Abstract
The proneural genes encode basic-helix–loop–helix (bHLH) proteins and promote the formation of distinct types of sensory organs. In Drosophila, two sets of proneural genes, atonal (ato) and members of the achaete–scute complex (ASC), are required for the formation of chordotonal (ch) organs and external sensory (es) organs, respectively. We assayed the production of sensory organs in transgenic flies expressing chimeric genes of ato and scute (sc), a member of ASC, and found that the information that specifies ch organs resides in the bHLH domain of ato; chimeras containing the b domain of ato and the HLH domain of sc also induced ch organ formation, but to a lesser extent than those containing the bHLH domain of ato. The b domains of ato and sc differ in seven residues. Mutations of these seven residues in the b domain of ato suggest that most or perhaps all of these residues are required for induction of ch organs. None of these seven residues is predicted to contact DNA directly by computer simulation using the structure of the myogenic factor MyoD as a model, implying that interaction of ato with other cofactors is likely to be involved in neuronal type specification.
Keywords: Drosophila melanogaster, sensory organ, neuronal type specification, atonal gene
The Drosophila peripheral nervous system consists of four major types of sensory organs: chordotonal (ch) organs, external sensory (es) organs, multiple dendritic (md) neurons, and photoreceptors (for review, see ref. 1). Despite apparent differences among these sensory organs, the basic developmental mechanisms for their formation are quite similar. The genesis of sensory organs begins with the expression of proneural genes in small patches of the epidermal cells (proneural clusters) (2–5). This step endows each cell of a proneural cluster with the competence to become a sensory organ precursor (sop) cell. Soon after, cells within a proneural cluster compete with one another; only one or a few cells increase expression of the proneural genes and are fated to become the sop cells (6–8). The neighboring cells receive inhibitory signals from the sop cells, cease expression of the proneural genes, and adopt the epidermal cell fate. The singled-out sop cells divide to give rise to several daughter cells, which then differentiate into neurons and associated support cells of the sensory organs (reviewed in refs. 9 and 10).
Two classes of proneural genes, atonal (ato) and members of the achaete–scute complex (ASC), including achaete (ac) and scute (sc), are required for the development of almost all the sensory organs in both embryos and imaginal discs. Loss-of-function mutations of ASC genes abolish es organs and most of the md neurons (11–13). When overexpressed in imaginal discs, ASC genes are able to induce ectopic bristles (one type of es organ) at many locations, including the wings and nota (14–19). ato is responsible for the formation of ch organs and photoreceptors. In ato− mutants, ch organs and photoreceptors are almost completely missing (20–22). Overexpression of ato induces ch organs, as expected from the ato mutant phenotype. It also induces bristles, even though no defects in es organs have been observed in ato mutants. This might arise from artefactual activation of endogenous ASC as a result of overexpression of ato.
While mutations in ato or ASC result in the loss of different subset of sensory organs, removal of the daughterless (da) gene product deletes the entire embryonic sensory peripheral nervous system. The gene products of ato and ASC belong to the basic-helix–loop–helix (bHLH) class of transcription factors, as does the product of da (20, 23–26). They form heterodimers with da and bind to E boxes (20, 27, 28), the cis-acting elements found in the promoters of downstream genes [e.g., scabrous and Enhancer of split (27)], thereby regulating gene transcription during neurogenesis. While the bHLH domains of the members of ASC share about 70% identity (not including the loop region) with one another, they share only 50% with the bHLH domain of ato. Outside the bHLH domains, no significant homology between ato and ASC was found. Given that only ato is capable of promoting ectopic ch organ formation, we tested chimeric genes of ato and sc for their ability to induce ch organs in transgenic flies, to identify structural elements responsible for neuronal specificity conferred by ato. On the basis of the structure of the myogenic factor MyoD, we have generated a model for the structure of the DNA-binding domain of the da/ato heterodimer, in an attempt to understand the possible roles of these structural elements in the specification of neuronal type.
MATERIALS AND METHODS
Chimeric ato/sc Genes and Transgenic Flies.
We dissected ato and sc into three separate domains: b domain, HLH domain, and the rest of the proteins (the boundaries of the domains are according to ref. 19 and see Fig. 3). Oligonucleotides were designed to anneal to the junction of those domains and have proper mutations at the 5′ end of these oligonucleotides [for example, with the sc-b(ato) construct, two oligonucleotides, in combination, have sequences on their 5′ ends annealing to the b domain of ato and sequences at their 3′ end annealing to the flanking regions from either side of the sc b domain]. Each of these oligonucleotides was used with another primer either from most 5′ (carrying an Asp718 site) or from 3′ (carrying an XbaI site) of ato or sc [for example, sc for sc-b(ato) construct] for doing the polymerase chain reaction (PCR). The PCR products, containing separate domains for a particular chimeric gene, were ligated into pUAST cut with Asp718 and XbaI. The same method was used for creating point mutations in the b domain of ato or sc. Those chimeric constructs were sequenced and then were injected into fly embryos to create transgenic flies (29). The transformants were balanced with either CyO or TM3 sb according to the chromosomal locations of the transgenes.
Figure 3.
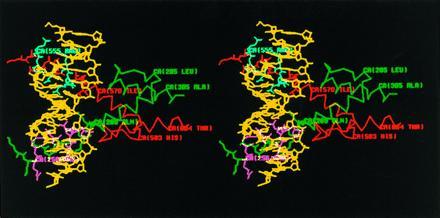
Stereo diagram of the predicted structure of ato/da bHLH domain heterodimer–DNA complex by computer modeling. The sequence of the DNA-binding site in this model is TCAACAGCTGTTGA, containing an E box. DNA is colored in yellow, ato in green, and da in red. Those residues of ato and da predicted to contact DNA are colored in magenta and cyan, respectively. Only the side chains of the residues in the b domains are shown.
Assay of ch Organs and Bristle Formation on the Wings with hs-GAL4.
Balanced male flies carrying UAS-ato/sc chimeras were crossed to hs-GAL4/CyO females at room temperature. White pupae (0–6 hr after puparium formation) from the crosses were collected and heat shocked at 39°C for 15 min. The heat-induced GAL4 then activated the expression of ato/sc proteins in various tissues, including the wing discs. The wings of adult flies were dissected and mounted with 80% glycerol. Ectopic bristles and ch organs were examined under the microscope. We scored the proximal quarter of the wings for the ch and es organs.
Assay for the Rescue of ch Neurons in ato1 Mutant Embryos.
UAS-ato/sc ato1/TM3 males were crossed to h-GAL4 ato1/TM3 females. The GAL4 is expressed in the odd-numbered segments (T2, A1, A3, A5, and A7) through the promoter of the pair-rule gene, hairy (h) (30). Embryos from these crosses were collected for an interval of 4 hr and aged for another 11 hr at 25°C. They were then fixed in 5% formaldehyde for 20–25 min, stained with monoclonal antibody 22C10 (31), and mounted in Permount (Fisher). One-quarter of the embryos with UAS-ato/sc ato1/h-GAL4 ato1 genotype were easily recognized due to lack of ch neurons in even-numbered abdominal segments. These embryos were scored for the number of ch neurons in the first four abdominal segments.
Computer Modeling.
To build a model for the ato/da bHLH domain heterodimer–DNA complex, the bHLH sequences of ato, da, and MyoD were aligned by a multiple-sequence alignment (32) to locate sequence-conserved regions under the quanta (Polygen, Waltham, MA) environment on a Silicon Graphics IRIS 4D/25 workstation. The protein models constructed are based on the x-ray structure of MyoD bHLH domain–DNA complex at 2.8-Å resolution. The different residues in ato and da were introduced by using the “mutate” technique in quanta. The program aligned the replacement residues to the backbone of the original residues and maintained the dihedral angles for the side chains which are in common between the two residues. The gap regions were determined by a fragment-search process. The “regularization modeling”’ technique was employed to build and relax the side-chain coordinates. The complete structure was obtained by the charmm energy minimization process with the charmm/quanta program package by 100 cycles of steepest descent, 200 cycles of Powell conjugate gradient, and 300 cycles of conjugate gradient minimization.
Statistics.
The data in Tables 1–3 were analyzed with the sigmastat software (Jandel) in which the program of descriptive statistics was used to calculate the mean and SEM and the Mann–Whitney rank sum test was used to calculate the P values described in the text. The rank sum test tests the hypothesis that two samples were not drawn from populations with different medians. We also did unpaired t tests on those comparisons, and the P values (not shown) from these tests gave the same conclusions as the rank sum test.
Table 1.
Ectopic ch and es organ induction by ato/sc chimeras in transgenic flies.
| Line | Chimera | Structure | No. of organs per
wing
|
ch/(ch + es), % | No. of wings | No. of lines | |
|---|---|---|---|---|---|---|---|
| Ectopic ch | Ectopic es | ||||||
| 1 | ato | |b| HLH | 31.6 ± 4.6 | 31.9 ± 2.2 | 50 | 16 | 5 |
| 2 | sc | |b| HLH| | 1.7 ± 0.7 | 57.0 ± 4.3 | 3 | 11 | 3 |
| 3 | ato-bHLH (sc) | |b| HLH | 0.5 ± 0.2 | 26.3 ± 1.8 | 2 | 34 | 7 |
| 4 | sc-bHLH (ato) | |b| HLH| | 23.5 ± 4.5 | 39 ± 2.1 | 37 | 18 | 4 |
| 5 | bHLH (sc) | b| HLH | 0 ± 0 | 13.5 ± 1.6 | 0 | 10 | 3 |
| 6 | bHLH (ato) | b| HLH | 0 ± 0 | 0 ± 0 | 10 | 3 | |
| 7 | sc-b (ato) | |b| HLH| | 8.4 ± 1.6 | 52.3 ± 3.2 | 14 | 17 | 6 |
| 8 | sc-HLH (ato) | |b| HLH| | 0 ± 0 | 12.3 ± 16.5 | 0 | 8 | 4 |
| 9 | ato-HLH (sc) | |b| HLH | 3.3 ± 0.5 | 31.0 ± 1.3 | 10 | 17 | 4 |
Shading in the chimeras indicates the ato domains. The numbers of ectopic ch (column 4) and es (column 5) organs in the basal one-quarter of the wing are given as mean ± SEM. The numbers of ectopic ch organs divided by the total number of ectopic sensory organs are given as percentages in column 6. The total number of wings and independent lines assayed for each construct are given in columns 7 and 8, respectively.
RESULTS
Phenotypes Due to Overexpression of ato and sc.
A targeted gene expression system (30) was used to express chimeras of proneural genes ato and sc (ato/sc) (see Table 1) in transgenic flies. These chimeric genes were placed under the control of UASG, the binding site for the yeast transcription factor GAL4. We first expressed ato and sc proteins in vivo by hs-GAL4 during wing development (see Materials and Methods). The wings from adult flies were then examined for the appearance of ectopic bristles (es organs) and ch organs. In wild-type wings, clusters of ch organs are normally found only in the ventral radius and tegula (Fig. 1B). Ectopic expression of sc did not increase the number of ch organs (1.7 ± 0.7 per wing; Table 1, line 2), but it did produce an abundance of ectopic bristles (57.0 ± 4.3 per wing; Table 1, line 2 and Fig. 1C). These ectopic bristles were solitary and evenly distributed on the wing. Ectopic expression of ato, on the other hand, resulted in numerous ch organs (31.6 ± 4.6 per wing; Table 1, line 1), which could be observed directly by using Nomarski optics, as shown in Fig. 1A. Most of these ectopic ch organs were distributed along the wing veins of the proximal quarter of the wing (Fig. 1B), and about 64% of them formed clusters, ranging from two to five in a group. Overexpression of ato also induced the formation of bristles (31.9 ± 2.2 per wing). Similar to those induced by overexpression of sc, they were solitary and evenly distributed, but not as numerous (Table 1, lines 1 and 2). The total numbers of ectopic sensory organs (es + ch organs) produced by overexpression of ato and sc are very similar (63.5 and 58.7 per wing).
Figure 1.
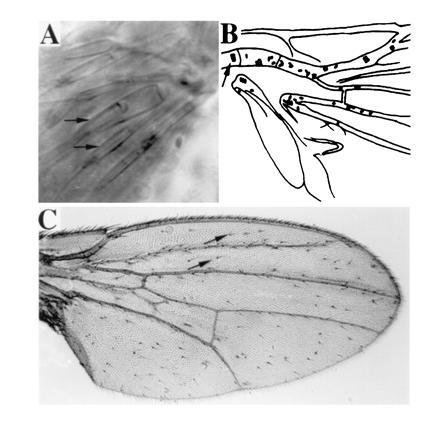
(A) In the scutellum of GAL4(109–68); UAS-ato fly, several scolopales, each formed as a part of an ectopic ch organ by its support cells (arrows mark two of the scolopales), are visible under Nomarski optics. (B) Camera lucida representation of the basal quarter region of the wing of an hs-GAL4; UAS-ato fly, examined directly with Nomarski optics. Each oval represents a ch organ. Wild-type ch organs on the ventral radius vein are indicated with an arrow; the rest of the ch organs are ectopic. (C) Numerous bristles are formed on the wing of an hs-GAL4; UAS-sc fly. Almost all the bristles, except for those on the wing margin, are ectopic ones (two of the ectopic bristles are marked with arrows).
Since the number of ectopic sensory organs induced by different chimeras is likely to vary with their expression levels, we also included the percentage of ectopic ch organs among the total ectopic sensory organs [ch % = 100 × ch/(ch + es)] as an index for the ability to induce ch organs. The ch % was 3% for sc and 50% for ato (Table 1, lines 1 and 2), indicating that ato, but not sc, induces a significant number of ectopic ch organs.
bHLH Domain of ato Specifies ch Organs.
To test whether the bHLH domain of ato is responsible for specification of ch organs, we replaced the bHLH domain of ato with that of sc [ato-bHLH(sc)], and we found that the ability to promote ch organ formation is essentially abolished (average number of ch organs per wing is 0.5, or 2% of ectopic sensory organs; Table 1, line 3). Conversely, transplanting the ato bHLH domain into sc [sc-bHLH(ato)] conferred the ability to induce ch organs (23.5 and 37%; Table 1, line 4). Thus the bHLH domain of ato accounts for the ability to specify precursors for ch organs.
To test if the bHLH domain of sc or ato alone is sufficient for inducing and specifying sop cells, we examined flies expressing bHLH(sc) or bHLH(ato) without the flanking sequences of ato and sc. We found that bHLH(sc) was able to promote es, but not ch, organ formation (Table 1, line 5). This is consistent with the observation that the bHLH domain of lethal of scute (l’sc), a member of ASC, is sufficient for the formation of es organs (18). However, bHLH(ato) did not induce either type of sensory organ (Table 1, line 6). Comparison of this result with the results due to ectopic expression of ato and sc-bHLH(ato) suggests that other sequences in ato or sc are necessary for the function or stability of bHLH(ato) in transgenic flies. Since the function of ato bHLH domain could be analyzed in the context of the full-length proteins of ato or sc, we subdivided the bHLH domain into smaller regions and generated additional chimeras, in an attempt to identify minimal structural elements that confer the specificity for the induction of ch organ precursors.
Involvement of b Domain and HLH Domain of ato in the Specification of ch Organ Precursors.
The bHLH domain of ato or sc is composed of a b domain, which is involved in DNA binding, and an HLH domain, which is involved in dimerization (33, 34). We have further tested some chimeras with either the b domain or the HLH domain swapped between ato and sc. Replacement of the b domain of sc with that of ato [sc-b(ato)] allowed this chimera to induce ch organs (Table 1, line 7). This is reflected by a statistically significant increase in the absolute number of ectopic ch organs (8.4 per wing) and the percentage of ch organs (ch % = 14%), as compared with the corresponding numbers of sc (1.7 and 3%; P = 0.001, see Materials and Methods). The only difference between sc and sc-b(ato) resides in the b domain. These results indicate that the b domain of ato contains the ability to induce ectopic ch organs. Consistently, ato-HLH(sc), consisting of the b domain of ato and HLH domain of sc, retained the ability to promote ectopic ch organ formation (3.3 and 10%; Table 1, line 9), in contrast to ato-bHLH(sc) [0.5 and 2%; P < 0.0001 for ato-HLH(sc) versus ato-bHLH(sc); Table 1, line 3], which contains both b and HLH domains of sc. On the other hand, the HLH domain appears insufficient for the specificity; transplanting the ato HLH domain into sc [sc-HLH(ato)] did not allow ch organ induction (ch % = 0%; Table 1, line 8). Thus the abilities of sc-b(ato) and ato-HLH(sc) in promoting ectopic ch organ formation indicate that the b domain of ato is able to confer the specificity for ch organ formation. However, these abilities of sc-b(ato) and ato-HLH(sc) are not as strong as sc-bHLH(ato) and ato, respectively [Table 1, P = 0.001 for sc-b(ato) versus sc-bHLH(ato) and P < 0.0001 for ato-HLH(sc) versus ato]. This indicates that, in the presence of ato b domain, the HLH domain of ato is more effective than the HLH domain of sc in promoting ch organ formation in this assay.
Rescue of ch Neurons in ato1 Embryos by sc-bHLH(ato) and sc-b(ato).
We further tested whether the b domain of ato also suffices to specify ch neurons in embryos lacking endogenous ato activity. In ato1 mutant embryos, almost all the ch neurons are eliminated (22), including the lateral five ch neurons (lch5) in each abdominal hemisegment (Fig. 2 B and G, compared with the wild-type embryos in Fig. 2 A and F and Table 2, lines 1 and 2). We used h-GAL4 (30) to express chimeras of ato and sc in alternate segments (T2, A1, A3, A5, and A7) of the embryos. As expected, segmental expression of ato in ato1 mutants restored most of the lch5 neurons in the odd-numbered abdominal segments (e.g., 82% in A1 and 100% in A3; Table 2, line 3, and Fig. 2 C and H) but not in even-numbered abdominal segments (such as A2 and A4). The rescue was also observed in T2, but not T3 segments (data not shown). By contrast, sc was incapable of rescuing the ato1 mutant phenotype in any of the segments (Fig. 2 D and I and Table 2, line 6). Replacing the bHLH domain of sc with that from ato [sc-bHLH(ato)] allowed the mutant sc protein to functionally substitute for ato in inducing ch neurons at their normal locations. The extent of rescue was about 84% (both A1 and A3 segments; Table 2, line 4), which is comparable to that by wild-type ato. The sc-b(ato), with only the b domain replaced by that of ato, also rescued ato1 mutants to a high degree (52% in A1 segments and 68% in A3 segments; P < 0.0001 for A1 and P = 0.0001 for A3 when compared with sc; Table 2, line 5 and Fig. 2 E and J). This is consistent with the results of overexpression of the sc-b(ato) in the wing discs and it further confirms that the b domain of ato is sufficient for the specification of ch organ precursors.
Figure 2.
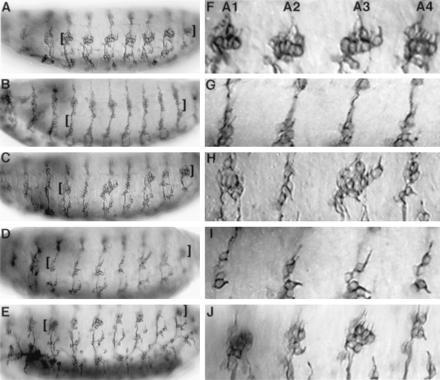
Rescue of ch organs in ato1 mutant embryos by UAS-ato/sc chimeric genes. (A and F) Wild-type embryo contains five ch neurons in the lateral region (lch5, marked by the brackets in A–E) in each abdominal hemisegment. Most of the ch neurons are eliminated in ato1 mutant embryo (B and G). The lch5 of ato1 embryos in alternate segments were rescued by h-GAL4; UAS-ato (C and H) and by h-GAL4; UAS-sc-b(ato) (E and J) combinations, but not by h-GAL4; UAS-sc (D and I). In F–J, the first four abdominal hemisegments of the embryos in A–E are shown, respectively.
Table 2.
Rescue of 1ch5 neurons in ato1 mutant embryos by ato/sc chimeras
| Line | Embryo | No. of ch organs in
hemisegment
|
No. of embryos | No. of lines | |||
|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | A4 | ||||
| 1 | Wild type | 5 | 5 | 5 | 5 | ||
| 2 | ato1/ato1 | 0.4 ± 0.1 | 0.4 ± 0.1 | 0.7 ± 0.1 | 0.4 ± 0.1 | 14 | 2 |
| 3 | ato | 4.1 ± 0.6 | 0.6 ± 0.2 | 5.0 ± 0.2 | 1.6 ± 0.4 | 12 | 2 |
| 4 | sc-bHLH(ato) | 4.2 ± 0.6 | 0.5 ± 0.2 | 4.2 ± 0.5 | 0.8 ± 0.3 | 6 | 3 |
| 5 | sc-b(ato) | 2.6 ± 0.3 | 1.4 ± 0.4 | 3.4 ± 0.4 | 1.1 ± 0.3 | 14 | 2 |
| 6 | sc | 0.7 ± 0.2 | 0.3 ± 0.1 | 0.8 ± 0.2 | 0.7 ± 0.2 | 15 | 3 |
The number (mean ± SEM) of ch organs in ato1 mutant and ato1 mutants expressing ato/sc chimeras is given for each of the first four abdominal hemisegments. The last columns indicate the number of embryos and the number of lines scored.
Mutagenesis Analysis of the ato b Domain.
To test whether specific residues of the b domain could endow the specificity for induction of ch organ precursors, we examined the phenotypes caused by point mutations in the b domains of ato or sc. The b domains of ato and sc differ in 7 of 15 residues (Table 3, lines 1 and 2). Of these 7 residues, 4 represent amino acid substitutions at homologous positions, and another 3 residues (Leu, Ala, and Ala) from the ato b domain correspond to a gap in the sc b domain. Of the various point mutations (Table 3, lines 3–7) tested, the substitution of Asn in sc with Arg, the corresponding residue in ato, increased the number of ch organs (compare lines 3–5 with lines 6 and 7 of Table 3). But none of these changes in the number of ectopic ch organs are statistically significant (P > 0.05 for every chimera compared with sc). On the other hand, substitution of the Arg in wild-type ato with Asn from sc, which is conserved among members of ASC, reduced the number of ectopic ch organs [17.1 organs per wing and 31%; Table 3, line 9, P = 0.01 for ato versus ato-(R-N)]. Insertion of the Leu, Ala, and Ala residues of ato into sc (sc-in; Table 3, line 8) did not increase the ability to induce ch organs (2.1 and 5%; P > 0.05 for sc versus sc-in). But the deletion of these three residues from ato (ato-del; Table 3, line 10) partially reduced ch organ induction in a statistically significant way (13.5 and 30%; P = 0.002 for ato versus ato-del). Therefore, our mutagenesis analysis indicates that the specificity for ch organ induction could arise from a combination of most or all of the 7 residues of the ato b domain that differ from their counterpart in sc.
Table 3.
Ectopic ch and es organs induced by various ato or sc proteins with mutations in the b domains
| Line | Protein | b domain sequence | No. of organs per wing
|
ch/(ch + es), % | No. of wings | No. of lines | |
|---|---|---|---|---|---|---|---|
| Ectopic ch | Ectopic es | ||||||
| ∗∗∗∗∗∗∗ ∗ | |||||||
| 1 | ato (WT) | KRKRRLAANARERRR | 31.6 ± 4.6 | 31.9 ± 2.2 | 50 | 16 | 5 |
| 2 | sc (WT) | SVQRR---NARERNR | 1.7 ± 0.7 | 57.0 ± 4.3 | 3 | 11 | 3 |
| 3 | sc-m14 | KRKRR---NARERRR | 2.9 ± 0.9 | 21.1 ± 1.0 | 12 | 8 | 4 |
| 4 | sc-m24 | SRQRR---NARERRR | 5.2 ± 1.9 | 24.7 ± 1.1 | 17 | 8 | 4 |
| 5 | sc-m34 | SVQRR---NARERRR | 4.0 ± 1.4 | 23.8 ± 1.3 | 14 | 10 | 4 |
| 6 | sc-m15 | KRKRR---NARERNR | 0.3 ± 0.2 | 20.8 ± 1.7 | 1 | 8 | 4 |
| 7 | sc-m25 | SRQRR---NARERNR | 0.5 ± 0.3 | 18.1 ± 1.9 | 3 | 6 | 2 |
| 8 | sc-in | SVQRRLAANARERNR | 2.1 ± 0.6 | 43.6 ± 2.2 | 5 | 21 | 8 |
| 9 | ato-(R-N) | KRKRRLAANARERNR | 17.1 ± 4.8 | 38.7 ± 1.7 | 31 | 16 | 4 |
| 10 | ato-del | KRKRR---NARERRR | 13.5 ± 2.6 | 31.8 ± 3.2 | 30 | 13 | 2 |
The amino acid sequences of the b domains start at Lys-253 of ato or Ser-100 of sc. WT, wild type. The ato sequence is in boldface type. The conserved residues are marked with ∗. Other columns are described in the legend of Table 1.
Model for the DNA-Binding Domain of ato/da Heterodimer.
To better understand the role of the b domain of ato in inducing ch organ precursors, we used the crystal structure of the MyoD bHLH domain (35) to predict the structure of ato/da bHLH heterodimer–DNA complex. In the model that we generated (Fig. 3), the b domain of ato forms an α-helical structure and is embedded in the major groove of DNA. Seven residues (Arg-256, Arg-257, Asn-261, Arg-263, Glu-264, Arg-265, and Arg-267, colored in magenta in Fig. 3) in the b domain of ato are predicted to contact DNA directly through hydrogen bonding (data not shown). Interestingly, these residues are conserved between ato and sc (Table 3, lines 1 and 2). Some of the nonconserved residues (Arg-254, Lys-255, Leu-258, Ala-259, and Arg-266, colored in green) and one of the conserved residues (Ala-262, in green) are on the other side of the α-helix. Their side chains are predicted to protrude away from DNA and are available for contact with other proteins. Thus, in this model, the residues that contact DNA are conserved between the b domains of ato and sc. In contrast, the seven nonconserved residues in the b domain of ato do not contact DNA and could be involved in interaction with other protein(s) that are also involved in the specification of ch organ precursors.
DISCUSSION
bHLH proteins play crucial roles in controlling cell fate in a variety of tissue types. In ectopic expression experiments, closely related bHLH proteins can often substitute for each other in promoting certain cell fates. For instance, each of the myogenic factors (MyoD, Myf-5, myogenin, and MRF4) is capable of transforming fibroblasts into myoblasts in a tissue culture system (for review, see refs. 35–37). Likewise, each member of the ASC (ac, sc, ase, and l’sc) can promote ectopic es organ development in wing discs (14–19). Those bHLH factors have a high degree of sequence homology in the bHLH domains (≈80% identity among members of myogenic factors and ≈70% among members of ASC) and are interchangeable in gain-of-function experiments, making it difficult to identify domains of these bHLH proteins required for specifying a particular cell type. ato is more distantly related to members of ASC (50% identity in bHLH domain) and is unique in its ability to promote ch organ formation. This has provided us with an opportunity to identify the structural elements that confer the neuronal type information. Previous work on myogenic factors has indicated that residues in the b domain contain the information needed to promote nonmuscle cell to differentiate into muscle cells (38, 39). From overexpression of chimeric proteins in the wing discs and in ato1 mutant embryos, our results indicate that the bHLH domain, particularly the b domain, of ato not only is able to promote neuronal cell fate but also confers the neuronal type information (in this case, ch neuron). The possible contributions of the b and HLH domains to neuronal type specification are discussed below.
Specificity of Sensory Organ Induction by ato and sc.
Each member of the ASC, when overexpressed, promotes ectopic es, but not ch, organ formation. On the other hand, overexpression of ato induces ch as well as es organs, even though normal es organ formation requires ASC genes, but not ato. This apparent discrepancy could be accounted for by one of the following possibilities. First, it is possible that overexpression of ato leads to fortuitous transcriptional activation of ASC genes. If overexpression of ato could cross-activate ac or sc in the regions where they are not normally expressed, it would consequently induce ectopic es organs. To test this possibility, one could examine the phenotypes of overexpressing ato in the ASC− mutants.
Second, ato could induce either type of sensory organs; it induces ch organs in the presence of a cofactor(s) and es organs in the absence of this cofactor(s). We have observed that most of the ectopic ch organs induced by ato were near the wing veins at the proximal regions, in contrast to the evenly distributed ectopic bristles induced by either sc or ato. This could result from the postulated cofactor(s) being restricted to the wing veins at the proximal regions. This hypothesis also presumes that sc, or any other member of ASC, is not able to interact with the cofactor(s) to direct ch organ formation.
Although we cannot distinguish among these possibilities at the moment, the unique ability of ato to induce ch organs supports the idea that the neuronal type information of ch organs arises from certain features of ato that are not present in sc or any other ASC proteins.
Possible Roles of the b Domain and the HLH Domain of ato in ch Organ Specification.
The ability to induce ch organs can be transferred to the sc protein by replacing its b domain with the corresponding domain of ato [sc-b(ato); Table 1, line 7]. Also, compared with ato-bHLH(sc), ato-HLH(sc) that has the ato b domain is more effective in promoting ch organ formation (Table 1, lines 3 and 9; P < 0.0001). Another experiment revealed that sc-b(ato) can restore ch neurons to a high degree in ato1 mutant embryos (Table 2, line 5). Thus, the b domain of ato harbors to some extent the neuronal type information.
The ability to induce ch organs is further enhanced when the ato b domain is accompanied with the ato HLH domain as in sc-bHLH(ato) (Table 1, line 4), even though presence of only the ato HLH domain in sc-HLH(ato) (Table 1, line 8) is insufficient to promote ch organ formation. The importance of the ato HLH domain is further shown with ato-HLH(sc) (Table 1, line 9) which is much less effective in promoting ch organ formation compared with wild-type ato (Table 1, line 1). These observations indicate that the difference between the HLH domains of ato and sc also contributes to the specification of neuronal type. This could arise if these HLH domains interact differently with positive or negative factors in a quantitative or qualitative way. It is also conceivable that these HLH domains affect the folding or stability of the chimeric proteins. Notwithstanding the quantitative differences in ectopic ch organ induction attributable to the HLH domains, the requirement of the ato b domain for ch organ formation indicates that the neuronal type information derives partially from the seven residues in the ato b domain that are different from those in sc.
The Seven Residues in the b Domains of ato That Differ from the Corresponding Residues in sc.
Strong conservation of ato b domain, including some of the seven residues that distinguish ato from sc b domain, has been observed among ato homologs from different species. For example, MATH1, a mammalian bHLH protein (40), has 12 residues that are identical to those in the b domain of ato, which contains 15 residues (Table 3, line 1). The gene product of Caenorhabditis elegans lin-32, which functions in an analogous manner as do the proneural genes of Drosophila (41), shares 11 residues with ato in the b domain. A Xenopus homolog of ato (M. Vetter, L.Y.J., and Y.N.J.) has 12 residues identical to those in the b domain of ato. It will be interesting to know whether the b domains of ato homologs in those species are involved in neuronal type specification.
Mutations, including the substitution of Asn of sc with Arg-266, the corresponding residue in ato, slightly increase the ability of sc to induce ch organs (Table 3, lines 3–5), but this increase is not statistically significant (P > 0.05). Insertion of three residues, Leu-258, Ala-259, and Ala-260 of ato, into sc does not confer on sc the ability to induce ch organ formation (Table 3, line 8). On the other hand, either deletion of the three residues Leu-258, Ala-259, and Ala-260 of ato or mutation of the Arg-266 of ato to the corresponding Asn of sc reduces the ability to induce ectopic ch organs in a statistically significant way (Table 3, lines 9 and 10, P = 0.002 or P = 0.01 compared with ato, respectively). Only the chimera sc-b(ato), in which seven mutations were introduced into the b domain of sc to convert it into the b domain of ato, acquires the ability to induce ch organs. Whereas it remains possible that fewer than seven mutations are necessary to endow sc with the ability to induce ch organs, it appears from the series of mutagenesis examined in this study (Table 3) that most, if not all, of the seven residues in the ato b domain contribute to the unique ability of ato to specify ch organ precursors.
In the computer model (Fig. 3), the b domain of ato forms α-helix. Most of the seven residues that differ from their counterparts in sc are on the opposite side of the surface that contacts DNA, suggesting that they might form an interface for protein–protein interaction. This interaction may affect the recognition and interaction of DNA by ato/da heterodimers and/or the subsequent effects on transcription, potentially leading to transcriptional regulation of downstream genes involved in the formation of ch organs.
Acknowledgments
We thank Dr. Sonsoles Campuzano for sc cDNA and I. Clark, D. Doherty, T. Doniach, M. Utset, M. Vetter, and W. Zhong for their comments on the manuscript. C.-t.C. is a fellow of The Jane Coffin Childs Memorial Fund for Medical Research. C.-D.H. was supported in part by a grant from the National Science Council, Taiwan. L.Y.J. and Y.N.J. are Investigators of the Howard Hughes Medical Institute.
Footnotes
Abbreviations: bHLH, basic-helix–loop–helix; ch, chordotonal; es, external sensory; md, multiple dendritic; sop, sensory organ precursor.
References
- 1.Jan Y N, Jan L Y. In: The Peripheral Nervous System. Bate M, Martinez-Arias A, editors. II. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 1207–1244. [Google Scholar]
- 2.Ghysen A, Dambly-Chaudiere C. Trends Genet. 1989;5:251–255. doi: 10.1016/0168-9525(89)90097-8. [DOI] [PubMed] [Google Scholar]
- 3.Jan Y N, Jan L Y. Trends Neurosci. 1990;13:493–498. doi: 10.1016/0166-2236(90)90083-m. [DOI] [PubMed] [Google Scholar]
- 4.Cabrera C V. Development (Cambridge, UK) 1992;15:893–901. doi: 10.1242/dev.115.4.893. [DOI] [PubMed] [Google Scholar]
- 5.Campuzano S, Modolell J. Trends Genet. 1992;8:202–208. doi: 10.1016/0168-9525(92)90234-u. [DOI] [PubMed] [Google Scholar]
- 6.Campos-Ortega J A. Trends Neurosci. 1988;11:400–405. doi: 10.1016/0166-2236(88)90077-x. [DOI] [PubMed] [Google Scholar]
- 7.Artavanis-Tsakonas S, Simpson P. Trends Genet. 1991;7:404–408. doi: 10.1016/0168-9525(91)90264-q. [DOI] [PubMed] [Google Scholar]
- 8.Ghysen A, Dambly-Chaudiere C, Jan L Y, Jan Y N. Genes Dev. 1993;7:723–733. doi: 10.1101/gad.7.5.723. [DOI] [PubMed] [Google Scholar]
- 9.Jan Y N, Jan L Y. Neuron. 1995;14:1–5. doi: 10.1016/0896-6273(95)90235-x. [DOI] [PubMed] [Google Scholar]
- 10.Posakony J W. Cell. 1994;76:415–418. doi: 10.1016/0092-8674(94)90105-8. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Bellido A, Santamaria P. Genetics. 1978;88:469–486. doi: 10.1093/genetics/88.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Bellido A. Genetics. 1979;91:491–520. doi: 10.1093/genetics/91.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dambly-Chaudiere C, Ghysen A. Genes Dev. 1987;1:297–306. [Google Scholar]
- 14.Campuzano S, Balcells L, Villares R, Garcia-Alonso L, Modolell J. Cell. 1986;44:303–312. doi: 10.1016/0092-8674(86)90764-6. [DOI] [PubMed] [Google Scholar]
- 15.Balcells L, Modolell J, Ruiz-Gomez M. EMBO J. 1988;7:3899–3906. doi: 10.1002/j.1460-2075.1988.tb03276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez I, Hernandez R, Modolell J, Ruiz-Gomez M. EMBO J. 1990;9:3583–3592. doi: 10.1002/j.1460-2075.1990.tb07569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brand M, Jarman A P, Jan L Y, Jan Y N. Development (Cambridge, UK) 1993;119:1–17. doi: 10.1242/dev.119.Supplement.1. [DOI] [PubMed] [Google Scholar]
- 18.Hinz U, Giebel B, Campos-Ortega J A. Cell. 1994;76:77–88. doi: 10.1016/0092-8674(94)90174-0. [DOI] [PubMed] [Google Scholar]
- 19.Dominguez M, Campuzano S. EMBO J. 1993;12:2049–2060. doi: 10.1002/j.1460-2075.1993.tb05854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarman A P, Grau Y, Jan L Y, Jan Y N. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- 21.Jarman A P, Grell E H, Ackerman L, Jan L Y, Jan Y N. Nature (London) 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- 22.Jarman A P, Sun Y, Jan L Y, Jan Y N. Development (Cambridge, UK) 1995;121:2019–2030. doi: 10.1242/dev.121.7.2019. [DOI] [PubMed] [Google Scholar]
- 23.Villares R, Cabrera C V. Cell. 1987;50:415–424. doi: 10.1016/0092-8674(87)90495-8. [DOI] [PubMed] [Google Scholar]
- 24.Alonso M C, Cabrera C V. EMBO J. 1988;7:2585–2591. doi: 10.1002/j.1460-2075.1988.tb03108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez F, Romani S, Cubas P, Modolell J, Campuzano S. EMBO J. 1989;8:3553–3562. doi: 10.1002/j.1460-2075.1989.tb08527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caudy M, Vaessin H, Brand M, Tuma R, Jan L Y, Jan Y N. Cell. 1988;55:1061–1067. doi: 10.1016/0092-8674(88)90250-4. [DOI] [PubMed] [Google Scholar]
- 27.Cabrera C V, Alonso M C. EMBO J. 1991;10:2965–2973. doi: 10.1002/j.1460-2075.1991.tb07847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singson A, Leviten M, Bang A G, Hua X H, Posakony J W. Genes Dev. 1994;8:2058–2071. doi: 10.1101/gad.8.17.2058. [DOI] [PubMed] [Google Scholar]
- 29.Spradling A C, Rubin G M. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 30.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 31.Zipursky S L, Venkatesh T R, Teplow D B, Benzer S. Cell. 1984;36:15–26. doi: 10.1016/0092-8674(84)90069-2. [DOI] [PubMed] [Google Scholar]
- 32.Feng D F, Doolittle R F. J Mol Evol. 1987;25:351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- 33.Murre C, Schonleber-McCaw P, Baltimore D. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 34.Murre C, Schonleber-McCaw P, Vaessin H, Caudy M, Jan L Y, Jan Y N, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, Weintraub H, Baltimore D. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 35.Ma P C M, Rould M A, Weintraub H, Pabo C O. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 36.Weintraub H. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- 37.Olson E N, Klein W H. Genes Dev. 1994;8:1–8. doi: 10.1101/gad.8.1.1. [DOI] [PubMed] [Google Scholar]
- 38.Brennan T J, Chakraborty T, Olson E N. Proc Natl Acad Sci USA. 1991;88:5675–5679. doi: 10.1073/pnas.88.13.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis R L, Weintraub H. Science. 1992;256:1027–1030. doi: 10.1126/science.1317057. [DOI] [PubMed] [Google Scholar]
- 40.Akazawa C, Ishibashi M, Shimizu C, Nakanishi S, Kageyama R. J Biol Chem. 1995;270:8730–8738. doi: 10.1074/jbc.270.15.8730. [DOI] [PubMed] [Google Scholar]
- 41.Zhao C, Emmons S W. Nature (London) 1995;373:74–78. doi: 10.1038/373074a0. [DOI] [PubMed] [Google Scholar]


