Abstract
Neuronal signaling requires that synaptic proteins be appropriately localized within the cell and regulated there. In mammalian neurons, polyribosomes are found not just in the cell body, but also in dendrites where they are concentrated within or beneath the dendritic spine. The α subunit of Ca2+-calmodulin-dependent protein kinase II (CaMKIIα) is one of only five mRNAs known to be present within the dendrites, as well as in the soma of neurons. This targeted subcellular localization of the mRNA for CaMKIIα provides a possible cell biological mechanism both for controlling the distribution of the cognate protein and for regulating independently the level of protein expression in individual dendritic spines. To characterize the cis-acting elements involved in the localization of dendritic mRNA we have produced two lines of transgenic mice in which the CaMKIIα promoter is used to drive the expression of a lacZ transcript, which either contains or lacks the 3′-untranslated region of the CaMKIIα gene. Although both lines of mice show expression in forebrain neurons that parallels the expression of the endogenous CaMKIIα gene, only the lacZ transcripts bearing the 3′-untranslated region are localized to dendrites. The β-galactosidase protein shows a variable level of expression along the dendritic shaft and within dendritic spines, which suggests that neurons can control the local biochemistry of the dendrite either through differential localization of the mRNA or variations in the translational efficiency at different sites along the dendrite.
Polyribosomes are localized within neurons to the cell soma, the proximal part of the axon, and throughout the full extent of the dendritic authorization (1). Within dendrites the polyribosomes are not distributed randomly, but rather seem to be concentrated within or beneath the dendritic spines (2, 3). Dendritic spines are elaborations of the dendrite on which excitatory synapses are formed. This concentration of the translational machinery at the site of synaptic input suggests the possibility that the local concentration of polyribosomes might function for the selective expression of certain gene products, which can be regulated in a synapse specific manner (4). Synapse specific gene expression might occur by the selective targeting of specific mRNAs to specific dendritic spines along with associated ribosomes. Alternatively, the mRNA for a given gene might be distributed uniformly to all dendritic spines in a neuron, but the translation of that mRNA might be differentially regulated at the individual spines.
Although the vast majority of neuronal mRNAs are restricted to the cell soma, a number of mRNAs have been found in the dendrite as well as the soma. These include the mRNAs for microtubule-associated protein 2 (MAP2), the Ca2+-calmodulin dependent protein kinase IIα subunit, the IP3 receptor type II, and two genes of unknown function designated L7 and ARC (5–10). The molecular mechanisms responsible for the localization of these mRNAs to dendrites are not known. However, dendritic mRNA appears to be associated with some component of the cytoskeleton (11, 12). The fact that only certain mRNAs are transported into dendrites suggests that a cis-acting signal, present only in those transcripts, mediates the targeting. That signal could be contained within the sequence or structure of the mRNA itself, or it could be carried within the nascent polypeptide chain. In the latter case, the entire complex consisting of polyribosome, mRNA, and nascent peptide would be transported into the dendrite.
The α subunit of Ca2+-calmodulin-dependent protein kinase II (CaMKIIα) subunit gene is expressed specifically in neurons of the forebrain where its mRNA is found within the dendrites as well as the soma of the neuron. We have characterized in transgenic mice cis-acting elements, which mediate the forebrain specific expression as well as the dendritic localization of mRNA. A dendritically localized lacZ gene shows an uneven expression along the length of the dendrite. This suggests that the expression of dendritically localized mRNA is regulated either at the level of the mRNA distribution or at the level of local translation.
MATERIALS AND METHODS
Transgene Constructs.
The CaMKIIα promoter was isolated from a cosmid library prepared from C57BL/6J mouse spleen using a 0.4-kb AvaI fragment comprising the transcription initiation region of the rat CaMKIIα gene (13) (provided by N. Sahyoun, Wellcome Research Labs, Research Triangle Park, NC). The full-length CaMKIIα cDNA was isolated from a mouse brain (C57BL/6J) cDNA library using a rat CaMKIIα cDNA probe. Constructs were assembled using standard techniques. The lacZ gene was obtained from a 3.5-kb HindIII/DraI fragment of pNSE lac (14) (provided by J. G. Sutcliffe, Scripps Research Institute, La Jolla, CA). The bovine polyadenylylation signal was from pRC/CMV (Invitrogen). The GFP gene was from pGFP-C1 (CLONTECH). The nuclear localization signal was from the simian virus 40 large t antigen. It was inserted using synthetic oligonucleotides and consisted of the sequence SSDDEATADSQHSTPPKKKRKVEDP. Transgenic mice were produced by DNA injection into B6CBA F2 or B6/SJL F2 embryos using standard techniques. (The lac-CMK transgenics were prepared by DNX, Princeton.)
Northern blot analysis was generated using 4 μg of A+ RNA isolated from the forebrain of the lac-A and lac-CMK transgenic lines. The blot was hybridized with a lacZ-specific cDNA probe and washed for 40 min at 68°C in 0.2 × SSC/0.1% SDS and exposed for 5 hr.
β-Galactosidase (β-gal) Histochemistry.
Brains were processed for histochemistry essentially as described (14). Animals were perfused with 2% paraformaldehyde/0.2% glutaraldehyde in PBS (pH 7.3) and cryoprotected in 30% sucrose. Fifty-micrometer horizontal sections were prepared and stained for β-gal activity for 4 hr at 37°C in 0.1 M sodium phosphate, pH 7.3/0.14 M NaCl/2 mM MgCl2/3 mM K3Fe (CN)6/3 mM K4Fe (CN)6/1 mg/ml 5-bromo-4-chloro-3-indoyl β-d-galactosidase (X-gal).
In Situ Hybridization.
For in situ hybridization, 20-μm coronal sections were taken from fresh frozen mouse brains. The slices were fixed for 10 min in 4% paraformaldehyde and dehydrated. Slices were probed with a pool of three oligonucleotides specific for the lacZ gene, which had been labeled by tailing with [α-35S]dATP and terminal transferase to a specific activity of >1 × 109 cpm/μg. Oligonucleotide sequences: lac 1, 5′-GTGCATCTGCCAGTTTGAGGGGACGACGACAGTAT-3′; lac 2, 5′-GCCGGAAACCAGGCAAAGCGCCATTCGCCATTCAGGCTGCGC-3′; lac 3, 5′-GTAACCGACCCAGCGCCCGTTGCACCACAGATGAAACGCCG-3′. Hybridization was overnight at 42°C in a solution containing 10% dextran sulfate, 50% formamide, 25 mM Hepes (pH 7.6), 600 mM NaCl, 100 mM DTT, 1 mM EDTA, 200 μg/ml denatured salmon sperm DNA, 200 μg/ml poly(A), 1× Denhardt’s solution, 107 cpm/ml probe. Slides were washed 2× 10′ at reverse transcriptase in 2× SSC and 2× 60′ at 55°C in 0.2× SSC. Slides were coated with Kodak NTB2 emulsion, exposed for 3 weeks, developed, counterstained with toluidine blue, and photographed under darkfield illumination.
Neuronal Cultures.
For neuronal cultures, hippocampi of P1–P3 mouse pups were dissected and treated for 30 min at 37°C with 0.25% trypsin (Sigma, type XI), and then gently titrated and the dissociated cells plated at a concentration of 2 × 105 per ml onto poly-d-lysine (Sigma, 0.1 mg/ml) and laminin (Collaborative Research, 10 μg/ml) coated glass coverslips as described (15).
Cells were plated in minimal essential Eagle’s medium (MEM) containing 10% heat inactivated fetal bovine serum (HyClone), 2 mM glutamine, and 0.76% glucose. On the following day, the medium was replaced with fresh SF1C medium, including B-27 supplements (GIBCO). For immunocytochemistry, cells were labeled as described (16). Briefly, cells were fixed for 10 min at room temperature with 2% paraformaldehyde and incubated overnight at 4°C with monoclonal antibody to MAP2 (Sigma, 5 μg/ml) and rabbit polyclonal antibody to β-gal (Cappel, 2 μg/ml) in PBS containing 10% goat serum. Cells were then stained with fluorescently conjugated secondary antibody (fluorescein isothiocyanate for β-gal detection and Cy3 for MAP2 detection). In several experiments a monoclonal antibody to β-gal (Promega) was used. Images were obtained using an MRC-1000 laser confocal microscope (Bio-Rad).
RESULTS
We have isolated two cis-acting elements of the CaMKIIα gene: one, the promoter, which controls the forebrain-specific expression of the gene and the other, the 3′-untranslated region or 3′ UTR, which controls the dendritic mRNA localization. Two DNA constructs (lac-CMK and lac-A, Fig. 1A) were prepared such that the lacZ reporter gene was placed downstream from an 8.5-kb fragment of CaMKIIα genomic DNA beginning at 84 kb following the transcription initiation site (13). In one construct we included the entire 3′-UTR of the CaMKIIα mRNA (3.2 kb) downstream from the lacZ coding region (lac-CMK) to determine whether the signal for dendritic RNA localization was contained in this region. As a control, the second construct contained a 3′ polyadenylylation signal provided by bovine growth hormone (lac-A). Transgenic mice were generated using the two DNA constructs. Of four founder animals obtained, two (one from each construct) expressed the lacZ gene and were analyzed in detail.
Figure 1.
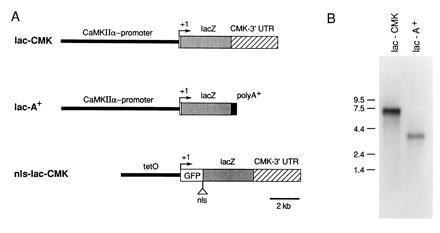
Expression of lacZ mRNA in mouse forebrain. (A) Schematic representation of the DNA constructs used for the generation of transgenic mice. lac-CMK: the 8.5-kbp CaMKIIα promoter region, as well as 84 nucleotides of the 5′ non-coding exon, was fused to the E. coli lacZ gene. The entire 3′-UTR of the CaMKIIα mRNA was placed downstream of the lacZ coding region. lac-A: identical to lac-CMK except that the bovine growth hormone polyadenylylation signal was substituted for the CaMKIIα 3′-UTR. nls-lac-CMK, the tet·O promoter (16) was linked to a modified lacZ gene with an in-phase fusion to the green fluorescent protein (GFP) and a nuclear localization sequence. (B) Northern blot analysis of poly(A)+ RNA isolated from the forebrain of the lac-CMK and lac-A mice.
To determine whether the two different transgenic lines expressed mRNAs of the expected size, we performed a Northern blot analysis of forebrain mRNA using a lacZ-specific probe. As expected, the lac-A and lac-CMK lines express lacZ-specific mRNAs of approximately 3.7 and 6.9 kb, respectively (Fig. 1B). In addition, the 6.9-kb transcript from the lac-CMK mice also hybridized to a probe specific for the CaMKII 3′-UTR (data not shown).
Histochemical detection of β-gal in brain sections revealed a similar pattern of expression in both lines (Fig. 2). With several exceptions, this expression was limited to those regions of the forebrain that normally express CaMKIIα. Notably, expression was absent in a medial layer of the cortex. Also, within the hippocampus, expression was much stronger in the dentate gyrus than in the CA3 and CA1 regions. Thus, the CaMKIIα promoter confers the expected cellular specificity on the expression of a heterologous transgene, with some variations in expression level.
Figure 2.
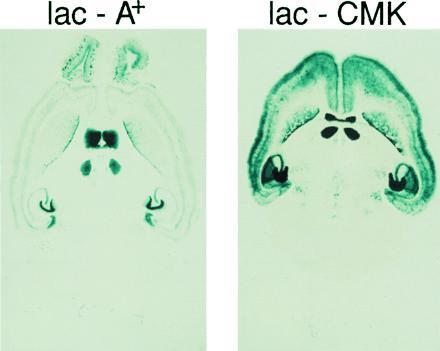
β-gal histochemistry.
The 3′-UTR of CaMKII Targets mRNA to Dendrites.
While the presence or absence of the CaMKIIα 3′-UTR seemed to have little effect on the regional distribution of transgene expression, in situ hybridization using a lacZ-specific oligonucleotide probe revealed a differential subcellular localization of the lacZ mRNA between the two transgenic lines. (Fig. 3A). To examine this subcellular localization in greater detail, we examined the hippocampus where the neuronal and dendritic layers are well differentiated. In the lac-CMK mice the hybridization signal covers not only the cell body layers of the dentate gyrus and CA1 region, but also extends into the corresponding dendritic layers. By contrast, the lac-A mice show strong hybridization in the cell body layer of the dentate gyrus and a weaker signal in CA1 cell bodies but no signal in the corresponding dendritic layers. The hybridization signal in the lac-CMK mice appears to be uniform throughout the dendritic layer and to extend into the most distal regions of the dendrite. This parallels the subcellular distribution of the endogenous CaMKIIα mRNA and differs from that of MAP 2, a dendritically-localized mRNA that is found only in the proximal portion of the dendrite (7). Thus, the presence of the CaMKIIα 3′ UTR is sufficient to localize the lacZ mRNA to dendrites and to yield a distribution of the mRNA within the dendritic layers that is indistinguishable from that of the endogenous CaMKIIα gene.
Figure 3.
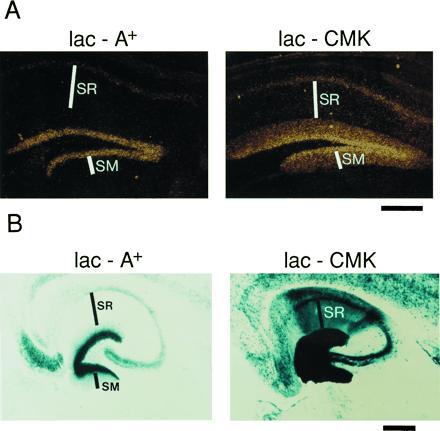
In situ localization of lacZ mRNA in hippocampus. (A) In situ hybridization using a lacZ-specific oligonucleotide probe. SM (stratum molecular), dendritic layer of the dentate gyrus granule cells; SR (stratum radiatum), dendritic layer of the CA1 pyramidal cells. (B) X-gal staining of hippocampus from 20-μm horizontal sections as described in Fig. 2. (Bar = 300 μm.)
In an attempt to identify a common sequence element, we compared the nucleotide sequence of the other known dendritically localized mRNAs [MAP2, Arc, IP-3R1, BC1 (6–9, 18)] to that of the CaMKIIα 3′-UTR. We found no major sequence homology. However, the critical determinant of the cis-acting element may not be reflected in its primary sequence but in the three-dimensional structure of the folded mRNA. This appears to be the case for the RNA localization elements important in early embryonic development (19–21). Alternately, the mechanism of localization for CaMKIIα may be different from that of the other dendritically localized RNAs. The difference in the extent of dendritic transport of the MAP2 and CaMKIIα mRNAs suggests some difference in the transport mechanism (5, 7).
Dendritically Localized mRNA Is Effectively Translated.
Is this dendritically targeted mRNA effectively translated? Previous studies have yielded conflicting results regarding basal protein synthesis in dendrites (22, 23). In mice bearing the dendritically localized lacZ mRNA, the level of β-gal protein in the dendrites of the hippocampal pyramidal cells is increased relative to controls, in which the lacZ mRNA is restricted to the cell body (Fig. 3B). This suggests that the dendritic mRNA is translationally active in the intact animal under basal conditions. However, when expressed at high levels, β-gal can diffuse into neuronal processes. It therefore is possible that some of the protein found in dendrites actually arose from translation in the cell body.
To clearly distinguish between protein synthesized locally within the dendrites and that synthesized in the cell body and diffusing into the dendrites, we generated mice in which the lacZ gene carried a nuclear localization signal (nls) so that β-gal synthesized in the neuronal cell body would be sequestered in the nucleus, thereby preventing it from diffusing into the dendrite. We generated mice using the nls-lac-CMK construct shown in Fig. 1A. In this case, we expressed the transgene off of the tet-O-promoter using the tTA system (16, 24). We obtained three lines of mice that expressed the nls-lac-CMK transgene in the hippocampus, one of which we examined in detail.
In situ hybridization revealed that the lacZ mRNA from the nls-lac-CMK line of mice was transported into dendrites (Fig. 4A). Histochemical detection of β-gal in this line revealed a pattern in which strong staining is found in the nucleus with little or no staining in the proximal dendrite and strong staining again in the more distal dendritic layer (Fig. 4B). Thus, the localization machinery is able to transport β-gal synthesized in the soma and proximal dendritic layer into the nucleus (compare Fig. 3B, lac-CMK, with Fig. 4B, nls-lac-CMK). Moreover, this expression pattern suggests that the β-gal found in the distal dendrite arises from local translation of the lacZ mRNA in the distal dendrite.
Figure 4.
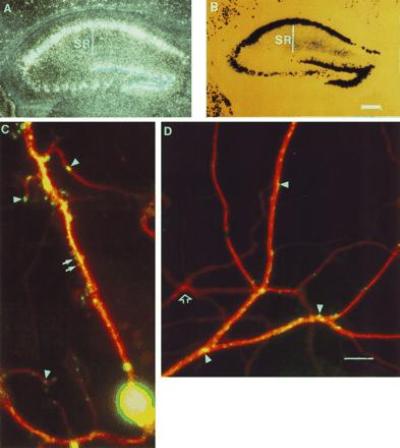
Differential expression of β-gal within dendrites. (A) In situ hybridization against the nls-lac-CMK mouse using a lacZ specific probe. (B) Histochemical detection of β-gal in the nls-lac-CMK mouse hippocampus (Bar = 300 μM.) (C) Immunofluorescent detection of MAP2 and β-gal expression in an nls-lac-CMK neuron in culture. The MAP2 antibody specifically labels microtubules along the dendritic shaft. MAP2 labeling is indicated in red. β-gal labeling is shown in green. Arrows denote β-gal in presumptive dendritic spines. Arrowheads indicate areas of punctate β-gal staining along the dendrite. (D) Expression of β-gal in a distal portion of the dendritic arbor. Arrowheads denote areas of punctate β-gal staining. Open arrow shows a dendrite arising from a neuron, which did not express the nls-lac-CMK transgene (Bar = 10 μM.)
β-Gal Is Unevenly Expressed Along the Dendrite.
To assess whether the β-gal is expressed evenly along the whole length of the dendrite, we cultured hippocampal neurons from nls-lac-CMK mice and used double-immunofluorescent detection of β-gal and MAP2. The MAP2 antibody, which labels microtubules specifically along the shaft of the dendrites, gives a smooth staining pattern. By contrast, the β-gal immunoreactivity is surprisingly patchy in its distribution with localized hot spots of staining along the dendritic shaft and within some presumptive dendritic spines (Fig. 4 C and D). This pattern of β-gal staining was observed using two different antibodies and was not detected in cultures from wild-type mice. This patchy, differential expression of β-gal along the dendrite suggests that the neuron is able to regulate expression of the transgene locally within the dendrite. This local regulation might occur through regulated distribution of the lacZ mRNA within the dendrite or through local differences in the rate of its translation.
DISCUSSION
Because neurons are highly polarized cells, a critical determinant of their function is the targeting of specific signaling molecules to their appropriate subcellular destination. In addition, neurons receive thousands of synaptic inputs and these can often be modulated independently in response to local differences in synaptic activity. For example, long-term potentiation or LTP is an activity-dependent form of synaptic plasticity that is synapse specific (25). The potentiation of synaptic strength with LTP occurs only at those synapses that are stimulated and not at other synapses onto the same cell. Thus, the LTP inducing stimulus must produce a biochemical change specific to the activated synapse. One mechanism for controlling the local biochemistry of a synapse is by regulating the distribution and translation of specific mRNAs at that synapse.
The CaMKIIα gene is expressed specifically in forebrain neurons, plays an essential role in LTP, and is one of the few mRNAs that are known to be targeted to dendrites (5, 26, 27). We therefore investigated the signals controlling both the forebrain specific expression and the dendritic mRNA localization. We found that an 8.5-kb fragment of the CaMKIIα gene is able to confer forebrain specific expression on a heterologous lacZ transgene. We have gone on to use this promoter element to express a number of other transgenes and find that in each case expression is limited to the forebrain neurons in a pattern similar to that shown in Fig. 2. The ability to direct transgene expression specifically to the forebrain neurons should prove useful in transgenic studies of neuronal function and its relation to behavior.
While the promoter targets CaMKIIα to the forebrain, we find that the 3′-UTR of CaMKIIα localizes the heterologous lacZ mRNA to dendrites. The mapping of the dendritic targeting signal of the CaMKIIα mRNA to the 3′-UTR demonstrates that the localization process is independent of the protein translated similar to the regulation of mRNA localization in other systems (10, 28, 29). We examined the expression of a lacZ gene in which the mRNA was targeted to dendrites, but the β-gal protein itself was targeted to the nucleus. We found strong staining for β-gal in the nucleus and distal dendrites with relatively little staining in the cytoplasm of the soma and proximal dendrite. These results suggest that the nuclear localization machinery may not function efficiently in the more distal regions of the dendrites. Within the dendrite, β-gal had an uneven distribution both along the shaft and in dendritic spines. This differential expression of the gene product provides a possible mechanism for the independent modulation of the biochemistry of individual synapses. The differential distribution could occur either through differential localization of the mRNA or differences in the translation of mRNA along the dendrite.
LTP is produced only at the appropriately stimulated synapses and its late phase is blocked by inhibitors of protein and mRNA synthesis (25, 30, 31). The requirement for new gene expression in LTP, coupled with the synapse specificity of the process, implies that the new gene products are targeted to or functionally used only at those synapses where the LTP is induced. One mechanism by which this might occur is for the LTP-inducing stimulus to convert the synapse from a translationally inactive to a translationally active state. This would lead to an immediate increase in the level of the gene product for those mRNA species localized to that synapse. In addition, newly transcribed mRNA species that were transported into dendrites would be expressed only at those translationally active synapses that received the LTP-inducing stimulation. Alternatively, an immediate increase in the translation of mRNA at the stimulated synapses might mark those synapses such that the newly induced gene products important for maintaining LTP would be targeted only to those marked synapses. It will be interesting to determine whether mislocalization of CaMKIIα mRNA, through deletion of the dendritic targeting signal, will interfere with the production or maintenance of a synapse-specific late phase for LTP.
Acknowledgments
We thank Richard Axel and Tom Jessell for critically reading this manuscript. We also thank Joseph Finkelstein for maintaining and genotyping the mice, Chuck Lam for help with the figures, Harriet Ayers and Irma Trumpet for typing the manuscript, and Mohammed Osman for helping with animal care. This research was supported by Howard Hughes Medical Institute and the National Institute of Mental Health.
Footnotes
Abbreviations: MAP, microtubule-associated protein; β-gal, β-galactosidase; UTR, untranslated region; LTP, long-term potentiation.
References
- 1.Steward O, Levy W B. J Neurosci. 1982;2:284–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steward O, Fass B. Prog Brain Res. 1983;58:131–136. doi: 10.1016/S0079-6123(08)60013-8. [DOI] [PubMed] [Google Scholar]
- 3.Steward O. Cold Spring Harbor Symp Quant Biol. 1983;48:745–759. doi: 10.1101/sqb.1983.048.01.077. [DOI] [PubMed] [Google Scholar]
- 4.Steward O. Trends Neurosci. 1992;15:180–186. doi: 10.1016/0166-2236(92)90170-d. [DOI] [PubMed] [Google Scholar]
- 5.Burgin K E, Waxham M N, Rickling S, Westgate S A, Mobley W C, Kelly P T. J Neurosci. 1990;10:1788–1798. doi: 10.1523/JNEUROSCI.10-06-01788.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furuichi T, Simon-Chazottes D, Fujino I, Yamada N, Hasegawa M, Miyawaki A, Yoshikawa S, Gueénet J-L, Mikoshiba K. Receptors Channels. 1993;1:11–24. [PubMed] [Google Scholar]
- 7.Garner C C, Tucker R P, Matus A. Nature (London) 1988;336:674–677. doi: 10.1038/336674a0. [DOI] [PubMed] [Google Scholar]
- 8.Lyford G L, Yamagata K, Kaufmann W E, Barnes C A, Sanders L K, Copeland N G, Gilbert D J, Jenkins N A, Lanahan A A, Worley P F. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- 9.Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, Frey U, Kuhl D. Proc Natl Acad Sci USA. 1995;92:5734–5738. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bian F, Chu T, Schilling K, Oberdick J. Mol Cell Neurosci. 1996;7:116–133. doi: 10.1006/mcne.1996.0009. [DOI] [PubMed] [Google Scholar]
- 11.Davis L, Banker G A, Steward O. Nature (London) 1987;330:477–479. doi: 10.1038/330477a0. [DOI] [PubMed] [Google Scholar]
- 12.Bassell G J, Singer R H, Kosik K S. Neuron. 1994;144:565–572. doi: 10.1016/0896-6273(94)90213-5. [DOI] [PubMed] [Google Scholar]
- 13.Sunyer T, Sahyoun N. Proc Natl Acad Sci USA. 1990;87:278–282. doi: 10.1073/pnas.87.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forss-Petter S, Danielson P E, Catsicas S,, Battenberg E, Price J, Nerenberg M, Sutcliffe J G. Neuron. 1990;5:187–197. doi: 10.1016/0896-6273(90)90308-3. [DOI] [PubMed] [Google Scholar]
- 15.Rayport S, Sulzer D, Shi W-X, Sawasdikosol S, Monaco J, Baston D, Rajendram G. J Neurosci. 1992;12:4264–4280. doi: 10.1523/JNEUROSCI.12-11-04264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig A M, Blackstone C D, Huganir R L, Banker G. Neuron. 1993;10:1055–1068. doi: 10.1016/0896-6273(93)90054-u. [DOI] [PubMed] [Google Scholar]
- 17.Feig S, Lipton P. J Neurosci. 1993;13:1010–1021. doi: 10.1523/JNEUROSCI.13-03-01010.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiedge H, Fremeau R T, Jr, Weinstock P H, Arancio O, Brosius J. Proc Natl Acad Sci USA. 1991;88:2093–2097. doi: 10.1073/pnas.88.6.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macdonald P M, Struhl G. Nature (London) 1988;336:595–598. doi: 10.1038/336595a0. [DOI] [PubMed] [Google Scholar]
- 20.Macdonald P M, Kerr K, Smith J L, Leask A. Development (Cambridge, UK) 1993;118:1233–1243. doi: 10.1242/dev.118.4.1233. [DOI] [PubMed] [Google Scholar]
- 21.Mowry K L, Melton D A. Science. 1992;255:991–994. doi: 10.1126/science.1546297. [DOI] [PubMed] [Google Scholar]
- 22.Torre E R, Steward O. J Neurosci. 1992;12:762–772. doi: 10.1523/JNEUROSCI.12-03-00762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayford, M., Wang, L., Podsypanina, K. & Kandel, E. R. (1995) Soc. Neurosci. Abstr., 433.15.
- 25.Bliss T V P, Collingridge G L. Nature (London) 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 26.Silva A J, Stevens C F, Tonegawa S, Wang Y. Science. 1992;257:201–206. doi: 10.1126/science.1378648. [DOI] [PubMed] [Google Scholar]
- 27.Mayford M, Wang J, Kandel E R, O’Dell T J. Cell. 1995;81:891–904. doi: 10.1016/0092-8674(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 28.Sundell C L, Singer R H. J Cell Biol. 1990;111:2397–2403. doi: 10.1083/jcb.111.6.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleiman R, Banker G, Steward O. Proc Natl Acad Sci USA. 1993;90:11192–11196. doi: 10.1073/pnas.90.23.11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frey U, Krug M, Reymann K, Matthies H. Brain Res. 1988;452:57–65. doi: 10.1016/0006-8993(88)90008-x. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen P V, Abel T, Kandel E R. Science. 1994;265:1104–1107. doi: 10.1126/science.8066450. [DOI] [PubMed] [Google Scholar]


