for excellent technical assistance and Jean Finnegan, Million Tadege, Candice Sheldon, Richard Richards, and Carol Andersson for helpful suggestions. We also thank Mohammad Shariflou for providing a full-length WAP1 cDNA clone (BE431095) and the Australian Winter Cereal Collection for providing the wheat and barley cultivars used in this study. B.T. was supported by a Commonwealth Scientific and Industrial Research Organisation Postdoctoral Fellowship.
Abbreviations: TDA, Triple Dirk A; TDC, Triple Dirk C.
References
- 1.Law, C. N. (2000) Flowering Newslett. 29 22-25. [Google Scholar]
- 2.Stelmakh, A. F. (1998) Euphytica 100 359-369. [Google Scholar]
- 3.Snape, J. W., Butterworth, K., Whitechurch, E. & Worland, A. J. (2001) Euphytica 119 185-190. [Google Scholar]
- 4.Pugsley, A. T. (1971) Aus. J. Agric. Res. 22 21-31. [Google Scholar]
- 5.Halloran, G. M. (1975) Can. J. Genet. Cytol. 17 365-373. [Google Scholar]
- 6.Law, C. N., Worland, A. J. & Giorgi, B. (1976) Heredity 36 49-58. [Google Scholar]
- 7.Galiba, G., Quarrie, S. A., Sutka, J., Morounov, A. & Snape, J. W. (1995) Theor. Appl. Genet. 90 1174-1179. [DOI] [PubMed] [Google Scholar]
- 8.Sarma, R. N., Gill, B. S., Sasaki, T., Galiba, G., Sutka, J., Laurie, D. A. & Snape, J. W. (1998) Theor. Appl. Genet. 97 103-109. [Google Scholar]
- 9.Dubcovsky, J., Lijavetzky, D., Appendino, L. & Tranquilli, G. (1998) Theor. Appl. Genet. 97 968-975. [Google Scholar]
- 10.Takahashi, R. & Yasuda, S. (1971) in Proceedings of the Second International Barley Genetics Symposium, Pullman, 1969, ed. Nilan, R. A. (Washington State Univ. Press, Pullman), pp. 388-408.
- 11.Laurie, D. A., Pratchett, N., Bezant, J. H. & Snape, J. W. (1995) Genome 38 575-585. [DOI] [PubMed] [Google Scholar]
- 12.Tranquilli, G. & Dubcovsky, J. (2000) J. Hered. 91 304-306. [DOI] [PubMed] [Google Scholar]
- 13.Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T. & Dubcovsky, J. (2003) Proc. Natl. Acad. Sci. USA 100 6263-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ausubel, F. M., Brenton, R., Kingston, R. E., Moore, D. D., Siedman, J. G., Smyth, J. A. & Struhl, K. (1994) Current Protocols in Molecular Biology (Wiley, New York).
- 15.Chang, S., Puryear, J. & Cairney, K. (1993) Plant Mol. Biol. Rep. 11 113-116. [Google Scholar]
- 16.Dolferus, R., Jacobs, M., Peacock, W. J. & Dennis, E. S. (1994) Plant Physiol. 105 1075-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pugsley, A. T. (1972) Euphytica 21 547-552. [Google Scholar]
- 18.Murai, K., Takumi, S., Koga, H. & Ogihara, Y. (2002) Plant J. 29 169-181. [DOI] [PubMed] [Google Scholar]
- 19.Schmitz, J., Franzen, R., Ngyuen, T. H., Garcia-Maroto, F., Pozzi, C., Salamini, F. & Rohde, W. (2000) Plant Mol. Biol. 42 899-913. [DOI] [PubMed] [Google Scholar]
- 20.Jeon, J. S., Lee, S., Jung, K. H., Yang, W. S., Yi, G. H., Oh, B. G. & An, G. (2000) Mol. Breeding 6 581-592. [Google Scholar]
- 21.Lee, H., Suh, S. S., Park, E., Cho, E., Ahn, J. H., Kim, S. G., Lee, J. S., Kwon, Y. M. & Lee, I. (2000) Genes Dev. 14 2366-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michaels, S. D., Ditta, G., Gustafson-Brown, C., Pelaz, S., Yanofsky, M. & Amasino, R. M. (2003) Plant J. 33 867-874. [DOI] [PubMed] [Google Scholar]
- 23.Gocal, G. F., King, R. W., Blundell, C. A., Schwartz, O. M., Andersen, C. H. & Weigel, D. (2001) Plant Physiol. 125 1788-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheldon, C. C., Burn, J. E., Perez, P. P., Metzger, J., Edwards, J. A., Peacock, W. J. & Dennis, E. S. (1999) Plant Cell 11 445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaels, S. D. & Amasino, R. M. (1999) Plant Cell 11 949-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandel, M. A. & Yanofsky, M. F. (1995) Nature 377 522-524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.