Abstract
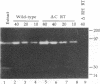
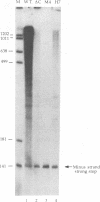
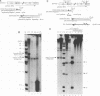
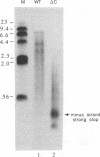
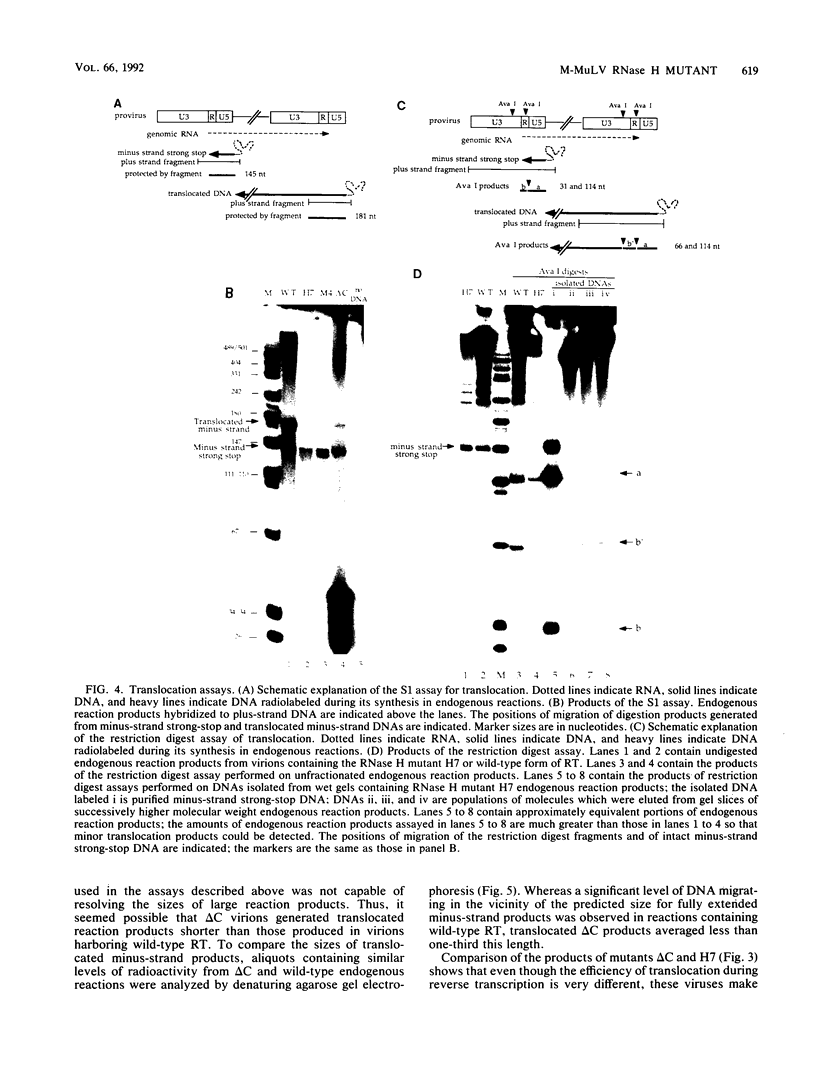
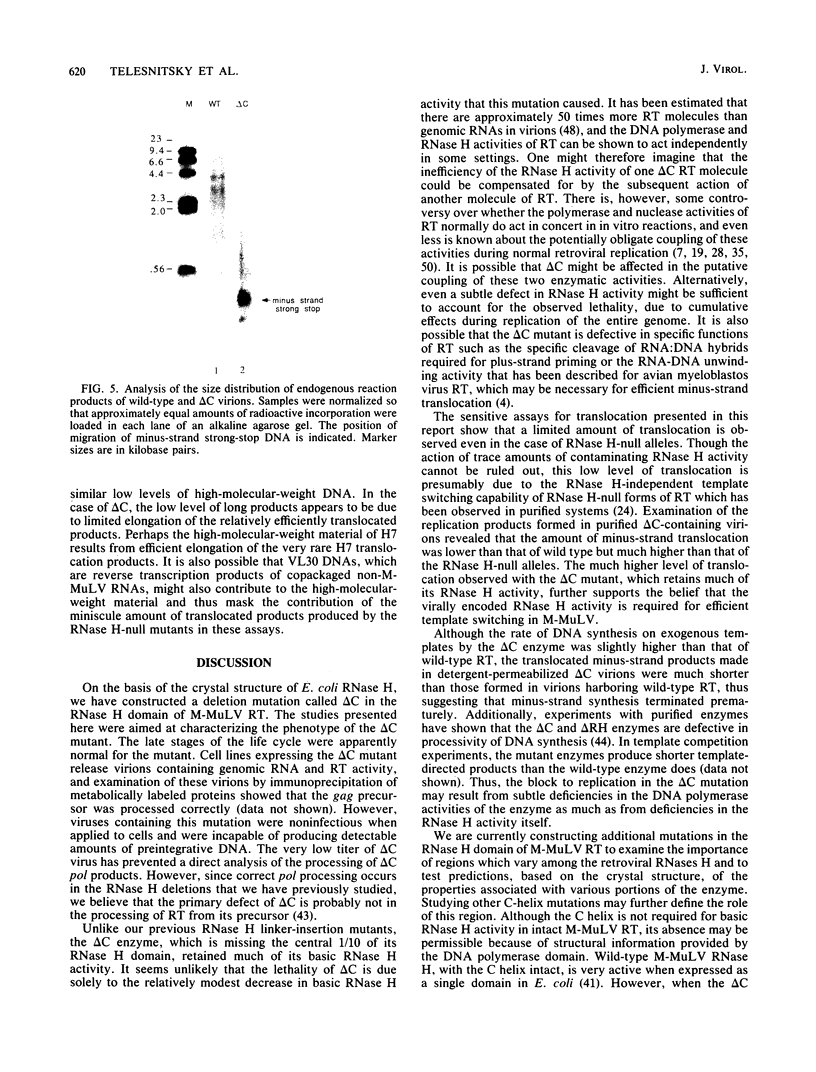
We have studied a mutant Moloney murine leukemia virus with a deletion in reverse transcriptase (RT) which is predicted to make its RNase H domain resemble structurally that of human immunodeficiency virus RT. This deletion was based on improved RNase H homology alignments made possible by the recently solved three-dimensional structure for Escherichia coli RNase H. This mutant Moloney murine leukemia virus RT was fully active in the oligo(dT)-poly(rA) DNA polymerase assay and retained nearly all of wild-type RT's RNase H activity in an in situ RNase H gel assay. However, proviruses reconstructed to include this deletion were noninfectious. Minus-strand strong-stop DNA was made by the deletion mutant, but the amount of minus-strand translocation was intermediate to the very low level measured with RNase H-null virions and the high level seen with wild-type RT. The average length of translocated minus-strand DNA was shorter for the deletion mutant than for wild type, suggesting that mutations in the RNase H domain of RT also affect DNA polymerase activity.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970 Jun 27;226(5252):1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- Colicelli J., Goff S. P. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988 Jan 5;199(1):47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- Collett M. S., Leis J. P., Smith M. S., Faras A. J. Unwinding-like activity associated with avian retrovirus RNA-directed DNA polymerase. J Virol. 1978 May;26(2):498–509. doi: 10.1128/jvi.26.2.498-509.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland T. D., Gerard G. F., Hixson C. W., Oroszlan S. Amino- and carboxyl-terminal sequence of Moloney murine leukemia virus reverse transcriptase. Virology. 1985 Jun;143(2):676–679. doi: 10.1016/0042-6822(85)90411-8. [DOI] [PubMed] [Google Scholar]
- Davies J. F., 2nd, Hostomska Z., Hostomsky Z., Jordan S. R., Matthews D. A. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991 Apr 5;252(5002):88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- DeStefano J. J., Buiser R. G., Mallaber L. M., Myers T. W., Bambara R. A., Fay P. J. Polymerization and RNase H activities of the reverse transcriptases from avian myeloblastosis, human immunodeficiency, and Moloney murine leukemia viruses are functionally uncoupled. J Biol Chem. 1991 Apr 25;266(12):7423–7431. [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989 Mar;64(1):1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- Gilboa E., Mitra S. W., Goff S., Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979 Sep;18(1):93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- Goff S. P. Retroviral reverse transcriptase: synthesis, structure, and function. J Acquir Immune Defic Syndr. 1990;3(8):817–831. [PubMed] [Google Scholar]
- Goff S., Traktman P., Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981 Apr;38(1):239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hansen J., Schulze T., Mellert W., Moelling K. Identification and characterization of HIV-specific RNase H by monoclonal antibody. EMBO J. 1988 Jan;7(1):239–243. doi: 10.1002/j.1460-2075.1988.tb02805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hizi A., Hughes S. H., Shaharabany M. Mutational analysis of the ribonuclease H activity of human immunodeficiency virus 1 reverse transcriptase. Virology. 1990 Apr;175(2):575–580. doi: 10.1016/0042-6822(90)90444-v. [DOI] [PubMed] [Google Scholar]
- Hizi A., McGill C., Hughes S. H. Expression of soluble, enzymatically active, human immunodeficiency virus reverse transcriptase in Escherichia coli and analysis of mutants. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1218–1222. doi: 10.1073/pnas.85.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostomsky Z., Hostomska Z., Hudson G. O., Moomaw E. W., Nodes B. R. Reconstitution in vitro of RNase H activity by using purified N-terminal and C-terminal domains of human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1148–1152. doi: 10.1073/pnas.88.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H. E., McCoy J. M., Seehra J. S., Richardson C. C. Human immunodeficiency virus 1 reverse transcriptase. Template binding, processivity, strand displacement synthesis, and template switching. J Biol Chem. 1989 Mar 15;264(8):4669–4678. [PubMed] [Google Scholar]
- Johnson M. S., McClure M. A., Feng D. F., Gray J., Doolittle R. F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayanagi K., Miyagawa M., Matsushima M., Ishikawa M., Kanaya S., Ikehara M., Matsuzaki T., Morikawa K. Three-dimensional structure of ribonuclease H from E. coli. Nature. 1990 Sep 20;347(6290):306–309. doi: 10.1038/347306a0. [DOI] [PubMed] [Google Scholar]
- Kotewicz M. L., Sampson C. M., D'Alessio J. M., Gerard G. F. Isolation of cloned Moloney murine leukemia virus reverse transcriptase lacking ribonuclease H activity. Nucleic Acids Res. 1988 Jan 11;16(1):265–277. doi: 10.1093/nar/16.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoote M. M., Coligan J. E., Folks T. M., Fauci A. S., Martin M. A., Venkatesan S. Structural characterization of reverse transcriptase and endonuclease polypeptides of the acquired immunodeficiency syndrome retrovirus. J Virol. 1986 Nov;60(2):771–775. doi: 10.1128/jvi.60.2.771-775.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G. X., Taylor J. Template switching by reverse transcriptase during DNA synthesis. J Virol. 1990 Sep;64(9):4321–4328. doi: 10.1128/jvi.64.9.4321-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure M. A., Johnson M. S., Feng D. F., Doolittle R. F. Sequence comparisons of retroviral proteins: relative rates of change and general phylogeny. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2469–2473. doi: 10.1073/pnas.85.8.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchan J. H., Pagano J. S. Enchancement of the infectivity of simian virus 40 deoxyribonucleic acid with diethylaminoethyl-dextran. J Natl Cancer Inst. 1968 Aug;41(2):351–357. [PubMed] [Google Scholar]
- Mölling K., Bolognesi D. P., Bauer H., Büsen W., Plassmann H. W., Hausen P. Association of viral reverse transcriptase with an enzyme degrading the RNA moiety of RNA-DNA hybrids. Nat New Biol. 1971 Dec 22;234(51):240–243. doi: 10.1038/newbio234240a0. [DOI] [PubMed] [Google Scholar]
- Oyama F., Kikuchi R., Crouch R. J., Uchida T. Intrinsic properties of reverse transcriptase in reverse transcription. Associated RNase H is essentially regarded as an endonuclease. J Biol Chem. 1989 Nov 5;264(31):18808–18817. [PubMed] [Google Scholar]
- Prasad V. R., Goff S. P. Linker insertion mutagenesis of the human immunodeficiency virus reverse transcriptase expressed in bacteria: definition of the minimal polymerase domain. Proc Natl Acad Sci U S A. 1989 May;86(9):3104–3108. doi: 10.1073/pnas.86.9.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repaske R., Hartley J. W., Kavlick M. F., O'Neill R. R., Austin J. B. Inhibition of RNase H activity and viral replication by single mutations in the 3' region of Moloney murine leukemia virus reverse transcriptase. J Virol. 1989 Mar;63(3):1460–1464. doi: 10.1128/jvi.63.3.1460-1464.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M. J., Tanese N., Goff S. P. Purification and characterization of murine retroviral reverse transcriptase expressed in Escherichia coli. J Biol Chem. 1985 Aug 5;260(16):9326–9335. [PubMed] [Google Scholar]
- Rowe W. P., Pugh W. E., Hartley J. W. Plaque assay techniques for murine leukemia viruses. Virology. 1970 Dec;42(4):1136–1139. doi: 10.1016/0042-6822(70)90362-4. [DOI] [PubMed] [Google Scholar]
- Schatz O., Cromme F. V., Grüninger-Leitch F., Le Grice S. F. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 1989 Nov 6;257(2):311–314. doi: 10.1016/0014-5793(89)81559-5. [DOI] [PubMed] [Google Scholar]
- Schatz O., Mous J., Le Grice S. F. HIV-1 RT-associated ribonuclease H displays both endonuclease and 3'----5' exonuclease activity. EMBO J. 1990 Apr;9(4):1171–1176. doi: 10.1002/j.1460-2075.1990.tb08224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzberg P., Colicelli J., Goff S. P. Construction and analysis of deletion mutations in the pol gene of Moloney murine leukemia virus: a new viral function required for productive infection. Cell. 1984 Jul;37(3):1043–1052. doi: 10.1016/0092-8674(84)90439-2. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Starnes M. C., Cheng Y. C. Human immunodeficiency virus reverse transcriptase-associated RNase H activity. J Biol Chem. 1989 Apr 25;264(12):7073–7077. [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Simon M. N., Dunn J. J. Genetic and physical mapping in the early region of bacteriophage T7 DNA. J Mol Biol. 1979 Dec 25;135(4):917–937. doi: 10.1016/0022-2836(79)90520-5. [DOI] [PubMed] [Google Scholar]
- Tanese N., Goff S. P. Domain structure of the Moloney murine leukemia virus reverse transcriptase: mutational analysis and separate expression of the DNA polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1777–1781. doi: 10.1073/pnas.85.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanese N., Roth M. J., Goff S. P. Analysis of retroviral pol gene products with antisera raised against fusion proteins produced in Escherichia coli. J Virol. 1986 Aug;59(2):328–340. doi: 10.1128/jvi.59.2.328-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanese N., Telesnitsky A., Goff S. P. Abortive reverse transcription by mutants of Moloney murine leukemia virus deficient in the reverse transcriptase-associated RNase H function. J Virol. 1991 Aug;65(8):4387–4397. doi: 10.1128/jvi.65.8.4387-4397.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesnitsky A. P., Chamberlin M. J. Sequences linked to prokaryotic promoters can affect the efficiency of downstream termination sites. J Mol Biol. 1989 Jan 20;205(2):315–330. doi: 10.1016/0022-2836(89)90343-4. [DOI] [PubMed] [Google Scholar]
- Temin H. M., Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970 Jun 27;226(5252):1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- Tisdale M., Schulze T., Larder B. A., Moelling K. Mutations within the RNase H domain of human immunodeficiency virus type 1 reverse transcriptase abolish virus infectivity. J Gen Virol. 1991 Jan;72(Pt 1):59–66. doi: 10.1099/0022-1317-72-1-59. [DOI] [PubMed] [Google Scholar]
- Wigler M., Sweet R., Sim G. K., Wold B., Pellicer A., Lacy E., Maniatis T., Silverstein S., Axel R. Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell. 1979 Apr;16(4):777–785. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]
- Wöhrl B. M., Moelling K. Interaction of HIV-1 ribonuclease H with polypurine tract containing RNA-DNA hybrids. Biochemistry. 1990 Nov 6;29(44):10141–10147. doi: 10.1021/bi00496a001. [DOI] [PubMed] [Google Scholar]
- Yang W., Hendrickson W. A., Crouch R. J., Satow Y. Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein. Science. 1990 Sep 21;249(4975):1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982 Oct 25;10(20):6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Marzo Veronese F., Copeland T. D., DeVico A. L., Rahman R., Oroszlan S., Gallo R. C., Sarngadharan M. G. Characterization of highly immunogenic p66/p51 as the reverse transcriptase of HTLV-III/LAV. Science. 1986 Mar 14;231(4743):1289–1291. doi: 10.1126/science.2418504. [DOI] [PubMed] [Google Scholar]